BRMS1 in Gliomas—An Expression Analysis
Abstract
Simple Summary
Abstract
1. Introduction
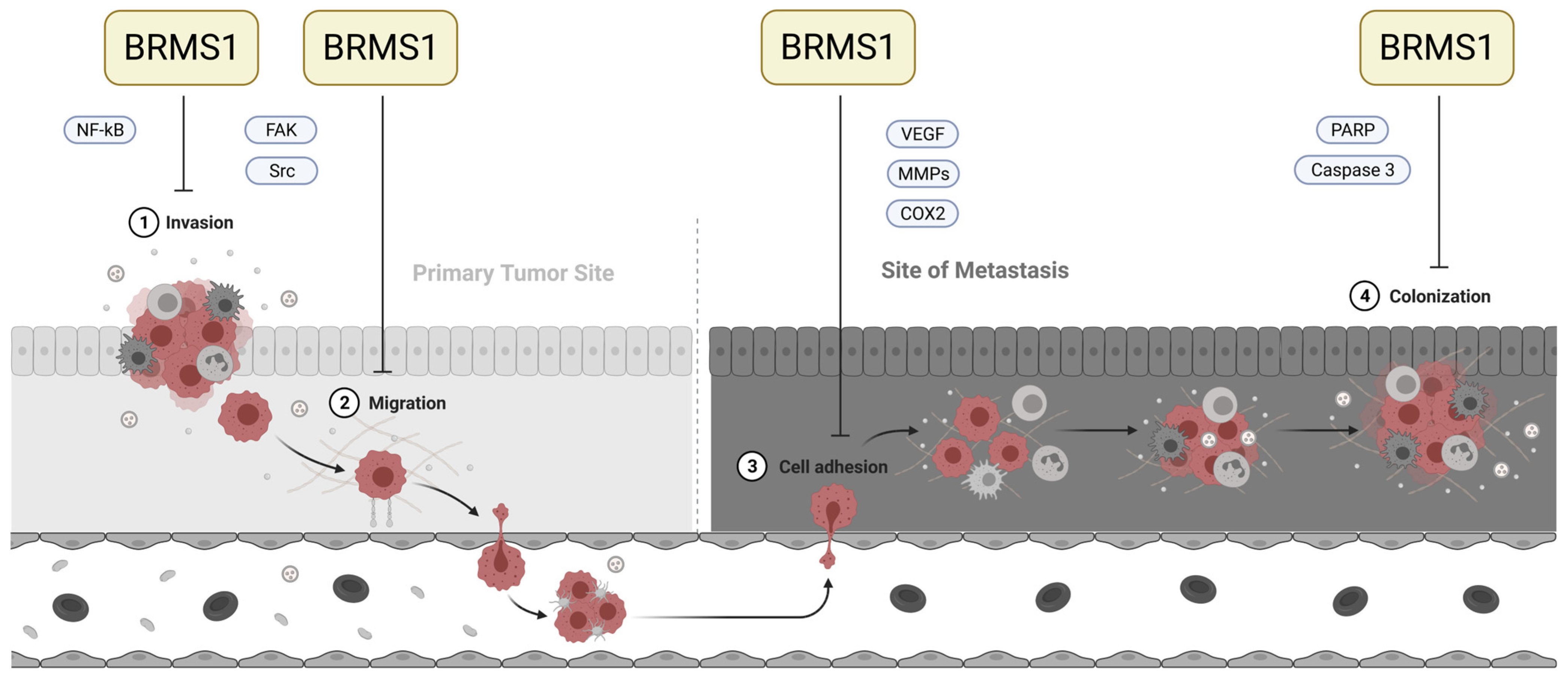
2. Materials and Methods
2.1. Tissue Samples and Clinical Data
2.2. Immunohistochemistry (IHC)
2.3. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.4. Software and Statistical Analysis
3. Results
3.1. Patient Cohort
3.2. BRMS1 Was Significantly Overexpressed in Gliomas Grade 2/3, Compared to NB, PA and GBM
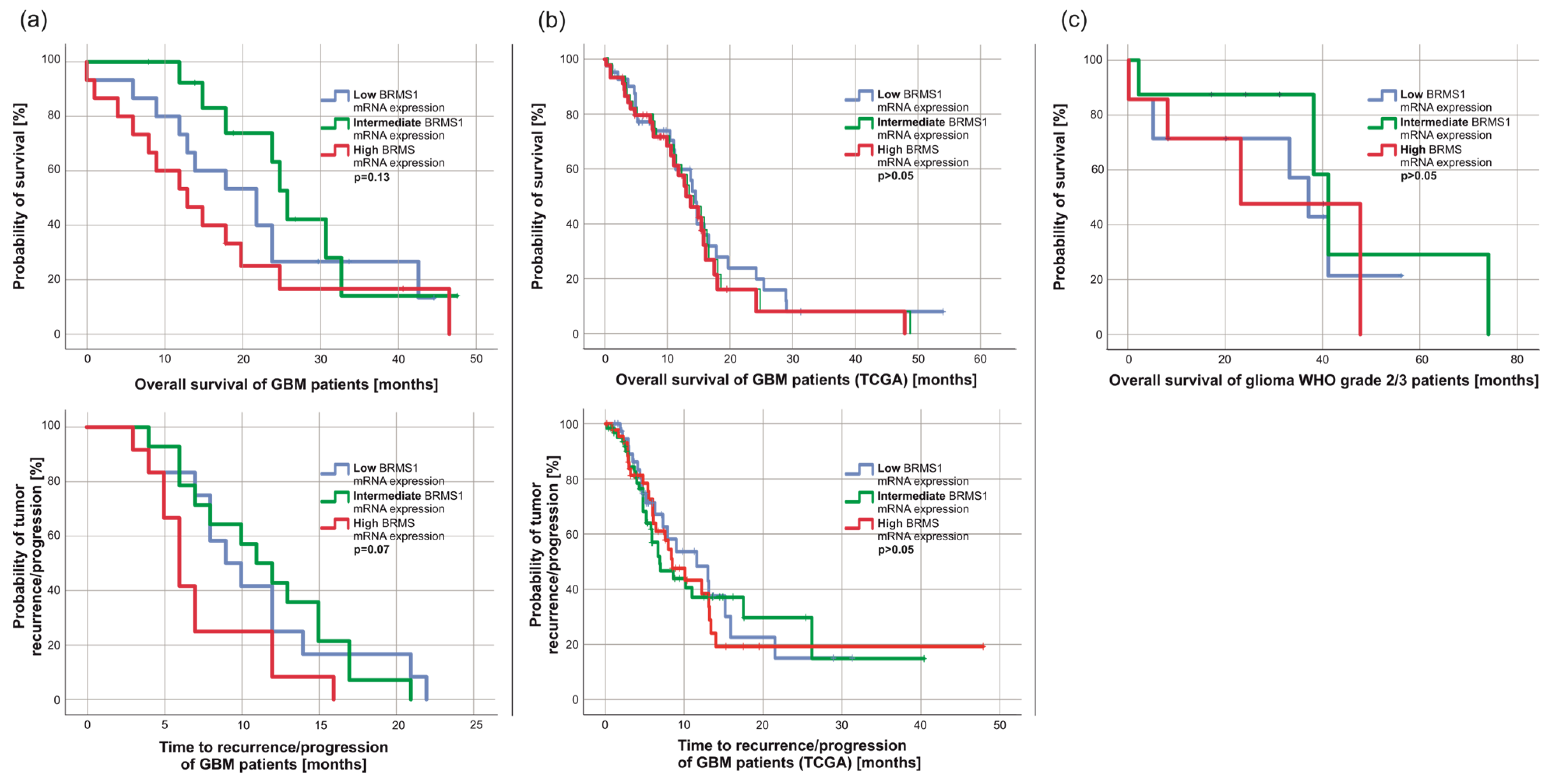
3.3. BRMS1 mRNA Expression Was Not Associated with Patients’ Survival but Correlated Weakly with Ki67 Staining
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 2018, 20, iv1–iv86. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Wick, W.; Osswald, M.; Wick, A.; Winkler, F. Treatment of glioblastoma in adults. Ther. Adv. Neurol. Disord. 2018, 11, 1756286418790452. [Google Scholar] [CrossRef]
- Wrensch, M.; Minn, Y.; Chew, T.; Bondy, M.; Berger, M.S. Epidemiology of primary brain tumors: Current concepts and review of the literature. Neuro-Oncology 2002, 4, 278–299. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.D.; Krex, D.; Grauer, O.; et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): A randomised, open-label, phase 3 trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef]
- Hottinger, A.F.; Pacheco, P.; Stupp, R. Tumor treating fields: A novel treatment modality and its use in brain tumors. Neuro-Oncology 2016, 18, 1338–1349. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance Therapy With Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA 2015, 314, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.; Wen, P.; Nishikawa, R.; Reardon, D.; Peters, K. Critical review of the addition of tumor treating fields (TTFields) to the existing standard of care for newly diagnosed glioblastoma patients. Crit. Rev. Oncol. Hematol. 2017, 111, 60–65. [Google Scholar] [CrossRef]
- Kessler, A.F.; Feldheim, J.; Schmitt, D.; Feldheim, J.J.; Monoranu, C.M.; Ernestus, R.I.; Löhr, M.; Hagemann, C. Monopolar Spindle 1 Kinase (MPS1/TTK) mRNA Expression is Associated with Earlier Development of Clinical Symptoms, Tumor Aggressiveness and Survival of Glioma Patients. Biomedicines 2020, 8, 192. [Google Scholar] [CrossRef]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Abacioglu, U.; Tavelin, B.; Lhermitte, B.; Hegi, M.E.; et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Monoranu, C.M.; Ernestus, R.I.; Lohr, M.; Hagemann, C. Changes of O(6)-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse-A Meta-Analysis Type Literature Review. Cancers 2019, 11, 1837. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Hasselblatt, M.; Jaber, M.; Reuss, D.; Grauer, O.; Bibo, A.; Terwey, S.; Schick, U.; Ebel, H.; Niederstadt, T.; Stummer, W.; et al. Diffuse Astrocytoma, IDH-Wildtype: A Dissolving Diagnosis. J. Neuropathol. Exp. Neurol. 2018, 77, 422–425. [Google Scholar] [CrossRef]
- Reuss, D.E.; Kratz, A.; Sahm, F.; Capper, D.; Schrimpf, D.; Koelsche, C.; Hovestadt, V.; Bewerunge-Hudler, M.; Jones, D.T.; Schittenhelm, J.; et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015, 130, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Brandner, S.; Jaunmuktane, Z. Neurological update: Gliomas and other primary brain tumours in adults. J. Neurol. 2018, 265, 717–727. [Google Scholar] [CrossRef]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef]
- Xie, Q.; Mittal, S.; Berens, M.E. Targeting adaptive glioblastoma: An overview of proliferation and invasion. Neuro-Oncology 2014, 16, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Giese, A.; Bjerkvig, R.; Berens, M.E.; Westphal, M. Cost of migration: Invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003, 21, 1624–1636. [Google Scholar] [CrossRef] [PubMed]
- Seraj, M.J.; Samant, R.S.; Verderame, M.F.; Welch, D.R. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000, 60, 2764–2769. [Google Scholar] [PubMed]
- Welch, D.R.; Manton, C.A.; Hurst, D.R. Breast Cancer Metastasis Suppressor 1 (BRMS1): Robust Biological and Pathological Data, But Still Enigmatic Mechanism of Action. Adv. Cancer Res. 2016, 132, 111–137. [Google Scholar] [CrossRef]
- Hedley, B.D.; Vaidya, K.S.; Phadke, P.; MacKenzie, L.; Dales, D.W.; Postenka, C.O.; MacDonald, I.C.; Chambers, A.F. BRMS1 suppresses breast cancer metastasis in multiple experimental models of metastasis by reducing solitary cell survival and inhibiting growth initiation. Clin. Exp. Metastasis 2008, 25, 727–740. [Google Scholar] [CrossRef]
- Shevde, L.A.; Samant, R.S.; Goldberg, S.F.; Sikaneta, T.; Alessandrini, A.; Donahue, H.J.; Mauger, D.T.; Welch, D.R. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp. Cell Res. 2002, 273, 229–239. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, J.; Sun, Y.; Chai, J.; Zou, T.; Gong, W.; Zhu, Z.; Liu, X.; Hou, Q.; Song, X. Effect of BRMS1 on tumorigenicity and metastasis of human rectal cancer. Cell Biochem. Biophys 2014, 70, 505–509. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, Q.D.; Di, W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int. J. Gynecol. Cancer 2006, 16, 522–531. [Google Scholar] [CrossRef]
- Smith, P.W.; Liu, Y.; Siefert, S.A.; Moskaluk, C.A.; Petroni, G.R.; Jones, D.R. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009, 276, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Seraj, M.J.; Harding, M.A.; Gildea, J.J.; Welch, D.R.; Theodorescu, D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin. Exp. Metastasis 2000, 18, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, S.; Selvam, S.P.; Mehrotra, S.; Kawamori, T.; Snider, A.J.; Obeid, L.M.; Shao, Y.; Sabbadini, R.; Ogretmen, B. Communication between host organism and cancer cells is transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate signalling to regulate tumour metastasis. EMBO Mol. Med. 2012, 4, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Qiao, Q.D.; Xie, H.F.; Wei, J.X. Breast cancer metastasis suppressor 1 (BRMS1) suppresses prostate cancer progression by inducing apoptosis and regulating invasion. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 68–75. [Google Scholar]
- Mei, P.; Bai, J.; Shi, M.; Liu, Q.; Li, Z.; Fan, Y.; Zheng, J. BRMS1 suppresses glioma progression by regulating invasion, migration and adhesion of glioma cells. PLoS ONE 2014, 9, e98544. [Google Scholar] [CrossRef]
- Khotskaya, Y.B.; Beck, B.H.; Hurst, D.R.; Han, Z.; Xia, W.; Hung, M.C.; Welch, D.R. Expression of metastasis suppressor BRMS1 in breast cancer cells results in a marked delay in cellular adhesion to matrix. Mol. Carcinog. 2014, 53, 1011–1026. [Google Scholar] [CrossRef] [PubMed]
- Phadke, P.A.; Vaidya, K.S.; Nash, K.T.; Hurst, D.R.; Welch, D.R. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am. J. Pathol. 2008, 172, 809–817. [Google Scholar] [CrossRef]
- You, J.; He, X.; Ding, H.; Zhang, T. BRMS1 regulates apoptosis in non-small cell lung cancer cells. Cell Biochem. Biophys. 2015, 71, 465–472. [Google Scholar] [CrossRef]
- Biorender. Overview of Metastatic Cascade. Available online: https://app.biorender.com/illustrations/646e3a1f37c184137741dd2c (accessed on 27 October 2022).
- Cicek, M.; Fukuyama, R.; Welch, D.R.; Sizemore, N.; Casey, G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-kappaB activity. Cancer Res. 2005, 65, 3586–3595. [Google Scholar] [CrossRef]
- Zimmermann, R.C.; Welch, D.R. BRMS1: A multifunctional signaling molecule in metastasis. Cancer Metastasis Rev. 2020, 39, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Salvador, E.; Monoranu, C.M.; Feldheim, J.J.; Ernestus, R.I.; Löhr, M.; Hagemann, C. Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma-A New Disease Biomarker? Cancers 2020, 12, 1085. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.I.; Hagemann, C.; Lohr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. Onco Targets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.W.; Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-Oncology 2017, 19, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Wend, D.; Lauer, M.J.; Monoranu, C.M.; Glas, M.; Kleinschnitz, C.; Ernestus, R.I.; Braunger, B.M.; Meybohm, P.; Hagemann, C.; et al. Protocadherin Gamma C3 (PCDHGC3) Is Strongly Expressed in Glioblastoma and Its High Expression Is Associated with Longer Progression-Free Survival of Patients. Int. J. Mol. Sci. 2022, 23, 8101. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef]
- Gravendeel, L.A.; Kouwenhoven, M.C.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.; Kloosterhof, N.K.; et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef]
- Murat, A.; Migliavacca, E.; Gorlia, T.; Lambiv, W.L.; Shay, T.; Hamou, M.F.; de Tribolet, N.; Regli, L.; Wick, W.; Kouwenhoven, M.C.; et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008, 26, 3015–3024. [Google Scholar] [CrossRef]
- Cancer Genome Atlas (TCGA) Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, B.M.; Wen, P.Y.; Cloughesy, T.F. Modified Criteria for Radiographic Response Assessment in Glioblastoma Clinical Trials. Neurotherapeutics 2017, 14, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J.; Kripke, M.L. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015, 34, 635–641. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer, 2nd ed.; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Brem, S.; Cotran, R.; Folkman, J. Tumor Angiogenesis: A Quantitative Method for Histologic Grading2. JNCI J. Natl. Cancer Inst. 1972, 48, 347–356. [Google Scholar] [CrossRef]
- Lefranc, F.; Le Rhun, E.; Kiss, R.; Weller, M. Glioblastoma quo vadis: Will migration and invasiveness reemerge as therapeutic targets? Cancer Treat. Rev. 2018, 68, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, V.P.; Moura Neto, V.; Mentlein, R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia 2018, 66, 1542–1565. [Google Scholar] [CrossRef]
- Torres, D.; Canoll, P. Alterations in the Brain Microenvironment in Diffusely Infiltrating Low-Grade Glioma. Neurosurg. Clin. N. Am. 2019, 30, 27–34. [Google Scholar] [CrossRef]
- Manini, I.; Caponnetto, F.; Bartolini, A.; Ius, T.; Mariuzzi, L.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D. Role of Microenvironment in Glioma Invasion: What We Learned from In Vitro Models. Int. J. Mol. Sci. 2018, 19, 147. [Google Scholar] [CrossRef]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune microenvironment of gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef]
- Sahm, F.; Capper, D.; Jeibmann, A.; Habel, A.; Paulus, W.; Troost, D.; von Deimling, A. Addressing diffuse glioma as a systemic brain disease with single-cell analysis. Arch. Neurol. 2012, 69, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, C.; Fuchs, S.; Monoranu, C.M.; Herrmann, P.; Smith, J.; Hohmann, T.; Grabiec, U.; Kessler, A.F.; Dehghani, F.; Löhr, M.; et al. Impact of MACC1 on human malignant glioma progression and patients’ unfavorable prognosis. Neuro-Oncology 2013, 15, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; Di Cristofano, C.; Capodanno, A.; Iorio, M.C.; Aretini, P.; Isola, P.; Tancredi, M.; Collecchi, P.; Naccarato, A.G.; Porta, R.P.; et al. High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int. J. Cancer 2007, 120, 1169–1178. [Google Scholar] [CrossRef]
- Cromer, A.; Carles, A.; Millon, R.; Ganguli, G.; Chalmel, F.; Lemaire, F.; Young, J.; Dembélé, D.; Thibault, C.; Muller, D.; et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene 2004, 23, 2484–2498. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Sun, X.; Wang, M.; Liu, H.; Wang, J. MicroRNA-423 enhances the invasiveness of hepatocellular carcinoma via regulation of BRMS1. Am. J. Transl. Res. 2017, 9, 5576–5584. [Google Scholar]
- Lin, J.; Huang, S.; Wu, S.; Ding, J.; Zhao, Y.; Liang, L.; Tian, Q.; Zha, R.; Zhan, R.; He, X. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis 2011, 32, 1641–1647. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, S.; Tu, Y.; Zhang, G.; Liu, Y.; Li, D.; Xu, S.; Le, Z.; Xiong, J.; Zou, W.; et al. MicroRNA-125a-5p inhibits invasion and metastasis of gastric cancer cells by targeting BRMS1 expression. Oncol. Lett. 2018, 15, 5119–5130. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, L.; Sun, Q.; Wu, J.; Ge, W.; Lu, G.; Cai, M. MicroRNA-3200-5p Promotes Osteosarcoma Cell Invasion via Suppression of BRMS1. Mol. Cells 2018, 41, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Amin, E.B.; Mayo, M.W.; Chudgar, N.P.; Bucciarelli, P.R.; Kadota, K.; Adusumilli, P.S.; Jones, D.R. CK2α’ Drives Lung Cancer Metastasis by Targeting BRMS1 Nuclear Export and Degradation. Cancer Res. 2016, 76, 2675–2686. [Google Scholar] [CrossRef]
- Cui, R.X.; Liu, N.; He, Q.M.; Li, W.F.; Huang, B.J.; Sun, Y.; Tang, L.L.; Chen, M.; Jiang, N.; Chen, L.; et al. Low BRMS1 expression promotes nasopharyngeal carcinoma metastasis in vitro and in vivo and is associated with poor patient survival. BMC Cancer 2012, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Slipicevic, A.; Holm, R.; Emilsen, E.; Ree Rosnes, A.K.; Welch, D.R.; Mælandsmo, G.M.; Flørenes, V.A. Cytoplasmic BRMS1 expression in malignant melanoma is associated with increased disease-free survival. BMC Cancer 2012, 12, 73. [Google Scholar] [CrossRef]


| Sex | Median Age (Quartiles) | Median Overall Survival (Quartiles) |
|---|---|---|
| female: 10/45.5% male: 12/54.5% | 38.5 years (33.8–48.8 years) | 31.0 months (8.0–40.3 months) |
| Patient Characteristics | ||||
| Sex | Female: 19/43.2% | Male: 25/56.8% | ||
| Median age | 58.5 years (49.0–69.7 years) | |||
| ECOG | 0: 24/54.5% | 1: 15/34.1% | >1: 5/11.4% | |
| Tumor characteristics | ||||
| Median tumor volume | 25.5 cm3 (15.9–54.3 cm3) | |||
| Tumor localization | left hemisphere: 25/56.8% | right hemisphere: 16/36.4% | both hemispheres: 3/6.8% | |
| Tumor localization (lobe) | frontal: 15/34.1% | temporal: 7/15.9% | multiple lobes: 11/25.0% | |
| occipital: 5/11.4% | parietal: 5/11.4% | cerebellar: 1/2.3% | ||
| MGMT promoter methylation | unmethylated: 10/31.3% | methylated: 22/68.8% | ||
| Median Ki67 staining | 25% (20–30%) | |||
| Therapy | ||||
| Time from diagnosis to surgery | 0–7 days: 26/59.1% | 8–14 days: 10/22.7% | >14 days: 8/18.2% | |
| Surgical intervention | biopsy: 6/13.6% | complete resection: 10/22.7% | incomplete resection: 28/63.6% | |
| Chemotherapy with TMZ | yes: 36/81.8% | no: 8/18.2% | ||
| Radiation therapy | yes: 41/93.2% | no: 3/6.8% | ||
| Treatment in relapse | Best supportive care: 14/36.8% | Systemic treatment (radiation and/or TMZ): 6/15.8% | Surgical resection and systemic treatment: 18/47.4% | |
| Relapse and outcome results | ||||
| PFS (quartiles) | 8.5 months (6.0–13.3 months) | |||
| Relapse | primarily multifocal: 6/13.6% | local relapse: 26/59.1% | multifocal relapse: 12/27.3% | |
| OS (quartiles) | 18.0 months (12.0–25.8 months) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feldheim, J.; Kessler, A.F.; Feldheim, J.J.; Schmitt, D.; Oster, C.; Lazaridis, L.; Glas, M.; Ernestus, R.-I.; Monoranu, C.M.; Löhr, M.; et al. BRMS1 in Gliomas—An Expression Analysis. Cancers 2023, 15, 2907. https://doi.org/10.3390/cancers15112907
Feldheim J, Kessler AF, Feldheim JJ, Schmitt D, Oster C, Lazaridis L, Glas M, Ernestus R-I, Monoranu CM, Löhr M, et al. BRMS1 in Gliomas—An Expression Analysis. Cancers. 2023; 15(11):2907. https://doi.org/10.3390/cancers15112907
Chicago/Turabian StyleFeldheim, Jonas, Almuth F. Kessler, Julia J. Feldheim, Dominik Schmitt, Christoph Oster, Lazaros Lazaridis, Martin Glas, Ralf-Ingo Ernestus, Camelia M. Monoranu, Mario Löhr, and et al. 2023. "BRMS1 in Gliomas—An Expression Analysis" Cancers 15, no. 11: 2907. https://doi.org/10.3390/cancers15112907
APA StyleFeldheim, J., Kessler, A. F., Feldheim, J. J., Schmitt, D., Oster, C., Lazaridis, L., Glas, M., Ernestus, R.-I., Monoranu, C. M., Löhr, M., & Hagemann, C. (2023). BRMS1 in Gliomas—An Expression Analysis. Cancers, 15(11), 2907. https://doi.org/10.3390/cancers15112907






