Epidermal Growth Factor Receptor T790M Mutation Testing in Non-Small Cell Lung Cancer: An International Collaborative Study to Assess Molecular EGFR T790M Testing in Liquid Biopsy
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Participants
2.2. Sample Panel Composition and Distribution
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Harrison, P.T.; Vyse, S.; Huang, P.H. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin. Cancer Biol. 2020, 61, 167–179. [Google Scholar] [CrossRef]
- Da Cunha Santos, G.; Shepherd, F.A.; Tsao, M.S. EGFR mutations and lung cancer. Annu. Rev. Pathol. 2011, 6, 49–69. [Google Scholar] [CrossRef]
- Remon, J.; Steuer, C.E.; Ramalingam, S.S.; Felip, E. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann. Oncol. 2018, 29, i20–i27. [Google Scholar] [CrossRef]
- Buder, A.; Hochmair, M.J.; Schwab, S.; Bundalo, T.; Schenk, P.; Errhalt, P.; Mikes, R.E.; Absenger, G.; Patocka, K.; Baumgartner, B.; et al. Cell-Free Plasma DNA-Guided Treatment With Osimertinib in Patients With Advanced EGFR-Mutated NSCLC. J. Thorac. Oncol. 2018, 13, 821–830. [Google Scholar] [CrossRef]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Buder, A.; Setinek, U.; Hochmair, M.J.; Schwab, S.; Kirchbacher, K.; Keck, A.; Burghuber, O.C.; Pirker, R.; Filipits, M. EGFR Mutations in Cell-free Plasma DNA from Patients with Advanced Lung Adenocarcinoma: Improved Detection by Droplet Digital PCR. Target. Oncol. 2019, 14, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Karlovich, C.; Goldman, J.W.; Sun, J.M.; Mann, E.; Sequist, L.V.; Konopa, K.; Wen, W.; Angenendt, P.; Horn, L.; Spigel, D.; et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin. Cancer Res. 2016, 22, 2386–2395. [Google Scholar] [CrossRef] [PubMed]
- Reungwetwattana, T.; Gray, J.; Markovets, A.; Nogami, N.; Lee, J.S.; Cho, B.C.; Chewaskulyong, B.; Majem, M.; Peled, N.; Vishwanathan, K.; et al. LBA17—Longitudinal circulating tumour DNA (ctDNA) monitoring for early detection of disease progression and resistance in advanced NSCLC in FLAURA. Ann. Oncol. 2019, 30 (Suppl. S9), ix199. [Google Scholar] [CrossRef]
- Schmid, S.; Li, J.J.; Leighl, N.B. Mechanisms of osimertinib resistance and emerging treatment options. Lung Cancer 2020, 147, 123–129. [Google Scholar] [CrossRef]
- Goss, G.; Tsai, C.M.; Shepherd, F.A.; Bazhenova, L.; Lee, J.S.; Chang, G.C.; Crino, L.; Satouchi, M.; Chu, Q.; Hida, T.; et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): A multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016, 17, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Jänne, P.A.; Yang, J.C.H.; Kim, D.W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ahn, M.J.; Kim, S.W.; Su, W.C.; Horn, L.; et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Ahn, M.J.; Kim, D.W.; Ramalingam, S.S.; Sequist, L.V.; Wc, S.; Kim, S.W.; Kim, J.H.; Planchard, D.; Felip, E.; et al. Osimertinib in Pretreated T790M-Positive Advanced Non-Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J. Clin. Oncol. 2017, 35, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.-C.; Griesing, S.; Yang, J.C.H. Second-line treatment of EGFR T790M-negative non-small cell lung cancer patients. Ther. Adv. Med. Oncol. 2019, 11, 1–16. [Google Scholar] [CrossRef]
- Nagasaka, M.; Zhu, V.W.; Lim, S.M.; Greco, M.; Wu, F.; Ou, S.H.I. Beyond Osimertinib: The Development of Third-Generation EGFR Tyrosine Kinase Inhibitors For Advanced EGFR+ NSCLC. J. Thorac. Oncol. 2021, 16, 740–763. [Google Scholar] [CrossRef]
- Buder, A.; Tomuta, C.; Filipits, M. The potential of liquid biopsies. Curr. Opin. Oncol. 2016, 28, 130–134. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Choo, J.R.E.; Tan, C.S.; Soo, R.A. Treatment of EGFR T790M-Positive Non-Small Cell Lung Cancer. Target. Oncol. 2018, 13, 141–156. [Google Scholar] [CrossRef]
- Hochmair, M.J.; Buder, A.; Schwab, S.; Burghuber, O.C.; Prosch, H.; Hilbe, W.; Cseh, A.; Fritz, R.; Filipits, M. Liquid-Biopsy-Based Identification of EGFR T790M Mutation-Mediated Resistance to Afatinib Treatment in Patients with Advanced EGFR Mutation-Positive NSCLC, and Subsequent Response to Osimertinib. Target. Oncol. 2019, 14, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Buder, A.; Hochmair, M.J.; Filipits, M. The Allele Frequency of EGFR Mutations Predicts Survival in Advanced EGFR T790M-Positive Non-small Cell Lung Cancer Patients Treated with Osimertinib. Target. Oncol. 2021, 16, 77–84. [Google Scholar] [CrossRef] [PubMed]
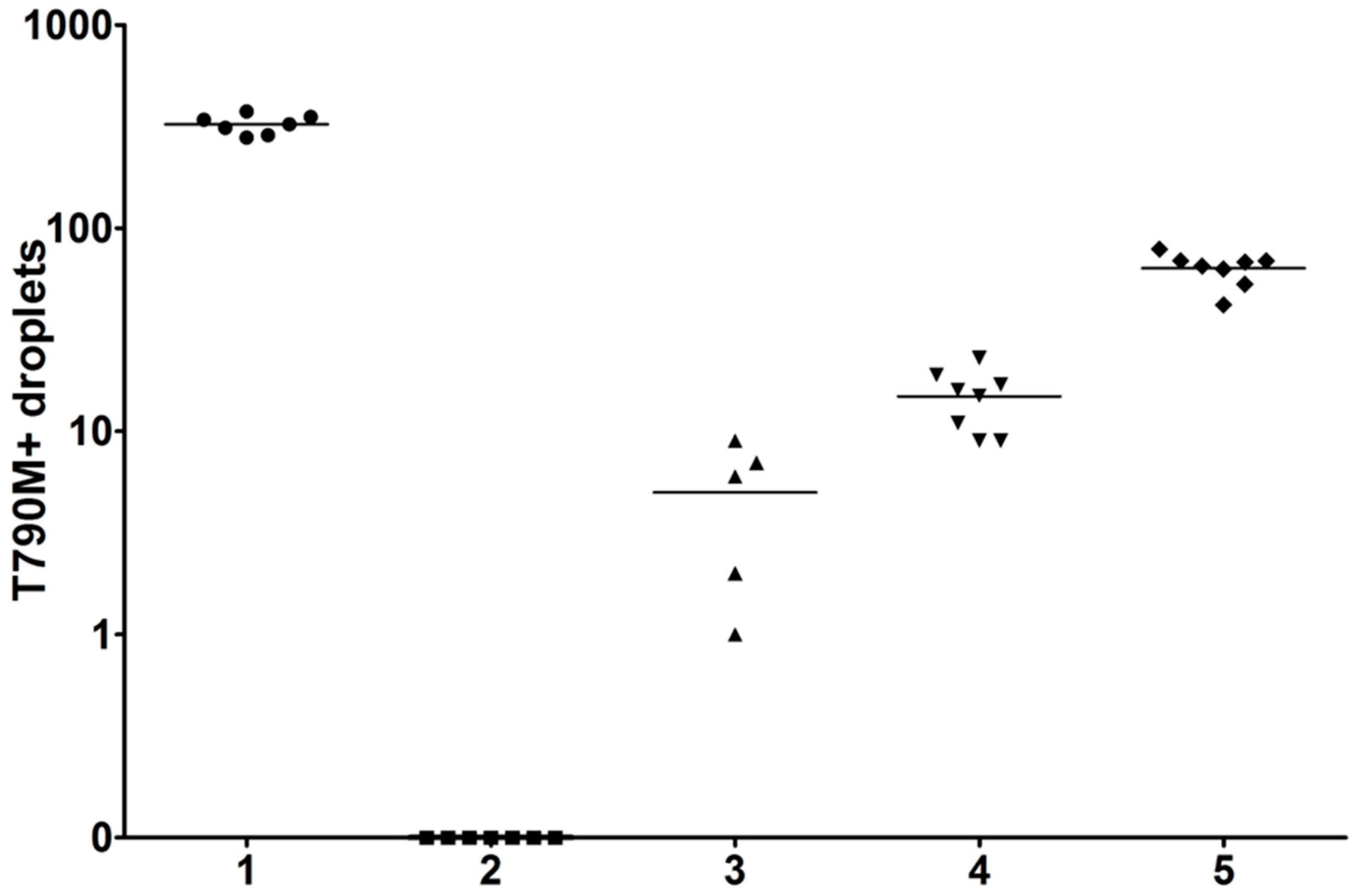
| Average Number of Positive Droplets | |||||
|---|---|---|---|---|---|
| Sample | Spiked DNA | EGFRT790M | EGFRDel19 | EGFRL858R | EGFRwt |
| 1 | HCC-8207 (30 ng) | 485 | 2279 | 0 | + |
| 2 | CRL-5908 (0.25 ng) | 5 | 0 | 7 | + |
| 3 | A549 (50 ng) | 0 | 0 | 0 | + |
| 4 | HCC-8207 (1.5 ng) | 34 | 139 | 0 | + |
| 5 | HCC-8207 (3 ng) | 49 | 465 | 0 | + |
| 6 | CRL-5908 (100 ng) | 780 | 0 | 951 | + |
| 7 | HCC-8207 (1.5 ng) | 25 | 132 | 0 | + |
| 8 | MCF-7 (27 ng) | 0 | 0 | 0 | + |
| 9 | HCC-8207 (1.5 ng) | 27 | 172 | 0 | + |
| 10 | HCC-8207 (3 ng) | 49 | 355 | 0 | + |
| Country | No. of Laboratories |
|---|---|
| (A) | |
| Austria | 5 |
| Bulgaria | 1 |
| Lithuania | 1 |
| Poland | 4 |
| Switzerland | 1 |
| 12 | |
| (B) | |
| Austria | 9 |
| Bulgaria | 1 |
| Croatia | 1 |
| Hungary | 3 |
| Lithuania | 1 |
| Poland | 2 |
| Russia | 3 |
| Slovakia | 5 |
| 25 | |
| (A) | |
| ctDNA Extraction Method | No. of Laboratories |
| Cobas® cfDNA Sample Preparation Kit | 6 |
| QIAamp Circulating Nucleic Acid Kit | 4 |
| IdyllaTM ctEGFR Mutation Assay | 2 |
| MagNA Pure 24 Total NA Isolation Kit | 2 |
| Maxwell® RSC ccfDNA Plasma Kit | 2 |
| EZ1 ccfDNA Midi Kit | 1 |
| AmoyDx® Circulating DNA Kit | 1 |
| QIAamp MinElute ccfDNA Kit | 1 |
| Maxwell® 16 FFPE Plus LEV DNA Purification Kit | 1 |
| QIAsymphony DSP Circulating DNA Kit | 1 |
| (B) | |
| Detection Method | No. of Laboratories |
| Cobas® EGFR Mutation Test v2 | 8 |
| AmoyDx® EGFR 29 Mutations Detection Kit | 4 |
| IdyllaTM | 2 |
| OncomineTM Lung cfDNA Assay | 2 |
| Avenio ctDNA Targeted Kit | 1 |
| Droplet Digital PCR | 1 |
| EasyPGX® ready EGFR | 1 |
| EGFR XL StripAssay® | 1 |
| QIAact Lung Plasma Track Panel | 1 |
| Center | Assay | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 |
|---|---|---|---|---|---|---|
| Reference | ddPCR | 324 | 0 | 5 | 17 | 72 |
| 1 | ddPCR | + | − | + | + | + |
| 2 | ddPCR | 512 | 0 | 7.4 | 26.2 | 49.4 |
| 3 | Genereader | + | − | + | + | + |
| 4 | Cobas v2 | + | − | + | + | + |
| 5 | NGS | + | invalid | + | + | + |
| 6 | Cobas v2 | + | invalid | invalid | invalid | + |
| 7 | Cobas/Entrogen | − | − | − | + | + |
| 8 | ddPCR | 200 | 0 | 2 | 9 | 41 |
| 9 | Cobas v2 | + | invalid | invalid | invalid | invalid |
| 10 | AmoyDx | + | − | + | + | + |
| 11/1 | NGS | 319 | +- | 8 | 22 | 65 |
| 11/2 | ddPCR | + | − | + | + | + |
| 12 | EasyPGX | + | − | + | + | + |
| A | ||||||||||||||||||||||||||
| Sample | Center | |||||||||||||||||||||||||
| No. | Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| 1 | T790M/Del19 | |||||||||||||||||||||||||
| 2 | T790M/L858R | |||||||||||||||||||||||||
| 3 | Wildtype | |||||||||||||||||||||||||
| 4 | T790M/Del19 | |||||||||||||||||||||||||
| 5 | T790M/Del19 | |||||||||||||||||||||||||
| 6 | T790M/L858R | |||||||||||||||||||||||||
| 7 | T790M/Del19 | |||||||||||||||||||||||||
| 8 | Wildtype | |||||||||||||||||||||||||
| 9 | T790M/Del19 | |||||||||||||||||||||||||
| 10 | T790M/Del19 | |||||||||||||||||||||||||
| Platforms | PCR | PCR | PCR | PCR | PCR | PCR | PCR | PCR | PCR | PCR | PCR | NGS | NGS | PCR | NGS | PCR | NGS | PCR | PCR | PCR | PCR | |||||
| Detection kits | A | I | C | C | C | A | C | E | A | D | S | N | G | C | O | I | O | A | C | C | C | |||||
| Extraction kits | A | I | C | C | C | Q1 | C | Q1 | C | Q1 | Q1 | M1 | Q2 | M1 | E | I | R | R | Q3 | M2 | C | |||||
| B | ||||||||||||||||||||||||||
| Sample | Center | |||||||||||||||||||||||||
| No. | Name | 4 | 5 | 6 | 8 | 18 | 23 | 24 | 25 | |||||||||||||||||
| 1 | T790M/Del19 | |||||||||||||||||||||||||
| 2 | T790M/L858R | |||||||||||||||||||||||||
| 3 | Wildtype | |||||||||||||||||||||||||
| 4 | T790M/Del19 | |||||||||||||||||||||||||
| 5 | T790M/Del19 | |||||||||||||||||||||||||
| 6 | T790M/L858R | |||||||||||||||||||||||||
| 7 | T790M/Del19 | |||||||||||||||||||||||||
| 8 | Wildtype | |||||||||||||||||||||||||
| 9 | T790M/Del19 | |||||||||||||||||||||||||
| 10 | T790M/Del19 | |||||||||||||||||||||||||
| Platforms | PCR | PCR | PCR | PCR | PCR | PCR | PCR | PCR | ||||||||||||||||||
| Detection kits | C | C | C | C | C | C | C | C | ||||||||||||||||||
| Extraction kits | C | C | C | C | M1 | Q3 | M2 | C | ||||||||||||||||||
| C | ||||||||||||||||||||||||||
| Sample | Center | |||||||||||||||||||||||||
| No. | Name | 1 | 7 | 10 | 22 | |||||||||||||||||||||
| 1 | T790M/Del19 | |||||||||||||||||||||||||
| 2 | T790M/L858R | |||||||||||||||||||||||||
| 3 | Wildtype | |||||||||||||||||||||||||
| 4 | T790M/Del19 | |||||||||||||||||||||||||
| 5 | T790M/Del19 | |||||||||||||||||||||||||
| 6 | T790M/L858R | |||||||||||||||||||||||||
| 7 | T790M/Del19 | |||||||||||||||||||||||||
| 8 | Wildtype | |||||||||||||||||||||||||
| 9 | T790M/Del19 | |||||||||||||||||||||||||
| 10 | T790M/Del19 | |||||||||||||||||||||||||
| Platforms | PCR | PCR | PCR | PCR | ||||||||||||||||||||||
| Detection kits | A | A | A | A | ||||||||||||||||||||||
| Extraction kits | A | Q1 | C | R | ||||||||||||||||||||||
| D | ||||||||||||||||||||||||||
| Sample | Center | |||||||||||||||||||||||||
| No. | Name | 16 | 17 | 19 | 21 | |||||||||||||||||||||
| 1 | T790M/Del19 | |||||||||||||||||||||||||
| 2 | T790M/L858R | |||||||||||||||||||||||||
| 3 | Wildtype | |||||||||||||||||||||||||
| 4 | T790M/Del19 | |||||||||||||||||||||||||
| 5 | T790M/Del19 | |||||||||||||||||||||||||
| 6 | T790M/L858R | |||||||||||||||||||||||||
| 7 | T790M/Del19 | |||||||||||||||||||||||||
| 8 | Wildtype | |||||||||||||||||||||||||
| 9 | T790M/Del19 | |||||||||||||||||||||||||
| 10 | T790M/Del19 | |||||||||||||||||||||||||
| Platforms | NGS | NGS | NGS | PCR | ||||||||||||||||||||||
| Detection kits | N | G | O | O | ||||||||||||||||||||||
| Extraction kits | M1 | Q2 | E | R | ||||||||||||||||||||||
| E | ||||||||||||||||||||||||||
| Sample | Center | |||||||||||||||||||||||||
| No. | Name | 3 | 20 | |||||||||||||||||||||||
| 1 | T790M/Del19 | |||||||||||||||||||||||||
| 2 | T790M/L858R | |||||||||||||||||||||||||
| 3 | Wildtype | |||||||||||||||||||||||||
| 4 | T790M/Del19 | |||||||||||||||||||||||||
| 5 | T790M/Del19 | |||||||||||||||||||||||||
| 6 | T790M/L858R | |||||||||||||||||||||||||
| 7 | T790M/Del19 | |||||||||||||||||||||||||
| 8 | Wildtype | |||||||||||||||||||||||||
| 9 | T790M/Del19 | |||||||||||||||||||||||||
| 10 | T790M/Del19 | |||||||||||||||||||||||||
| Platforms | PCR | PCR | ||||||||||||||||||||||||
| Detection kits | I | I | ||||||||||||||||||||||||
| Extraction kits | I | I | ||||||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filipits, M.; Kainz, V.; Sebek, V.; Zach, H.; on behalf of the Liquid Biopsy Collaborative Study Group. Epidermal Growth Factor Receptor T790M Mutation Testing in Non-Small Cell Lung Cancer: An International Collaborative Study to Assess Molecular EGFR T790M Testing in Liquid Biopsy. Cancers 2023, 15, 3528. https://doi.org/10.3390/cancers15133528
Filipits M, Kainz V, Sebek V, Zach H, on behalf of the Liquid Biopsy Collaborative Study Group. Epidermal Growth Factor Receptor T790M Mutation Testing in Non-Small Cell Lung Cancer: An International Collaborative Study to Assess Molecular EGFR T790M Testing in Liquid Biopsy. Cancers. 2023; 15(13):3528. https://doi.org/10.3390/cancers15133528
Chicago/Turabian StyleFilipits, Martin, Verena Kainz, Viktor Sebek, Herwig Zach, and on behalf of the Liquid Biopsy Collaborative Study Group. 2023. "Epidermal Growth Factor Receptor T790M Mutation Testing in Non-Small Cell Lung Cancer: An International Collaborative Study to Assess Molecular EGFR T790M Testing in Liquid Biopsy" Cancers 15, no. 13: 3528. https://doi.org/10.3390/cancers15133528
APA StyleFilipits, M., Kainz, V., Sebek, V., Zach, H., & on behalf of the Liquid Biopsy Collaborative Study Group. (2023). Epidermal Growth Factor Receptor T790M Mutation Testing in Non-Small Cell Lung Cancer: An International Collaborative Study to Assess Molecular EGFR T790M Testing in Liquid Biopsy. Cancers, 15(13), 3528. https://doi.org/10.3390/cancers15133528





