Predicting Lymph Node Metastasis Status from Primary Muscle-Invasive Bladder Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
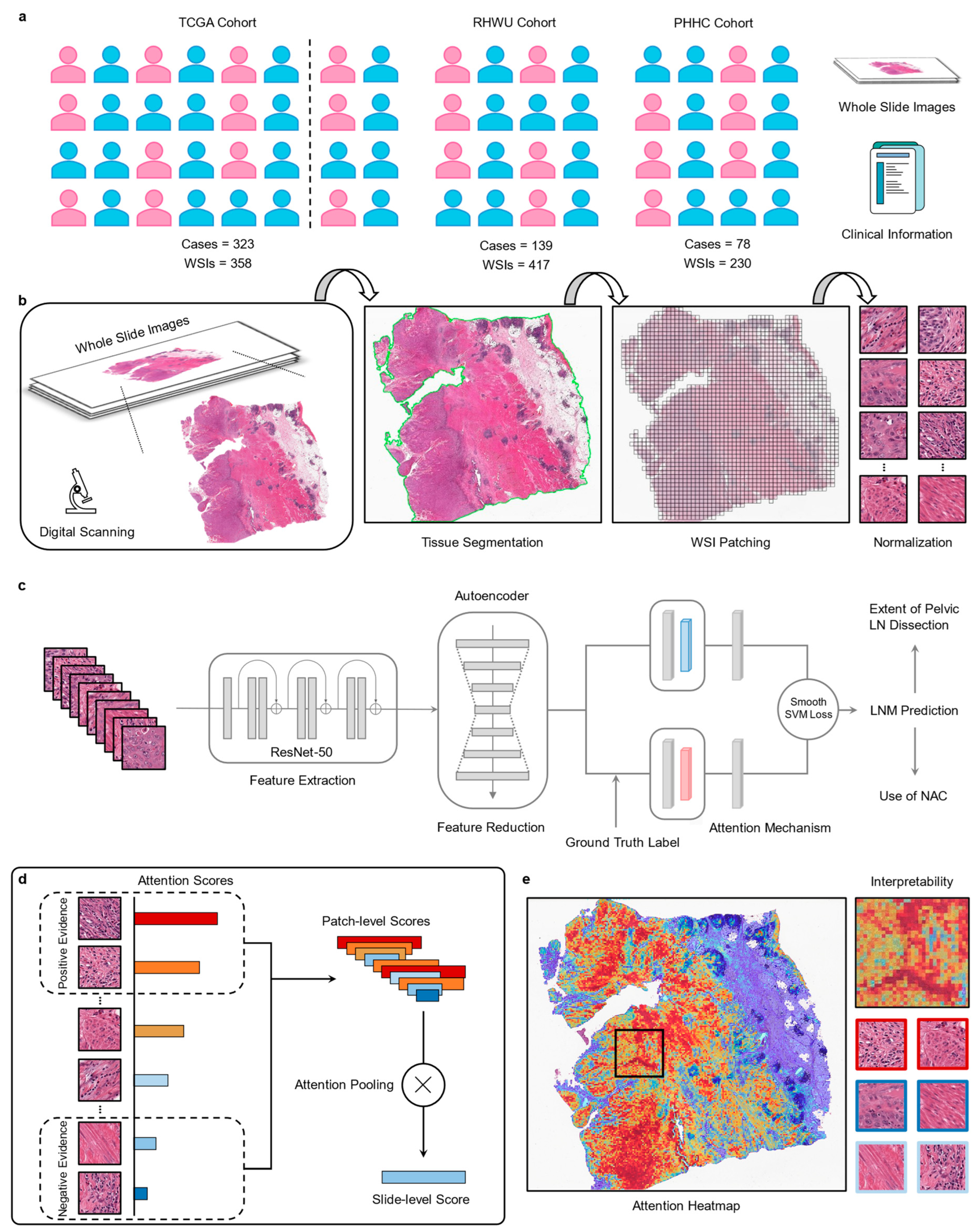
2.1. Patient Cohorts
2.2. Image Preprocessing
2.3. Feature Extraction and Reduction
2.4. Slide-Based Lymph Node Predictor (SBLNP)
2.5. Interpreting Predictions via Attention Heatmap
2.6. Quantification of Histopathological Features
2.7. Clinical Classifier and Combined Classifier (Clinical Classifier + SBLNP)
2.8. Statistical Analysis
3. Results
3.1. Patient Characteristics
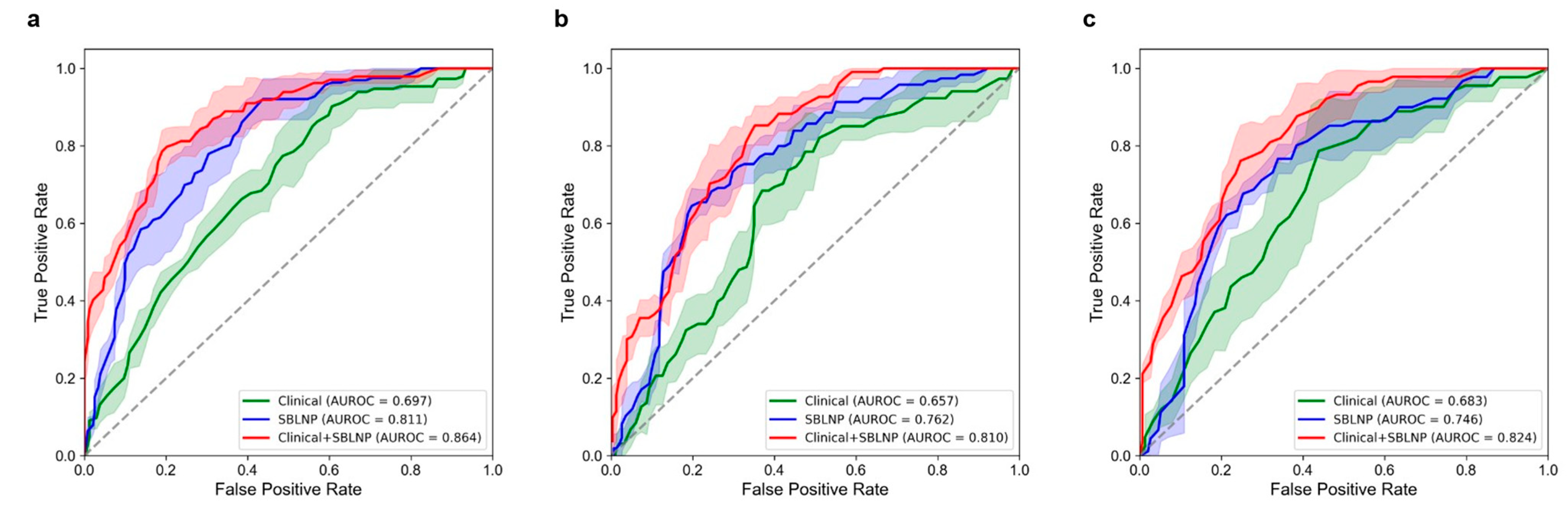
3.2. Performance of the SBLNP
3.3. Performance of the Clinical Classifier
3.4. Performance of the Combined Classifier (Clinical Classifier + SBLNP)
3.5. Visualizing Deep Learning-Based Predictions
3.6. Quantitative Assessment of Histopathological Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder cancer. Nat. Rev. Dis. Primers 2017, 3, 17022. [Google Scholar] [CrossRef]
- Patel, V.G.; Oh, W.K.; Galsky, M.D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J. Clin. 2020, 70, 404–423. [Google Scholar] [CrossRef]
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malmström, P.U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.; Mcgrath, S.; Sengupta, S.; Crozier, J.; Bolton, D.; Lawrentschuk, N. Pelvic lymph node dissection during radical cystectomy for muscle-invasive bladder cancer. Nat. Rev. Urol. 2018, 15, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Yafi, F.A.; Aprikian, A.G.; Chin, J.L.; Fradet, Y.; Izawa, J.; Estey, E.; Fairey, A.; Rendon, R.; Cagiannos, I.; Lacombe, L.; et al. Contemporary outcomes of 2287 patients with bladder cancer who were treated with radical cystectomy: A canadian multicentre experience. BJU Int. 2011, 108, 539–545. [Google Scholar] [CrossRef]
- Stein, J.P.; Lieskovsky, G.; Cote, R.; Groshen, S.; Feng, A.C.; Boyd, S.; Skinner, E.; Bochner, B.; Thangathurai, D.; Mikhail, M.; et al. Radical cystectomy in the treatment of invasive bladder cancer: Long-term results in 1054 patients. J. Clin. Oncol. 2001, 19, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Lobo, N.; Mount, C.; Omar, K.; Nair, R.; Thurairaja, R.; Khan, M.S. Landmarks in the treatment of muscle-invasive bladder cancer. Nat. Rev. Urol. 2017, 14, 565–574. [Google Scholar] [CrossRef]
- Motterle, G.; Andrews, J.R.; Morlacco, A.; Karnes, R.J. Predicting response to neoadjuvant chemotherapy in bladder cancer. Eur. Urol. Focus 2020, 6, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Linares, E.E.; Lorch, A.; Neuzillet, Y.; et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2020 guidelines. Eur. Urol. 2021, 79, 82–104. [Google Scholar] [CrossRef]
- Wagner, J.; Simon, R.; Büchler, J.W.; Kirchhoff, F.; Kehl, V.; Retz, M.; Gschwend, J.E.; Sauter, A.; Horn, T. Both radiographical and pathological lymph node statuses are independent predictors for survival following neoadjuvant chemotherapy and radical cystectomy for ct3/4 or cn+ bladder cancer. World J. Urol. 2022, 41, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, G.; Maes, A.; Pottel, H.; Vanneste, A.; Billiet, I.; Lesage, K.; Werbrouck, P. Fdg-pet/ct for the preoperative lymph node staging of invasive bladder cancer. Eur. Urol. 2010, 57, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Lodde, M.; Lacombe, L.; Friede, J.; Morin, F.; Saourine, A.; Fradet, Y. Evaluation of fluorodeoxyglucose positron-emission tomography with computed tomography for staging of urothelial carcinoma. BJU Int. 2010, 106, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Meng, L.; Wang, X.; Diao, T.; Hu, M.; Wang, M.; Zhang, Y.; Liu, M. Predictive nomogram and risk factors for lymph node metastasis in bladder cancer. Front. Oncol. 2021, 11, 690324. [Google Scholar] [CrossRef]
- Zhang, C.; Hu, J.; Li, H.; Ma, H.; Othmane, B.; Ren, W.; Yi, Z.; Qiu, D.; Ou, Z.; Chen, J.; et al. Emerging biomarkers for predicting bladder cancer lymph node metastasis. Front. Oncol. 2021, 11, 648968. [Google Scholar] [CrossRef]
- Seiler, R.; Lam, L.L.; Erho, N.; Takhar, M.; Mitra, A.P.; Buerki, C.; Davicioni, E.; Skinner, E.C.; Daneshmand, S.; Black, P.C. Prediction of lymph node metastasis in patients with bladder cancer using whole transcriptome gene expression signatures. J. Urol. 2016, 196, 1036–1041. [Google Scholar] [CrossRef]
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Nojima, S.; Terayama, K.; Shimoura, S.; Hijiki, S.; Nonomura, N.; Morii, E.; Okuno, Y.; Fujita, K. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021, 129, 984–995. [Google Scholar] [CrossRef]
- Zheng, Q.; Jiang, Z.; Ni, X.; Yang, S.; Jiao, P.; Wu, J.; Xiong, L.; Yuan, J.; Wang, J.; Jian, J.; et al. Machine learning quantified tumor-stroma ratio is an independent prognosticator in muscle-invasive bladder cancer. Int. J. Mol. Sci. 2023, 24, 2746. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Jiao, P.; Wu, J.; Xiong, L.; Wang, J.; Jian, J.; Jiang, Z.; et al. Quantitative assessment of tumor-infiltrating lymphocytes using machine learning predicts survival in muscle-invasive bladder cancer. J. Clin. Med. 2022, 11, 7081. [Google Scholar] [CrossRef]
- Woerl, A.C.; Eckstein, M.; Geiger, J.; Wagner, D.C.; Daher, T.; Stenzel, P.; Fernandez, A.; Hartmann, A.; Wand, M.; Roth, W.; et al. Deep learning predicts molecular subtype of muscle-invasive bladder cancer from conventional histopathological slides. Eur. Urol. 2020, 78, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Xiong, L.; Yan, D.; Xia, L.; Yuan, J.; Wang, J.; Jiao, P.; et al. Accurate diagnosis and survival prediction of bladder cancer using deep learning on histological slides. Cancers 2022, 14, 5807. [Google Scholar] [CrossRef]
- Qaiser, T.; Lee, C.Y.; Vandenberghe, M.; Yeh, J.; Gavrielides, M.A.; Hipp, J.; Scott, M.; Reischl, J. Usability of deep learning and h&e images predict disease outcome-emerging tool to optimize clinical trials. NPJ Precis. Oncol. 2022, 6, 37. [Google Scholar]
- Xu, F.; Zhu, C.; Tang, W.; Wang, Y.; Zhang, Y.; Li, J.; Jiang, H.; Shi, Z.; Liu, J.; Jin, M. Predicting axillary lymph node metastasis in early breast cancer using deep learning on primary tumor biopsy slides. Front. Oncol. 2021, 11, 759007. [Google Scholar] [CrossRef]
- Kiehl, L.; Kuntz, S.; Höhn, J.; Jutzi, T.; Krieghoff-Henning, E.; Kather, J.N.; Holland-Letz, T.; Kopp-Schneider, A.; Chang-Claude, J.; Brobeil, A.; et al. Deep learning can predict lymph node status directly from histology in colorectal cancer. Eur. J. Cancer 2021, 157, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Brockmoeller, S.; Echle, A.; Ghaffari, L.N.; Eiholm, S.; Malmstrøm, M.L.; Plato, K.T.; Levic, K.; Grabsch, H.I.; West, N.P.; Saldanha, O.L.; et al. Deep learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J. Pathol. 2022, 256, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Hong, Y.; Kim, E.R.; Kim, S.H.; Sohn, I. Utility of artificial intelligence with deep learning of hematoxylin and eosin-stained whole slide images to predict lymph node metastasis in t1 colorectal cancer using endoscopically resected specimens; Prediction of lymph node metastasis in t1 colorectal cancer. J. Gastroenterol. 2022, 57, 654–666. [Google Scholar]
- Wessels, F.; Schmitt, M.; Krieghoff-Henning, E.; Jutzi, T.; Worst, T.S.; Waldbillig, F.; Neuberger, M.; Maron, R.C.; Steeg, M.; Gaiser, T.; et al. Deep learning approach to predict lymph node metastasis directly from primary tumour histology in prostate cancer. BJU Int. 2021, 128, 352–360. [Google Scholar] [CrossRef]
- Harmon, S.A.; Sanford, T.H.; Brown, G.T.; Yang, C.; Mehralivand, S.; Jacob, J.M.; Valera, V.A.; Shih, J.H.; Agarwal, P.K.; Choyke, P.L.; et al. Multiresolution application of artificial intelligence in digital pathology for prediction of positive lymph nodes from primary tumors in bladder cancer. JCO Clin. Cancer Info. 2020, 4, 367–382. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Abel, J.T.; Balis, U.; Mcclintock, D.S.; Pantanowitz, L. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology. Am. J. Pathol. 2021, 191, 1684–1692. [Google Scholar] [CrossRef]
- Vahadane, A.; Peng, T.Y.; Albarqouni, S.; Baust, M.; Steiger, K.; Schlitter, A.M.; Sethi, A.; Esposito, I.; Navab, N. Structure-preserved color normalization for histological images. In Proceedings of the IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 1012–1015. [Google Scholar]
- Anand, D.; Ramakrishnan, G.; Sethi, A. Fast GPU-enabled color normalization for digital pathology. In Proceedings of the 2019 International Conference on Systems, Signals and Image Processing (IWSSIP 2019), Osijek, Croatia, 5–7 June 2019; pp. 219–224. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Leng, L.; Li, M.; Kim, C.; Bi, X. Dual-source discrimination power analysis for multi-instance contactless palmprint recognition. Multimed. Tools Appl. 2017, 76, 333–354. [Google Scholar] [CrossRef]
- Leng, L.; Zhang, J. Palmhash code vs. Palmphasor code. Neurocomputing 2013, 108, 1–12. [Google Scholar] [CrossRef]
- Ilse, M.; Tomczak, J.M.; Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; Volume 80. [Google Scholar]
- Berrada, L.; Zisserman, A.; Kumar, M.P. Smooth loss functions for deep top-k classification. arXiv 2018, arXiv:1802.07595. [Google Scholar]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, P.; Studer, U.E.; Daneshmand, S.; Birkhäuser, F.D.; Skinner, E.C.; Roth, B.; Miranda, G.; Burkhard, F.C.; Cai, J.; Skinner, D.G.; et al. Outcomes of radical cystectomy with extended lymphadenectomy alone in patients with lymph node-positive bladder cancer who are unfit for or who decline adjuvant chemotherapy. BJU Int. 2014, 113, 554–560. [Google Scholar] [CrossRef]
- Bassi, P.; Ferrante, G.D.; Piazza, N.; Spinadin, R.; Carando, R.; Pappagallo, G.; Pagano, F. Prognostic factors of outcome after radical cystectomy for bladder cancer: A retrospective study of a homogeneous patient cohort. J. Urol. 1999, 161, 1494–1497. [Google Scholar] [CrossRef]
- Abdel-Latif, M.; Abol-Enein, H.; El-Baz, M.; Ghoneim, M.A. Nodal involvement in bladder cancer cases treated with radical cystectomy: Incidence and prognosis. J. Urol. 2004, 172, 85–89. [Google Scholar] [CrossRef]
- Karl, A.; Carroll, P.R.; Gschwend, J.E.; Knüchel, R.; Montorsi, F.; Stief, C.G.; Studer, U.E. The impact of lymphadenectomy and lymph node metastasis on the outcomes of radical cystectomy for bladder cancer. Eur. Urol. 2009, 55, 826–835. [Google Scholar] [CrossRef]
- Darwish, C.; Sparks, A.; Amdur, R.; Reddy, A.; Whalen, M. Trends in treatment strategies and comparison of outcomes in lymph node positive bladder cancer: An analysis of the national cancer database. Urology 2020, 146, 168–176. [Google Scholar] [CrossRef]
- Mckibben, M.J.; Woods, M.E. Preoperative imaging for staging bladder cancer. Curr. Urol. Rep. 2015, 16, 22. [Google Scholar] [CrossRef]
- Baltaci, S.; Resorlu, B.; Yagci, C.; Turkolmez, K.; Gogus, C.; Beduk, Y. Computerized tomography for detecting perivesical infiltration and lymph node metastasis in invasive bladder carcinoma. Urol. Int. 2008, 81, 399–402. [Google Scholar] [CrossRef]
- Shariat, S.F.; Rink, M.; Ehdaie, B.; Xylinas, E.; Babjuk, M.; Merseburger, A.S.; Svatek, R.S.; Cha, E.K.; Tagawa, S.T.; Fajkovic, H.; et al. Pathologic nodal staging score for bladder cancer: A decision tool for adjuvant therapy after radical cystectomy. Eur. Urol. 2013, 63, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Werntz, R.P.; Smith, Z.L.; Packiam, V.T.; Smith, N.; Steinberg, G.D. The impact of lymphovascular invasion on risk of upstaging and lymph node metastasis at the time of radical cystectomy. Eur. Urol. Focus 2020, 6, 292–297. [Google Scholar] [CrossRef]
- Sherif, A.; Holmberg, L.; Rintala, E.; Mestad, O.; Nilsson, J.; Nilsson, S.; Malmström, P.U. Neoadjuvant cisplatinum based combination chemotherapy in patients with invasive bladder cancer: A combined analysis of two nordic studies. Eur. Urol. 2004, 45, 297–303. [Google Scholar] [CrossRef]
- Griffiths, G.; Hall, R.; Sylvester, R.; Raghavan, D.; Parmar, M.K. International phase iii trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: Long-term results of the ba06 30894 trial. J. Clin. Oncol. 2011, 29, 2171–2177. [Google Scholar] [PubMed]
- Hanna, N.; Trinh, Q.D.; Seisen, T.; Vetterlein, M.W.; Sammon, J.; Preston, M.A.; Lipsitz, S.R.; Bellmunt, J.; Menon, M.; Choueiri, T.K.; et al. Effectiveness of neoadjuvant chemotherapy for muscle-invasive bladder cancer in the current real world setting in the usa. Eur. Urol. Oncol. 2018, 1, 83–90. [Google Scholar] [CrossRef]
- Mertens, L.S.; Meijer, R.P.; Meinhardt, W.; van der Poel, H.G.; Bex, A.; Kerst, J.M.; van der Heijden, M.S.; Bergman, A.M.; Horenblas, S.; van Rhijn, B.W. Occult lymph node metastases in patients with carcinoma invading bladder muscle: Incidence after neoadjuvant chemotherapy and cystectomy vs after cystectomy alone. BJU Int. 2014, 114, 67–74. [Google Scholar] [CrossRef]
- Wang, L.; Mudaliar, K.; Mehta, V.; Barkan, G.A.; Quek, M.L.; Flanigan, R.C.; Picken, M.M. Seeking a standard for adequate pathologic lymph node staging in primary bladder carcinoma. Virchows Arch. 2014, 464, 595–602. [Google Scholar] [CrossRef]
- Dorin, R.P.; Daneshmand, S.; Eisenberg, M.S.; Chandrasoma, S.; Cai, J.; Miranda, G.; Nichols, P.W.; Skinner, D.G.; Skinner, E.C. Lymph node dissection technique is more important than lymph node count in identifying nodal metastases in radical cystectomy patients: A comparative mapping study. Eur. Urol. 2011, 60, 946–952. [Google Scholar] [CrossRef]
- Bruins, H.M.; Veskimae, E.; Hernandez, V.; Imamura, M.; Neuberger, M.M.; Dahm, P.; Stewart, F.; Lam, T.B.; N’Dow, J.; van der Heijden, A.G.; et al. The impact of the extent of lymphadenectomy on oncologic outcomes in patients undergoing radical cystectomy for bladder cancer: A systematic review. Eur. Urol. 2014, 66, 1065–1077. [Google Scholar] [CrossRef]
- Wu, S.X.; Huang, J.; Liu, Z.W.; Chen, H.G.; Guo, P.; Cai, Q.Q.; Zheng, J.J.; Qin, H.D.; Zheng, Z.S.; Chen, X.; et al. A genomic-clinicopathologic nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Ebiomedicine 2018, 31, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zheng, J.; Li, Y.; Wu, Z.; Shi, S.; Huang, M.; Yu, H.; Dong, W.; Huang, J.; Lin, T. Development and validation of an mri-based radiomics signature for the preoperative prediction of lymph node metastasis in bladder cancer. Ebiomedicine 2018, 34, 76–84. [Google Scholar] [CrossRef]
- Wu, S.; Zheng, J.; Li, Y.; Yu, H.; Shi, S.; Xie, W.; Liu, H.; Su, Y.; Huang, J.; Lin, T. A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin. Cancer Res. 2017, 23, 6904–6911. [Google Scholar] [CrossRef] [PubMed]
- Fontanet, S.; Basile, G.; Baboudjian, M.; Gallioli, A.; Huguet, J.; Territo, A.; Parada, R.; Gavrilov, P.; Aumatell, J.; Sanz, I.; et al. Robot-assisted vs. open radical cystectomy: Systematic review and meta-analysis of randomized controlled trials. Actas Urol. Esp. 2023. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Dupré, A.; Malik, H.Z. Inflammation and cancer: What a surgical oncologist should know. EJSO 2018, 44, 566–570. [Google Scholar] [CrossRef]
- Chittezhath, M.; Dhillon, M.K.; Lim, J.Y.; Laoui, D.; Shalova, I.N.; Teo, Y.L.; Chen, J.; Kamaraj, R.; Raman, L.; Lum, J.; et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014, 41, 815–829. [Google Scholar] [CrossRef]
- Wei, W.F.; Chen, X.J.; Liang, L.J.; Yu, L.; Wu, X.G.; Zhou, C.F.; Wang, Z.C.; Fan, L.S.; Hu, Z.; Liang, L.; et al. Periostin(+) cancer-associated fibroblasts promote lymph node metastasis by impairing the lymphatic endothelial barriers in cervical squamous cell carcinoma. Mol. Oncol. 2021, 15, 210–227. [Google Scholar] [CrossRef]
- Li, X.; Zhai, J.; Shen, Y.; Zhang, T.; Wang, Y.; He, Y.; You, Q.; Shen, L. Tumor-derived il-8 facilitates lymph node metastasis of gastric cancer via pd-1 up-regulation in cd8(+) T cells. Cancer Immunol. Immunother. 2022, 71, 3057–3070. [Google Scholar] [CrossRef]



| TCGA (n = 323) | RHWU (n = 139) | PHHC (n = 78) | |
|---|---|---|---|
| Age (years) | 69 (34, 90) | 66 (26, 87) | 70 (45, 90) |
| Gender | |||
| Female | 88 (27.24%) | 21 (15.11%) | 17 (21.79%) |
| Male | 235 (72.76%) | 118 (84.89%) | 61 (78.21%) |
| pT stage | |||
| pT2 | 98 (30.34%) | 67 (48.20%) | 30 (38.46%) |
| pT3 | 175 (54.18%) | 44 (31.65%) | 28 (35.90%) |
| pT4 | 50 (15.48%) | 28 (20.14%) | 20 (25.64%) |
| pM stage | |||
| pM0 | 146 (45.20%) | 104 (74.82%) | 57 (73.08%) |
| pM1 | 7 (2.17%) | 35 (35.18%) | 21 (26.92%) |
| pMx | 170 (52.63%) | 0 (0%) | 0 (0%) |
| pTNM stage | |||
| Stage II | 83 (25.70%) | 58 (41.73%) | 74 (32.18%) |
| Stage III | 122 (37.77%) | 45 (32.37%) | 78 (33.91%) |
| Stage IV | 118 (36.53%) | 36 (25.90%) | 78 (33.91%) |
| Histologic grade | |||
| High grade | 303 (93.81%) | 129 (92.81%) | 74 (94.87%) |
| Low grade | 18 (5.57%) | 10 (7.19%) | 4 (5.13%) |
| Missing | 2 (0.62%) | 0 (0%) | 0 (0%) |
| LVI | |||
| No | 104 (32.20%) | 81 (58.27%) | 47 (60.26%) |
| Yes | 127 (39.32%) | 58 (41.73%) | 31 (39.74%) |
| Missing | 92 (28.48%) | 0 (0%) | 0 (0%) |
| LN status | |||
| Negative (pN0) | 207 (64.09%) | 102 (73.38%) | 53 (67.95%) |
| Positive (pN1-3) | 116 (35.91%) | 37 (26.62%) | 25 (32.05%) |
| LNs examined number | 18 (1, 170) | 21 (1, 64) | 16 (1, 47) |
| Positive LNs number | 2 (1, 97) | 2 (1, 20) | 3 (1, 31) |
| Survival status | |||
| Alive | 178 (55.11%) | - | - |
| Dead | 145 (44.89%) | - | - |
| OS time (months) | 17.4 (0, 165.6) | - | - |
| Model | TCGA Cohort | RHWU Cohort | PHHC Cohort |
|---|---|---|---|
| AUROC (95% CI) | AUROC (95% CI) | AUROC (95% CI) | |
| Clinical | 0.697 (0.661, 0.728) | 0.657 (0.595, 0.713) | 0.683 (0.537, 0.829) |
| SBLNP | 0.811 (0.771, 0.855) | 0.762 (0.725, 0.801) | 0.746 (0.687, 0.799) |
| Clinical + SBLNP | 0.864 (0.827, 0.906) | 0.810 (0.780, 0.844) | 0.824 (0.788, 0.861) |
| Characteristic | Coefficient | p Value | Odds Ratio (95% CI) |
|---|---|---|---|
| Age | 0.0034 | 0.120 | 1.003 (0.999–1.008) |
| Gender | 0.0591 | 0.256 | 1.061 (0.958–1.175) |
| LVI | 0.3211 | <0.001 | 1.379 (1.252–1.518) |
| pT stage | 0.1072 | 0.005 | 1.113 (1.033–1.199) |
| Histologic grade | −0.0808 | 0.474 | 0.922 (0.739–1.151) |
| SBLNP | 0.7285 | <0.001 | 2.072 (1.694–2.535) |
| Comparisons | TCGA Cohort | RHWU Cohort | PHHC Cohort |
|---|---|---|---|
| Clinical vs. SBLNP | p = 0.028 | p = 0.632 | p = 0.703 |
| Clinical vs. Clinical + SBLNP | p = 0.001 | p = 0.004 | p = 0.021 |
| SBLNP vs. Clinical + SBLNP | p = 0.093 | p = 0.014 | p = 0.005 |
| Positive LNM Status | Negative LNM Status | |||
|---|---|---|---|---|
| Histological Features | n Patches | % Patches | n Patches | % Patches |
| Tumor cells | 45 | 6 | 654 | 87.2 |
| Normal transitional epithelium | 23 | 3.07 | 27 | 3.6 |
| Muscle tissue | 82 | 10.93 | 24 | 3.2 |
| Adipose tissue | 8 | 1.07 | 1 | 0.13 |
| Immune cells | 218 | 29.06 | 3 | 0.4 |
| Necrotic tissue | 29 | 3.87 | 18 | 2.4 |
| Stroma | 336 | 44.8 | 16 | 2.13 |
| Out of focus | 9 | 1.2 | 7 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Jian, J.; Wang, J.; Wang, K.; Fan, J.; Xu, H.; Ni, X.; Yang, S.; Yuan, J.; Wu, J.; et al. Predicting Lymph Node Metastasis Status from Primary Muscle-Invasive Bladder Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study. Cancers 2023, 15, 3000. https://doi.org/10.3390/cancers15113000
Zheng Q, Jian J, Wang J, Wang K, Fan J, Xu H, Ni X, Yang S, Yuan J, Wu J, et al. Predicting Lymph Node Metastasis Status from Primary Muscle-Invasive Bladder Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study. Cancers. 2023; 15(11):3000. https://doi.org/10.3390/cancers15113000
Chicago/Turabian StyleZheng, Qingyuan, Jun Jian, Jingsong Wang, Kai Wang, Junjie Fan, Huazhen Xu, Xinmiao Ni, Song Yang, Jingping Yuan, Jiejun Wu, and et al. 2023. "Predicting Lymph Node Metastasis Status from Primary Muscle-Invasive Bladder Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study" Cancers 15, no. 11: 3000. https://doi.org/10.3390/cancers15113000
APA StyleZheng, Q., Jian, J., Wang, J., Wang, K., Fan, J., Xu, H., Ni, X., Yang, S., Yuan, J., Wu, J., Jiao, P., Yang, R., Chen, Z., Liu, X., & Wang, L. (2023). Predicting Lymph Node Metastasis Status from Primary Muscle-Invasive Bladder Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study. Cancers, 15(11), 3000. https://doi.org/10.3390/cancers15113000








