WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens and Immunohistochemistry (IHC)
2.2. Cell Lines and Cultures
2.3. Plasmids
2.4. Plasmid Transduction
2.5. Total Nuclear and Histone Protein Extraction and Immunoblotting
2.6. In Silico Public Dataset Analysis
2.7. RNA-seq
2.8. ChIP-seq
2.9. Cell Proliferation Assay
2.10. In Vivo Studies
2.11. Statistical Analysis
3. Results
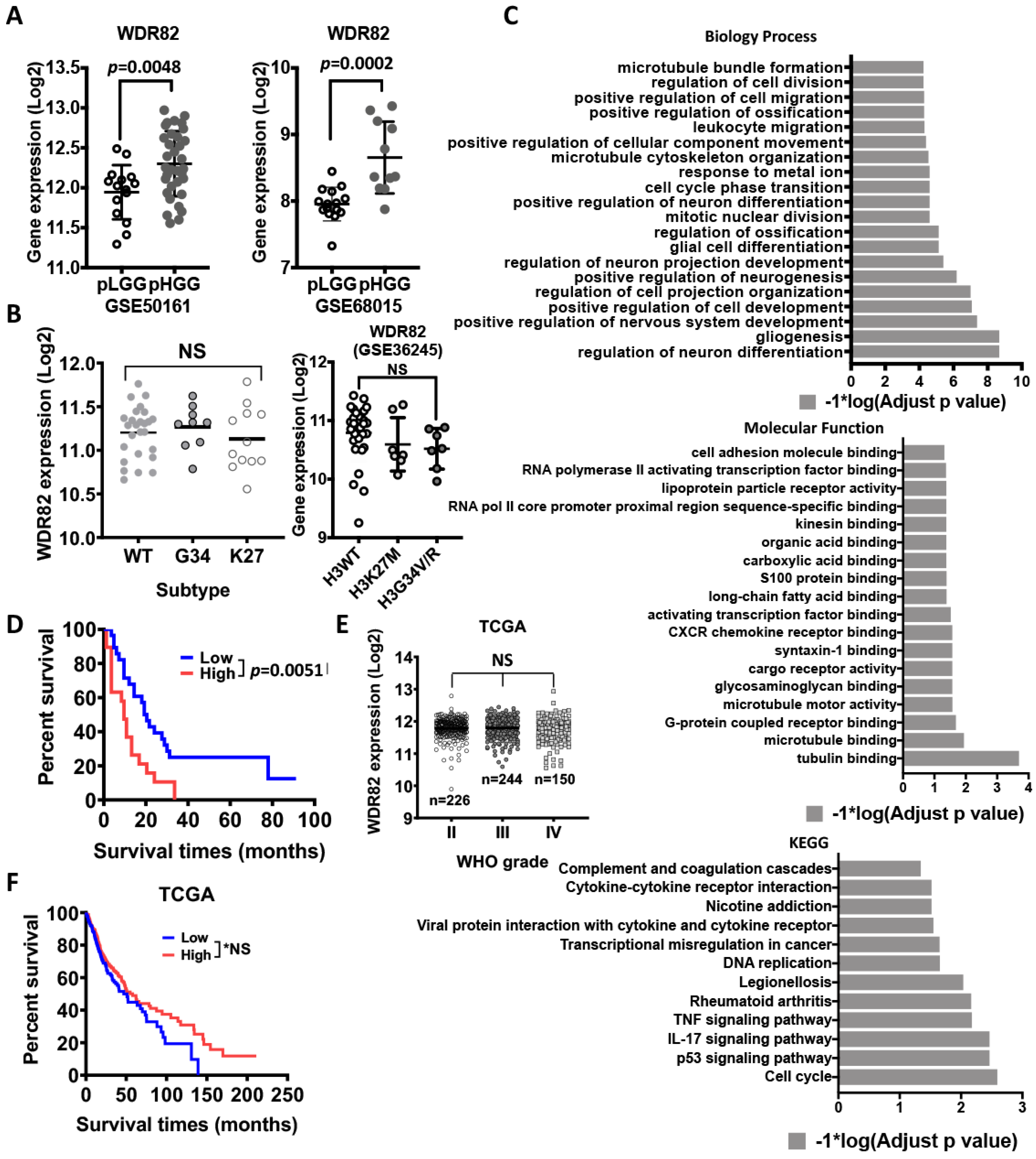
3.1. H3K4me3 Levels Increase with Histopathological Malignancy in Pediatric Gliomas
3.2. WDR82 Plays a Distinct Role in the Tumor Biology of Pediatric Glioma
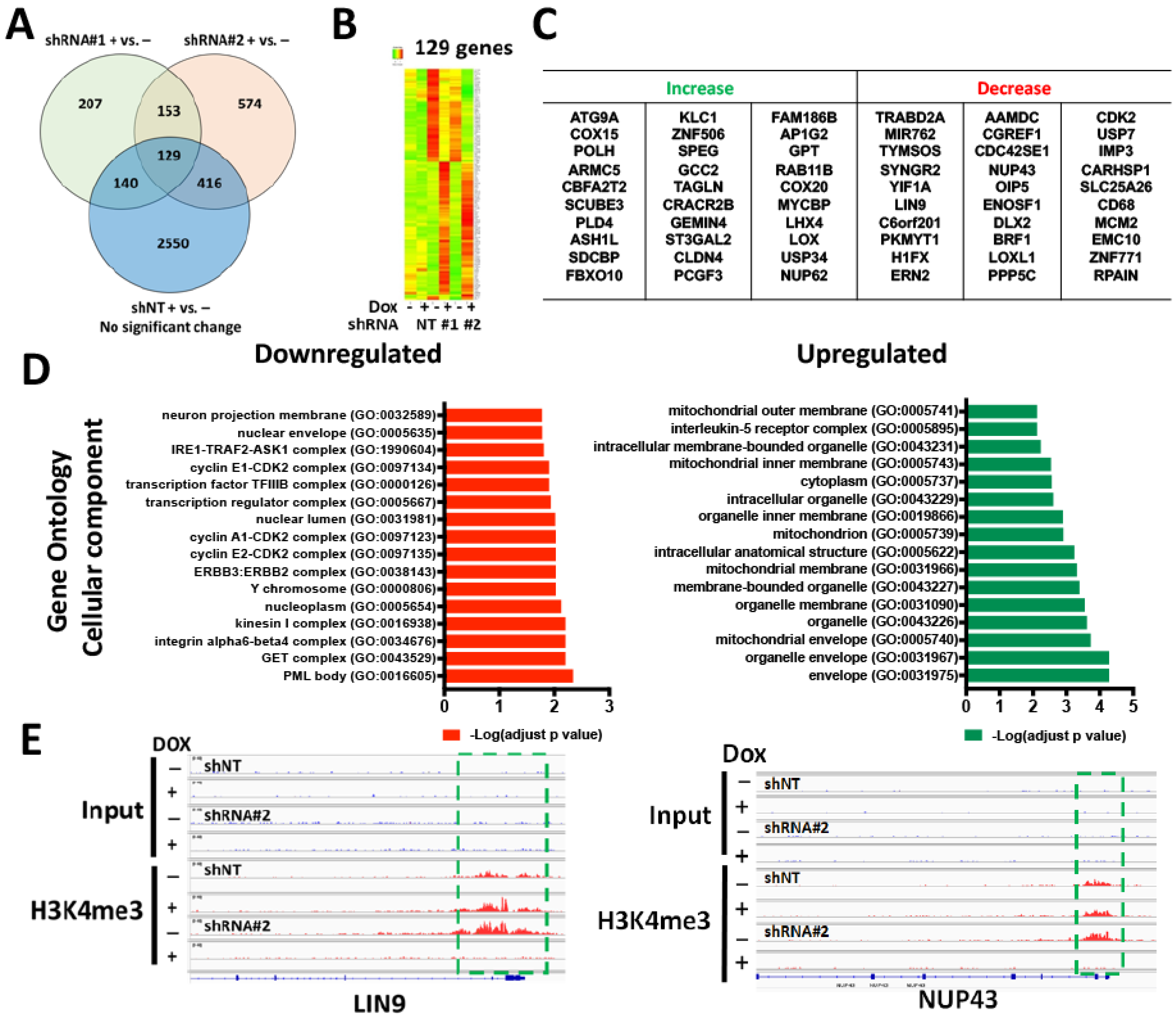
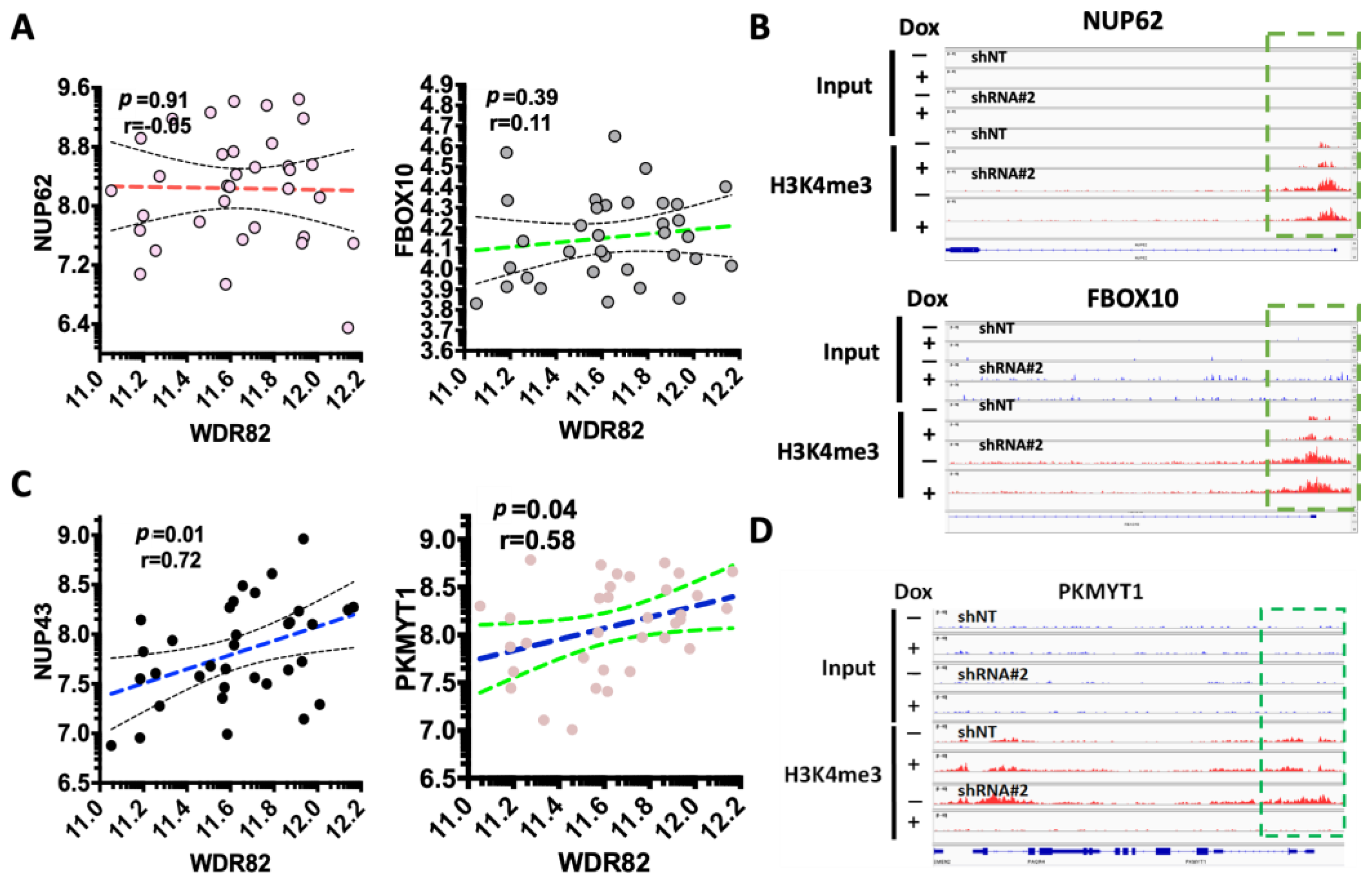
3.3. WDR82-Mediated H3K4me3 Is Associated with Mitotic and Cell-Cycle-Related Gene Regulation
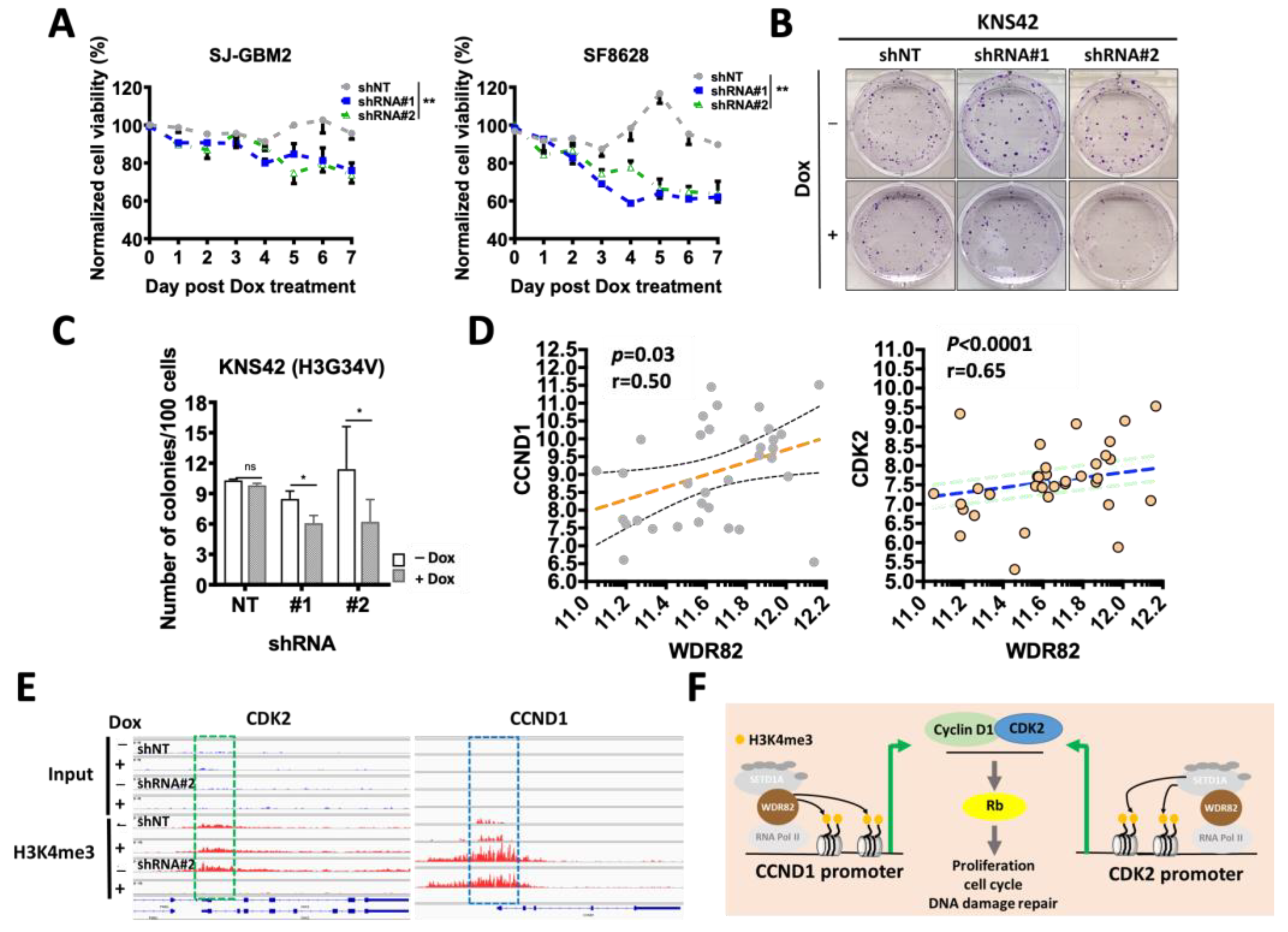
3.4. WDR82 Knockdown Decreases In Vitro Cell Viability and DNA Damage Repair in pHGG Cells
3.5. WDR82 Knockdown Increases Therapeutic Sensitivity against DNA-Damaging Agents and Radiation Sensitivity
3.6. WDR82 Knockdown Decreases In Vivo pHGG Tumor Growth and Extends Survival of pHGG-Tumor-Bearing Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Egan, K.M.; Nabors, L.B.; Gerke, T.; Thompson, R.C.; Olson, J.J.; Olson, J.J.; LaRocca, R.; Chowdhary, S.; Eckel-Passow, E.J.; et al. Glioma risk associated with extent of estimated European genetic ancestry in African Americans and Hispanics. Int. J. Cancer 2020, 146, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, M.A.; Quang, D.K.; Tonjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar]
- Fang, J.; Huang, Y.; Mao, G.; Yang, S.; Rennert, G.; Gu, L.; Li, H.; Li, G.M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSα interaction. Proc. Natl. Acad. Sci. USA 2018, 115, 9598–9603. [Google Scholar] [CrossRef] [PubMed]
- Jani, K.S.; Jain, S.U.; Ge, E.J.; Diehl, K.L.; Lundgren, S.M.; Muller, M.M.; Lewis, P.W.; Muir, T.W. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl. Acad. Sci. USA 2019, 116, 8295–8300. [Google Scholar] [CrossRef]
- Shi, L.; Shi, J.; Shi, X.; Li, W.; Wen, H. Histone H3.3 G34 Mutations Alter Histone H3K36 and H3K27 Methylation. In Cis. J. Mol. Biol. 2018, 430, 1562–1565. [Google Scholar] [CrossRef]
- Piunti, A.; Smith, E.R.; Morgan, M.A.J.; Ugarenko, M.; Khaltyan, N.; Helmin, K.A.; Ryan, C.A.; Murray, D.C.; Rickels, R.A.; Yilmaz, B.D.; et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 2019, 5, eaax2887. [Google Scholar] [CrossRef]
- Lewis, P.W.; Muller, M.M.; Koletsky, M.S.; Cordero, F.; Lin, S.; Banaszynski, L.A.; Garcia, B.A.; Muir, T.W.; Becher, O.J.; Allis, C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013, 340, 857–861. [Google Scholar] [CrossRef]
- Chan, K.M.; Fang, D.; Gan, H.; Hashizume, R.; Yu, C.; Schroeder, M.; Gupata, N.; Mueller, S.; James, C.D.; Jenkins, R.; et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013, 27, 985–990. [Google Scholar] [CrossRef]
- Lu, C.; Jain, S.U.; Hoelper, D.; Bechet, D.; Molden, R.C.; Ran, L.; Murphy, D.; Venneti, S.; Hameed, M.; Pawel, B.R.; et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016, 352, 844–849. [Google Scholar] [CrossRef]
- Gu, B.; Lee, M.G. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Xu, J.; Zhang, J.; Xie, D.; Ye, H.; Xiao, Z.; Cai, M.; Xu, K.; Zeng, Y.; Li, H.; et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 2012, 43, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.S.; Qu, Y.; Rostad, K.; Li, W.C.; Lin, B.; Halvorsen, O.J.; Haukaas, S.A.; Jonassen, I.; Petersen, K.; Goldfinger, N.; et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE 2009, 4, e4687. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.; Li, Y.D.; Hoffman, L.; Hashizume, R.; Gravohac, G.; Rice, G.; Wadhwani, N.R.; Jie, C.; Pundy, T.; Mania-Farnell, B.; et al. Global Reduction of H3K4me3 Improves Chemotherapeutic Efficacy for Pediatric Ependymomas. Neoplasia 2019, 21, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Schneider, R.; Bannister, A.J.; Myers, F.A.; Thorne, A.W.; Crane-Robinson, C.; Kouzarides, T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004, 6, 73–77. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Lee, J.H.; Skalnik, D.G. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005, 280, 41725–41731. [Google Scholar] [CrossRef]
- Lee, J.H.; Tate, C.M.; You, J.S.; Skalnik, D.G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007, 282, 13419–13428. [Google Scholar] [CrossRef]
- Wu, M.; Wang, P.F.; Lee, J.S.; Martin-Brown, S.; Florens, L.; Washburn, M.; Shilatifard, A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell Biol. 2008, 28, 7337–7344. [Google Scholar] [CrossRef]
- Lee, J.H.; Skalnik, D.G. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell Biol. 2008, 28, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Lv, Z.; Wang, Y.; Hai, T.; Huo, R.; Zhou, Z.; Zhou, Q.; Sha, J. WDR82, a key epigenetics-related factor, plays a crucial role in normal early embryonic development in mice. Biol. Reprod. 2011, 84, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; You, J.; Dobrota, E.; Skalnik, D.G. Identification and characterization of a novel human PP1 phosphatase complex. J. Biol. Chem. 2010, 285, 24466–24476. [Google Scholar] [CrossRef]
- Chapman-Rothe, N.; Curry, E.; Zeller, C.; Liber, D.; Stronach, E.; Gabra, H.; Ghaem-Maghami, S.; Brown, R. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene 2013, 32, 4586–4592. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Lee, M.H.; Lee, M.O. Histone methyltransferases regulate the transcriptional expression of ERα and the proliferation of tamoxifen-resistant breast cancer cells. Breast Cancer Res. Treat. 2020, 180, 45–54. [Google Scholar] [CrossRef]
- Hasan, T.; Caragher, S.P.; Shireman, J.M.; Park, C.H.; Atashi, F.; Baisiwala, S.; Lee, G.; Guo, D.; Wang, J.Y.; Dey, M.; et al. Interleukin-8/CXCR2 signaling regulates therapy-induced plasticity and enhances tumorigenicity in glioblastoma. Cell Death Dis. 2019, 10, 292. [Google Scholar] [CrossRef]
- Bounaix Morand du Puch, C.; Barbier, E.; Kraut, A.; Coute, Y.; Fuchs, J.; Buhot, A.; Livache, T.; Seve, M.; Favier, A.; Douki, T.; et al. TOX4 and its binding partners recognize DNA adducts generated by platinum anticancer drugs. Arch. Biochem. Biophys. 2011, 507, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Cawthorne, C.; Swindell, R.; Stratford, I.J.; Dive, C.; Welman, A. Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. J. Biomol. Tech. JBT 2007, 18, 120–123. [Google Scholar]
- Volm, M.; Koomägi, R.; Stammler, G.; Rittgen, W.; Zintl, F.; Sauerbrey, A. Prognostic implications of cyclins (D1, E, A), cyclin-dependent kinases (CDK2, CDK4) and tumor-suppressor genes (pRB, p16INK4A) in childhood acute lymphoblastic leukemia. Int. J. Cancer 1997, 74, 508–512. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Weng, Z.; Li, P.; Hu, F.; Zhang, Y.; Guo, Z.; Shen, W.; Zhao, C.; Dai, S. BATF3 promotes malignant phenotype of colorectal cancer through the S1PR1/p-STAT3/miR-155-3p/WDR82 axis. Cancer Gene Ther. 2021, 28, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Chen, P.; Zhang, F.; Zhang, N.; Zhu, J.; Wang, X.; Jiang, T. M2 macrophages-derived exosomal microRNA-501-3p promotes the progression of lung cancer via targeting WD repeat domain 82. Cancer Cell Int. 2021, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Camarillo, J.M.; Huang, T.Y.; Li, D.; Morris, J.A.; Zoltek, M.A.; Qi, J.; Behbahani, M.; Kambhampati, M.; Kelleher, N.; et al. Histone tail analysis reveals H3K36me2 and H4K16ac as epigenetic signatures of diffuse intrinsic pontine glioma. J. Exp. Clin. Cancer Res. 2020, 39, 261. [Google Scholar] [CrossRef] [PubMed]
- Larson, J.D.; Kasper, L.H.; Paugh, B.S.; Jin, H.; Wu, G.; Kwon, C.H.; Fan, Y.; Shaw, T.I.; Silveira, A.B.; Qu, C.; et al. Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell. 2019, 35, 140–155.e7. [Google Scholar] [CrossRef]
- Silveira, A.B.; Kasper, L.H.; Fan, Y.; Jin, H.; Wu, G.; Shaw, T.I.; Zhe, X.; Larson, J.D.; Easton, J.; Shao, Y.; et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019, 137, 637–655. [Google Scholar] [CrossRef]
- Sweha, S.R.; Chung, C.; Natarajan, S.K.; Panwalkar, P.; Pun, M.; Ghali, A.; Bayliss, J.; Pratt, D.; Shankar, A.; Ravikumar, V.; et al. Epigenetically defined therapeutic targeting in H3.3G34R/V high-grade gliomas. Sci. Transl. Med. 2021, 13, eabf7860. [Google Scholar] [CrossRef]
- Golbourn, B.J.; Halbert, M.E.; Halligan, K.; Varadharajan, S.; Krug, B.; Mbah, N.E.; Kabir, N.; Stanton, A.J.; Locke, A.L.; Casillo, S.M.; et al. Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome. Nat. Cancer 2022, 3, 629–648. [Google Scholar] [CrossRef]
- Dai, B.; Xiao, Z.; Zhu, G.; Mao, B.; Huang, H.; Guan, F.; Lin, Z.; Peng, W.; Liang, X.; Zhang, B.; et al. WD Repeat Domain 5 Promotes Invasion, Metastasis and Tumor Growth in Glioma Through Up-Regulated Zinc Finger E-Box Binding Homeobox 1 Expression. Cancer Manag. Res. 2020, 12, 3223–3235. [Google Scholar] [CrossRef]
- Taylor, J.T.; Ellison, S.; Pandele, A.; Wood, S.; Nathan, E.; Forte, G.; Parker, H.; Zindy, E.; Elvin, M.; Dickson, A.; et al. Actinomycin D downregulates Sox2 and improves survival in preclinical models of recurrent glioblastoma. Neuro-oncology 2020, 22, 1289–1301. [Google Scholar] [CrossRef]
- Franks, T.M.; McCloskey, A.; Shokirev, M.N.; Benner, C.; Rathore, A.; Hetzer, M.W. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 2017, 31, 2222–2234. [Google Scholar] [CrossRef]
- Zuccolo, M.; Alves, A.; Galy, V.; Bolhy, S.; Formstecher, E.; Racine, V.; Sibarita, J.; Fukagawa, T.; Shiekhattar, R.; Yen, T.; et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007, 26, 1853–1864. [Google Scholar] [CrossRef]
- Esterlechner, J.; Reichert, N.; Iltzsche, F.; Krause, M.; Finkernagel, F.; Gaubatz, S. LIN9, a subunit of the DREAM complex, regulates mitotic gene expression and proliferation of embryonic stem cells. PLoS ONE 2013, 8, e62882. [Google Scholar] [CrossRef]
- Davis, A.J.; Tsinkevich, M.; Rodencal, J.; Abbas, H.A.; Su, X.H.; Gi, Y.J.; Fang, B.; Rajapakshe, K.; Coarfa, C.; Gunaratne, P.H.; et al. TAp63-Regulated miRNAs Suppress Cutaneous Squamous Cell Carcinoma through Inhibition of a Network of Cell-Cycle Genes. Cancer Res. 2020, 80, 2484–2497. [Google Scholar] [CrossRef]
- Tassinari, V.; Cesarini, V.; Tomaselli, S.; Ianniello, Z.; Silvestris, D.A.; Ginistrelli, L.C.; Martini, M.; Angelis, B.D.; Luca, G.D.; Vitiani, L.R.; et al. ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol. 2021, 22, 51. [Google Scholar] [CrossRef]
- Min, C.; Yu, Z.; Kirsch, K.H.; Zhao, Y.; Vora, S.R.; Trackman, P.C.; Spicer, D.B.; Rosenberg, L.; Palmer, J.R.; Sonenshein, G.E. A loss-of-function polymorphism in the propeptide domain of the LOX gene and breast cancer. Cancer Res. 2009, 69, 6685–6693. [Google Scholar] [CrossRef] [PubMed]
- Gasset-Rosa, F.; Lu, S.; Yu, H.; Chen, C.; Melamed, Z.; Guo, L.; Shorter, J.; Cruz, S.D.; Cleveland, D.W. Cytoplasmic TDP-43 De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron 2019, 102, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Veeranki, S.N.; Tyagi, S. A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia. Nucleic Acids Res. 2014, 42, 7611–7624. [Google Scholar] [CrossRef] [PubMed]
- Tajima, K.; Yae, T.; Javaid, S.; Tam, O.; Comaills, V.; Morris, R.; Wittner, B.S.; Liu, M.; Engstrom, A.; Takahashi, F.; et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat Commun. 2015, 6, 8257. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wadhwani, N.; Nayak, S.; Wang, Y.; Hashizume, R.; Jie, C.; Mania-Farnell, B.; James, C.D.; Xi, G.; Tomita, T. WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas. Cancers 2023, 15, 3429. https://doi.org/10.3390/cancers15133429
Wadhwani N, Nayak S, Wang Y, Hashizume R, Jie C, Mania-Farnell B, James CD, Xi G, Tomita T. WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas. Cancers. 2023; 15(13):3429. https://doi.org/10.3390/cancers15133429
Chicago/Turabian StyleWadhwani, Nitin, Sonali Nayak, Yufen Wang, Rintaro Hashizume, Chunfa Jie, Barbara Mania-Farnell, Charles David James, Guifa Xi, and Tadanori Tomita. 2023. "WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas" Cancers 15, no. 13: 3429. https://doi.org/10.3390/cancers15133429
APA StyleWadhwani, N., Nayak, S., Wang, Y., Hashizume, R., Jie, C., Mania-Farnell, B., James, C. D., Xi, G., & Tomita, T. (2023). WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas. Cancers, 15(13), 3429. https://doi.org/10.3390/cancers15133429





