Current Model Systems for Investigating Epithelioid Haemangioendothelioma
Abstract
Simple Summary
Abstract
1. Introduction
2. Epithelioid Haemangioendothelioma: Clinical Characteristics and Molecular Alterations
3. An Overview of the Hippo Pathway
4. EHE Model Systems
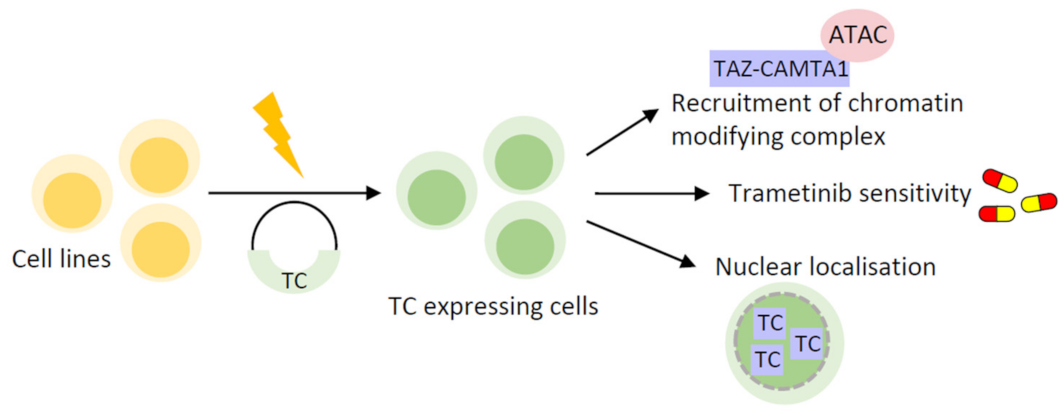
4.1. Cell Line-Based Models
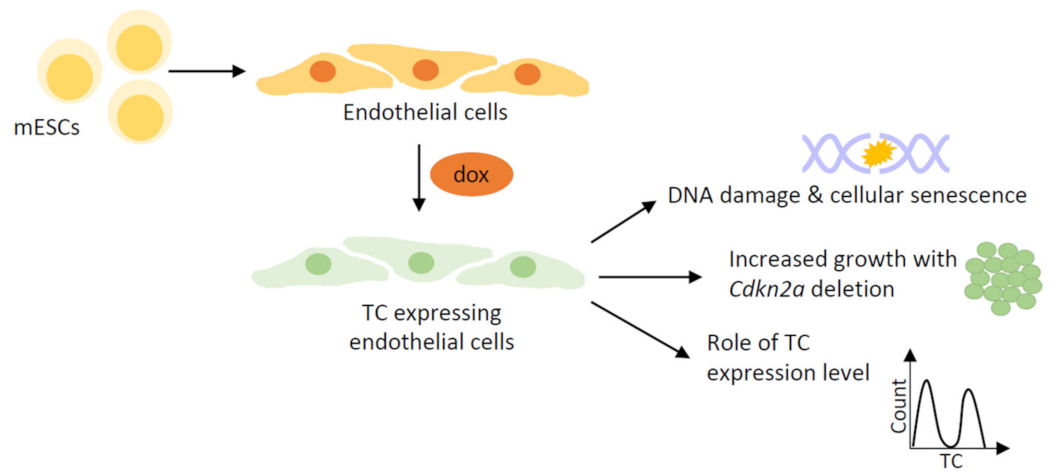
4.2. Stem Cell-Based Models
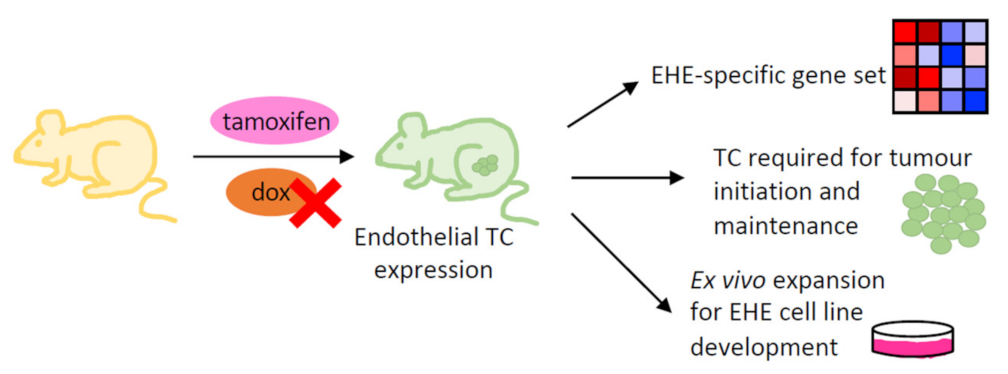
4.3. Genetically Engineered Mouse Models
4.4. Development of an EHE Cell Line
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan:, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Guan, K.-L. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012, 23, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Chen, L.-F.; Shigesada, K.; Murakami, Y.; Ito, Y. A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999, 18, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Kanai, F.; Marignani, P.A.; Sarbassova, D.; Yagi, R.; Hall, R.; Donowitz, M.; Hisaminato, A.; Fujiwara, T.; Ito, Y.; Cantley, L.; et al. TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000, 19, 6778–6791. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef]
- Vassilev, A.; Kaneko, K.J.; Shu, H.; Zhao, Y.; DePamphilis, M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001, 15, 1229–1241. [Google Scholar] [CrossRef]
- Mahoney, W.M.; Hong, J.-H.; Yaffe, M.B.; Farrance, I.K.G. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem. J. 2005, 388, 217–225. [Google Scholar] [CrossRef]
- Garcia, K.; Gingras, A.-C.; Harvey, K.F.; Tanas, M.R. TAZ/YAP fusion proteins: Mechanistic insights and therapeutic opportunities. Trends Cancer 2022, 8, 1033–1045. [Google Scholar] [CrossRef]
- Merritt, N.; Garcia, K.; Rajendran, D.; Lin, Z.-Y.; Zhang, X.; Mitchell, K.A.; Borcherding, N.; Fullenkamp, C.; Chimenti, M.S.; Gingras, A.-C.; et al. TAZ-CAMTA1 and YAP1-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex. eLife 2021, 10, e62857. [Google Scholar] [CrossRef]
- Szulzewsky, F.; Arora, S.; Hoellerbauer, P.; King, C.; Nathan, E.; Chan, M.; Cimino, P.J.; Ozawa, T.; Kawauchi, D.; Pajtler, K.W.; et al. Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev. 2020, 34, 1051–1064. [Google Scholar] [CrossRef]
- Seavey, C.N.; Pobbati, A.V.; Rubin, B.P. Unraveling the Biology of Epithelioid Hemangioendothelioma, a TAZ-CAMTA1 Fusion Driven Sarcoma. Cancers 2022, 14, 2980. [Google Scholar] [CrossRef]
- Torrence, D.; Antonescu, C.R. The genetics of vascular tumours: An update. Histopathology 2022, 80, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Le Loarer, F.; Mosquera, J.-M.; Sboner, A.; Zhang, L.; Chen, C.-L.; Chen, H.-W.; Pathan, N.; Krausz, T.; Dickson, B.C.; et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosom. Cancer 2013, 52, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Tanas, M.R.; Sboner, A.; Oliveira, A.M.; Erickson-Johnson, M.R.; Hespelt, J.; Hanwright, P.J.; Flanagan, J.; Luo, Y.; Fenwick, K.; Natrajan, R.; et al. Identification of a Disease-Defining Gene Fusion in Epithelioid Hemangioendothelioma. Sci. Transl. Med. 2011, 3, 98ra82. [Google Scholar] [CrossRef]
- Weiss, S.W.; Enzinger, F.M. Epithelioid hemangioendothelioma a vascular tumor often mistaken for a carcinoma. Cancer 1982, 50, 970–981. [Google Scholar] [CrossRef]
- Theurillat, J.P.; Vavricka, S.R.; Went, P.; Weishaupt, D.; Perren, A.; Leonard-Meier, C.; Bachli, E.B. Morphologic changes and altered gene expression in an epithelioid hemangioendothelioma during a ten-year course of disease. Pathol. Res. Pract. 2003, 199, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, E.; Jadeja, B.; Xu, B.; Zhang, L.; Agaram, N.P.; Travis, W.; Singer, S.; Tap, W.D.; Antonescu, C.R. Prognostic stratification of clinical and molecular epithelioid hemangioendothelioma subsets. Mod. Pathol. 2020, 33, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, S.; Miah, A.; Frezza, A.; Messiou, C.; Morosi, C.; Caraceni, A.; Antonescu, C.; Bajpai, J.; Baldini, E.; Bauer, S.; et al. Epithelioid hemangioendothelioma, an ultra-rare cancer: A consensus paper from the community of experts. ESMO Open 2021, 6, 100170. [Google Scholar] [CrossRef] [PubMed]
- Sardaro, A.; Bardoscia, L.; Petruzzelli, M.F.; Portaluri, M. Epithelioid hemangioendothelioma: An overview and update on a rare vascular tumor. Oncol. Rev. 2014, 8, 82–91. [Google Scholar] [CrossRef]
- Lau, K.; Massad, M.; Pollak, C.; Rubin, C.; Yeh, J.; Wang, J.; Edelman, G.; Yeh, J.; Prasad, S.; Weinberg, G. Clinical patterns and outcome in epithelioid hemangioendothelioma with or without pulmonary involvement: Insights from an internet registry in the study of a rare cancer. Chest 2011, 140, 1312–1318. [Google Scholar] [CrossRef]
- Mendlick, M.R.; Nelson, M.; Pickering, D.; Johansson, S.L.; Seemayer, T.A.; Neff, J.R.; Vergara, G.; Rosenthal, H.; Bridge, J.A. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in Epithelioid Hemangioendothelioma. Am. J. Surg. Pathol. 2001, 25, 684–687. [Google Scholar] [CrossRef]
- Errani, C.; Zhang, L.; Sung, Y.S.; Hajdu, M.; Singer, S.; Maki, R.G.; Healey, J.H.; Antonescu, C.R. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosom. Cancer 2011, 50, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Szulzewsky, F.; Holland, E.C.; Vasioukhin, V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev. Biol. 2021, 475, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A Novel Family of Calmodulin-binding Transcription Activators in Multicellular Organisms. J. Biol. Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef]
- La Spina, M.; Contreras, P.S.; Rissone, A.; Meena, N.K.; Jeong, E.; Martina, J.A. MiT/TFE Family of Transcription Factors: An Evolutionary Perspective. Front. Cell. Dev. Biol. 2020, 8, 609683. [Google Scholar] [CrossRef] [PubMed]
- Henrich, K.O.; Bauer, T.; Schulte, J.; Ehemann, V.; Deubzer, H.; Gogolin, S.; Muth, D.; Fischer, M.; Benner, A.; König, R.; et al. CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res. 2011, 71, 3142–3151. [Google Scholar] [CrossRef]
- Barbashina, B.; Salazar, P.; Holland, E.C.; Rosenblum, M.K.; Ladanyi, M. Allelic losses at 1p36 and 19q13 in gliomas; correlation with histologic classification, definition of a 150 kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin. Cancer Res. 2005, 11, 1119–1128. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.H.; Dickson, B.C.; Swanson, D.; Sung, Y.S.; Zhang, L.; Antonescu, C.R. Variant WWTR1 gene fusions in epithelioid hemangioendothelioma-A genetic subset associated with cardiac involvement. Genes Chromosom. Cancer 2020, 59, 389–395. [Google Scholar] [CrossRef]
- Seligson, N.D.; Awasthi, A.; Millis, S.Z.; Turpin, B.K.; Meyer, C.F.; Grand’Maison, A.; Liebner, D.A.; Hays, J.L.; Chen, J.L. Common Secondary Genomic Variants Associated With Advanced Epithelioid Hemangioendothelioma. JAMA Netw. Open 2019, 2, e1912416. [Google Scholar] [CrossRef]
- Collado, M.; Serrano, M. Senescence in tumours: Evidence from mice and humans. Nat. Rev. Cancer 2010, 10, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, K.; Natsume, A.; Ohka, F.; Motomura, K.; Alim, A.; Tanaka, I.; Senga, T.; Harada, I.; Fukuyama, R.; Sumiyoshi, N.; et al. Activation of Yes associated protein in low grade meningiomas is regulated by merlin, cell density, and ECM stiffness. J. Neuropathol. Exp. Neurol. 2015, 7, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Miyoshi, Y.; Takahata, C.; Irahara, N.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Downregulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 2005, 11, 1380–1385. [Google Scholar] [CrossRef]
- Jeong, W.; Kim, S.-B.; Sohn, B.H.; Park, Y.-Y.; Park, E.S.; Kim, S.C.; Kim, S.S.; Johnson, R.L.; Birrer, M.; Bowtell, D.S.; et al. Activation of YAP1 is associated with poor prognosis and response to taxanes in ovarian cancer. Anticancer Res. 2014, 34, 811–814. [Google Scholar] [PubMed]
- Wang, Q.; Xu, Z.; An, Q.; Jiang, D.; Wang, L.; Liang, B.; Li, Z. TAZ promotes epithelial to mesenchymal transition via the upregulation of connective tissue growth factor expression in neuroblastoma cells. Mol. Med. Rep. 2015, 11, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Tanas, M.R.; Ma, S.; Jadaan, F.O.; Ng, C.K.Y.; Weigelt, B.; Reis-Filho, J.S.; Rubin, B.P. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 2016, 35, 929–938. [Google Scholar] [CrossRef]
- Driskill, J.H.; Zheng, Y.; Wu, B.-K.; Wang, L.; Cai, J.; Rakheja, D.; Dellinger, M.; Pan, D. WWTR1(TAZ)-CAMTA1 reprograms endothelial cells to drive epithelioid haemangioendothelioma. Genes Dev. 2021, 35, 495–511. [Google Scholar] [CrossRef]
- Harvey, K.F.; Pfleger, C.M.; Hariharan, I.K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 2003, 114, 457–467. [Google Scholar] [CrossRef]
- Halder, G.; Johnson, R.L. Hippo signaling: Growth control and beyond. Development 2011, 138, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Justice, R.W.; Zilian, O.; Woods, D.F.; Noll, M.; Bryant, P.J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995, 9, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Poon, C.L.; Lin, J.I.; Zhang, X.; Harvey, K. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 2011, 21, 896–906. [Google Scholar] [CrossRef]
- Hergovich, A.; Schmitz, D.; Hemmings, B.A. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 2006, 345, 50–58. [Google Scholar] [CrossRef]
- Callus, B.A.; Verhagen, A.M.; Vaux, D.L. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 2006, 273, 4264–4276. [Google Scholar] [CrossRef]
- Lai, Z.-C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.-L.; Li, Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell 2005, 120, 675–685. [Google Scholar] [CrossRef]
- Lim, S.; Hermance, N.; Mudianto, T.; Mustaly, H.M.; Mauricio, I.P.M.; Vittoria, M.A.; Quinton, R.J.; Howell, B.W.; Cornils, H.; Manning, A.L.; et al. Identification of the kinase STK25 as an upstream activator of LATS signaling. Nat. Commun. 2019, 10, 1547. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Moroishi, T.; Mottier-Pavie, V.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.-S.; Lu, W.; Lu, S.; et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6, 8357. [Google Scholar] [CrossRef]
- Kapoor, A.; Yao, W.; Ying, H.; Hua, S.; Liewen, A.; Wang, Q.; Zhong, Y.; Wu, C.-J.; Sadanandam, A.; Hu, B.; et al. Yap1 Activation Enables Bypass of Oncogenic Kras Addiction in Pancreatic Cancer. Cell 2014, 158, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Varelas, X.; Sakuma, R.; Samavarchi-Tehrani, P.; Peerani, R.; Rao, B.M.; Dembowy, J.; Yaffe, M.B.; Zandstra, P.W.; Wrana, J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008, 10, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.B.; Cooper, L.F.; Yang, X.; Karsenty, G.; Aukhil, I. Transcriptional Coactivation of Bone-Specific Transcription Factor Cbfa1 by TAZ. Mol. Cell Biol. 2003, 23, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Strano, S.; Munarriz, E.; Rossi, M.; Castagnoli, L.; Shaul, Y.; Sacchi, A.; Oren, M.; Sudol, M.; Cesareni, G.; Blandino, G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001, 276, 15164–15173. [Google Scholar] [CrossRef]
- Park, H.W.; Kim, Y.C.; Yu, B.; Moroishi, T.; Mo, J.-S.; Plouffe, S.W.; Meng, Z.; Lin, K.C.; Yu, F.-X.; Alexander, C.M.; et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 2015, 162, 780–794. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 2014, 158, 157–170. [Google Scholar] [CrossRef]
- Holmes, B.; Benavides-Serrato, A.; Saunders, J.T.; Kumar, S.; Nishimura, R.N.; Gera, J. mTORC2-mediated direct phosphorylation regulates YAP activity promoting glioblastoma growth and invasive characteristics. Neoplasia 2021, 23, 951–965. [Google Scholar] [CrossRef]
- Sciarretta, S.; Zhai, P.; Maejima, Y.; Del Re, D.P.; Nagarajan, N.; Yee, D.; Liu, T.; Magnuson, M.A.; Volpe, M.; Frati, G.; et al. mTORC2 Regulates Cardiac Response to Stress by Inhibiting MST1. Cell Rep. 2015, 11, 125–136. [Google Scholar] [CrossRef]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef]
- Ma, S.; Kanai, R.; Pobbati, A.V.; Li, S.; Che, K.; Seavey, C.N.; Hallett, A.; Burtscher, A.; Lamar, J.M.; Rubin, B.P. The TAZ-CAMTA1 Fusion Protein Promotes Tumorigenesis via Connective Tissue Growth Factor and Ras–MAPK Signaling in Epithelioid Hemangioendothelioma. Clin. Cancer Res. 2022, 28, 3116–3126. [Google Scholar] [CrossRef]
- Seavey, C.N.; Pobbati, A.V.; Hallett, A.; Ma, S.; Reynolds, J.P.; Kanai, R.; Lamar, J.M.; Rubin, B.P. WWTR1(TAZ)-CAMTA1 gene fusion is sufficient to dysregulate YAP/TAZ signalling and drive epithelioid haemangioendothelioma tumorigenesis. Genes Dev. 2021, 35, 512–527. [Google Scholar] [CrossRef]
- Seavey, C.N.; Hallett, A.; Li, S.; Che, K.; Pobbati, A.V.; Ma, S.; Burtscher, A.; Kanai, R.; Lamar, J.M.; Rubin, B.P. Loss of CDKN2A cooperates with WWTR1(TAZ)-CAMTA1 gene fusion to promote tumor progression in epithelioid hemangioendothelioma. Clin. Cancer Res. 2023, OF1–OF14. [Google Scholar] [CrossRef] [PubMed]
- Neil, E.; Rubin, B.; Kouskoff, V. The oncogenic fusion protein TAZ-CAMTA1 promotes genomic instability and senescence through hypertranscription. BioRxiv 2022. [Google Scholar] [CrossRef]
- Pajtler, K.W.; Wei, Y.; Okonechnikov, K.; Silva, P.B.G.; Vouri, M.; Zhang, L.; Brabetz, S.; Sieber, L.; Gulley, M.; Mauermann, M.; et al. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat. Commun. 2019, 10, 3914. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Kiyono, T.; Ryo, E.; Ogawa, R.; Wakai, S.; Ichikawa, H.; Suzuki, K.; Arai, S.; Tsuta, K.; Ishida, M.; et al. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J. Clin. Investig. 2019, 129, 3827–3832. [Google Scholar] [CrossRef]
- Pocaterra, A.; Romani, P.; Dupont, S. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020, 133, jcs230425. [Google Scholar] [CrossRef]
- Chang, L.; Azzolin, L.; Di Biagio, D.; Zanconato, F.; Battilana, G.; Xiccato, R.L.; Aragona, M.; Giulitti, S.; Panciera, T.; Gandin, A.; et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 2018, 563, 265–269. [Google Scholar] [CrossRef]
- Brien, G.L.; Stegmaier, K.; Armstrong, S.A. Targeting chromatin complexes in fusion protein-driven malignancies. Nat. Rev. Cancer 2019, 19, 255–269. [Google Scholar] [CrossRef]
- Cottini, F.; Hideshima, T.; Xu, C.; Sattler, M.; Dori, M.; Agnelli, L.; Hacken, E.T.; Bertilaccio, M.T.; Antonini, E.; Neri, A.; et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat. Med. 2014, 20, 599–606. [Google Scholar] [CrossRef]
- Pearson, J.D.; Huang, K.; Pacal, M.; McCurdy, S.R.; Lu, S.; Aubry, A.; Yu, T.; Wadosky, K.M.; Zhang, L.; Wang, T.; et al. Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell 2021, 39, 1115–1134e12. [Google Scholar] [CrossRef]
- Avior, Y.; Sagi, I.; Benvenisty, N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Moleular Cell Biol. 2016, 17, 170–182. [Google Scholar] [CrossRef]
- Bai, X. Stem Cell-Based Disease Modeling and Cell Therapy. Cells 2020, 9, 2193. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Karanam, K.; Kafri, R.; Loewer, A.; Lahav, G. Quantitative Live Cell Imaging Reveals a Gradual Shift between DNA Repair Mechanisms and a Maximal Use of HR in Mid S Phase. Mol. Cell 2012, 47, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Gorthi, A.; Romero, J.C.; Loranc, E.; Cao, L.; Lawrence, L.A.; Goodale, E.; Iniguez, A.B.; Bernard, X.; Masamsetti, V.P.; Roston, S.; et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018, 555, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, Q.; Tang, M.; Barthel, F.; Amin, S.; Yoshihara, K.; Lang, F.M.; Martinez-Ledesma, E.; Lee, S.H.; Zheng, S.; et al. TumorFusions: An integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018, 46, D1144–D1149. [Google Scholar] [CrossRef]
- Pobbati, A.V.; Kumar, R.; Rubin, B.P.; Hong, W. Therapeutic targeting of TEAD transcription factors in cancer. Trends Biochem. Sci. 2023, 48, 450–462. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP and TAZ: A signalling hub of the tumour microenvironment. Nat. Rev. Cancer 2019, 19, 454–464. [Google Scholar] [CrossRef]
- Guo, X.; Zhao, Y.; Yan, H.; Yang, Y.; Shen, S.; Dai, X.; Ji, X.; Ji, F.; Gong, X.-G.; Li, L.; et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017, 31, 247–259. [Google Scholar] [CrossRef]
- Wang, G.; Lu, X.; Dey, P.; Deng, P.; Wu, C.C.; Jiang, S.; Fang, Z.; Zhao, K.; Konaparthi, R.; Hua, S.; et al. Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discov. 2016, 6, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Shahbazian, D.; Surana, R.; Zhang, W.; Chen, H.; Graham, G.T.; White, S.M.; Weiner, L.M.; Yi, C. Yes-associated protein mediates immune reprogramming in pancreatic ductal adenocarcinoma. Oncogene 2017, 36, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Calvo, F.; Ege, N.; Grande-Garcia, A.; Hooper, S.; Jenkins, R.P.; Chaudhry, S.I.; Harrington, K.; Williamson, P.; Moeendarbary, E.; Charras, G.; et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013, 15, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Oldham, W.M.; Grasset, E.M.; Bourget, I.; Boulter, E.; Pisano, S.; Hofman, P.; Bellvert, F.; Meneguzzi, G.; Bulavin, D.V.; et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019, 29, 124–140.e10. [Google Scholar] [CrossRef]
- Nakajima, H.; Yamamoto, K.; Agarwala, S.; Terai, K.; Fukui, H.; Fukuhara, S.; Ando, K.; Miyazaki, T.; Yokota, Y.; Schmelzer, E.; et al. Flow-Dependent Endothelial YAP Regulation Contributes to Vessel Maintenance. Dev. Cell 2017, 40, 523–536.e6. [Google Scholar] [CrossRef]
| Type | Cell Lines | Mouse Model | Fusion | Reference |
|---|---|---|---|---|
| In vitro | HEK293, NIH3T3 | - | TC | Tanas [38] |
| In vitro and in vivo | HEK293 | GEMM | YT | Szulzewsky [11] |
| In vitro and in vivo | NIH3T3, SW872 | Xenograft | TC/YT | Merritt [10] |
| In vitro and in vivo | MS1 | Xenograft and GEMM | TC | Driskill [39] |
| In vitro and in vivo | NIH3T3 | Xenograft | TC | Ma [60] |
| In vivo | - | GEMM | TC | Seavey [61] |
| In vitro and in vivo | EHE cell line | GEMM | TC | Seavey [62] |
| In vitro | mESCs | - | TC | Neil [63] |
| Cell Line-Based | Stem Cell-Based | GEMM | EHE Cell Line | |
|---|---|---|---|---|
| Drug testing | ✓ | - | ✓ | ✓ |
| Generate large amount of sample | ✓ | ✓ | - | ✓ |
| Reduce the use of animals | - | ✓ | - | ✓ |
| Transfer to in vivo by xenograft | ✓ | - | - | ✓ |
| Correct cell context | - | ✓ | - | ✓ |
| Easy to study other driver fusion proteins | ✓ | - | - | - |
| Easy to establish and simple culture conditions | ✓ | - | - | - |
| Representative of disease course and tumour histology | - | - | ✓ | - |
| Study initiating events | - | ✓ | - | - |
| Study TC at various stages of development | - | ✓ | - | - |
| Study metastasis | - | - | ✓ | - |
| Determine tumour microenvironmental contributions | - | - | ✓ | - |
| EHE cell line development | - | - | ✓ | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neil, E.; Kouskoff, V. Current Model Systems for Investigating Epithelioid Haemangioendothelioma. Cancers 2023, 15, 3005. https://doi.org/10.3390/cancers15113005
Neil E, Kouskoff V. Current Model Systems for Investigating Epithelioid Haemangioendothelioma. Cancers. 2023; 15(11):3005. https://doi.org/10.3390/cancers15113005
Chicago/Turabian StyleNeil, Emily, and Valerie Kouskoff. 2023. "Current Model Systems for Investigating Epithelioid Haemangioendothelioma" Cancers 15, no. 11: 3005. https://doi.org/10.3390/cancers15113005
APA StyleNeil, E., & Kouskoff, V. (2023). Current Model Systems for Investigating Epithelioid Haemangioendothelioma. Cancers, 15(11), 3005. https://doi.org/10.3390/cancers15113005







