HIF-1-Induced hsa-miR-429: Understanding Its Direct Targets as the Key to Developing Cancer Diagnostics and Therapies
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Literature Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
2.4. Prediction of mRNA Targets and Gene Ontology Analysis
3. Experimental Approaches to Define the Molecular Network of hsa-miR-429
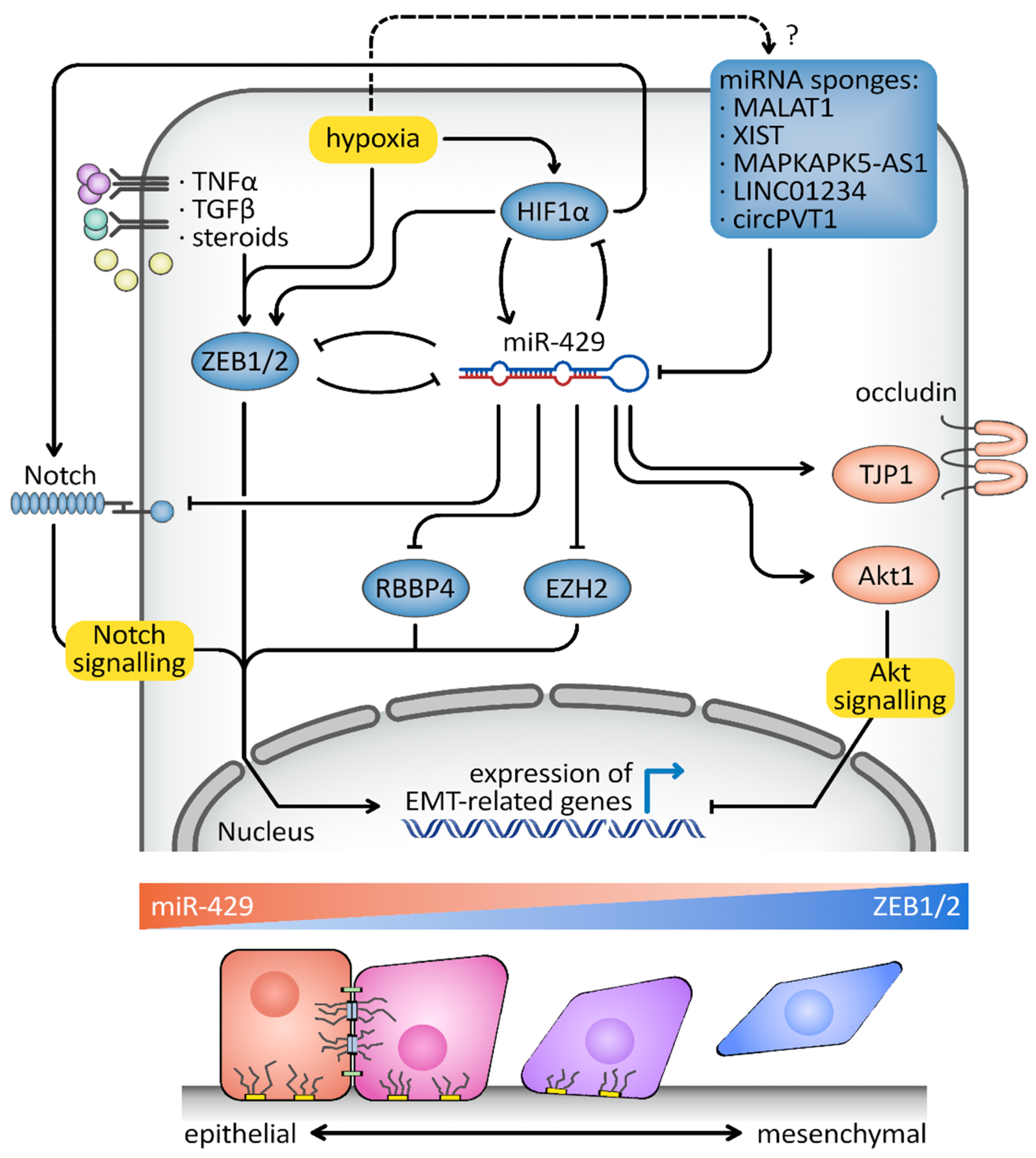
4. hsa-miR-429 and Cancer
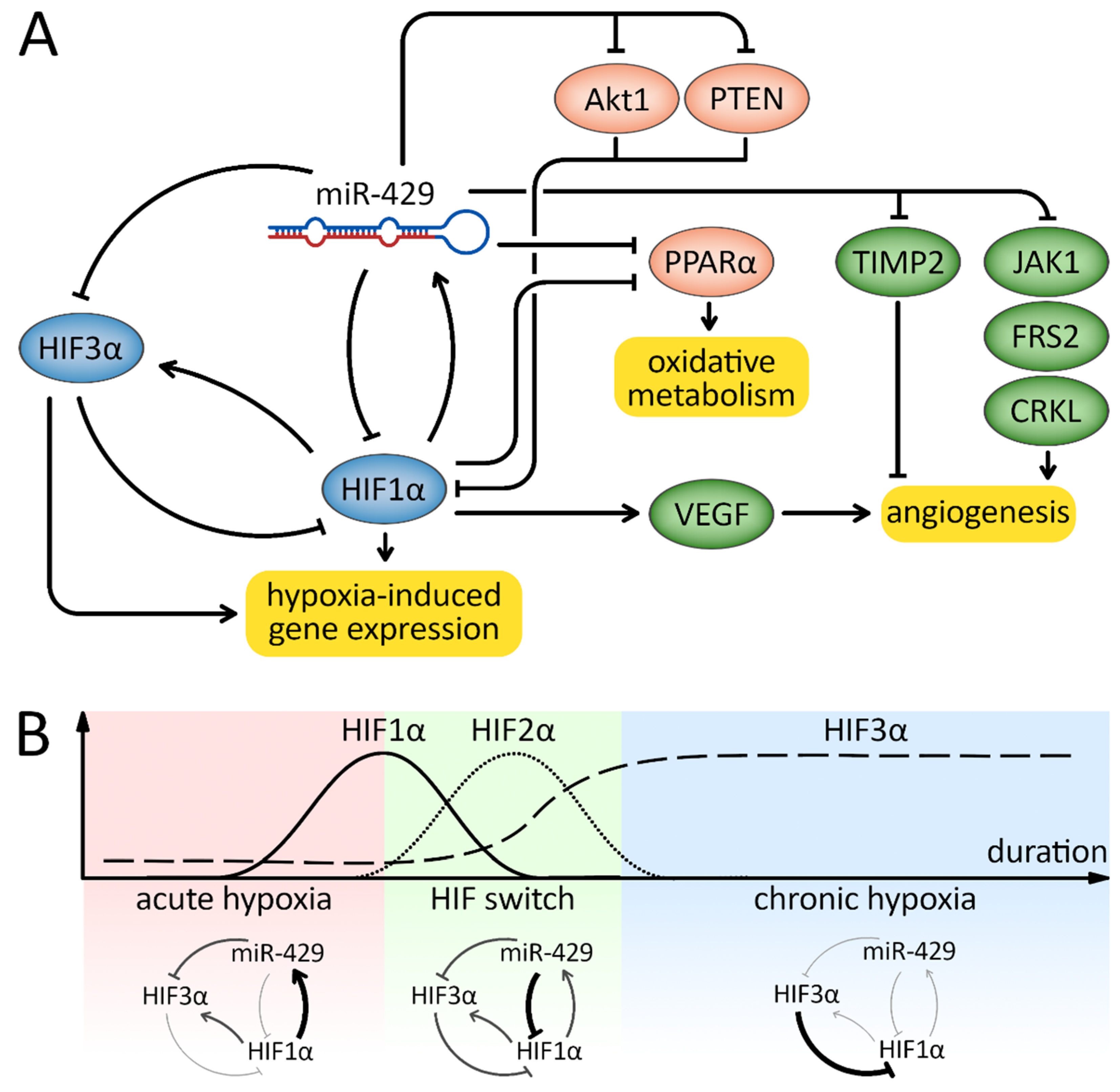
5. Responses to Hypoxia
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef]
- Caplen, N.J.; Parrish, S.; Imani, F.; Fire, A.; Morgan, R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 2001, 98, 9742–9747. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther.—Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Bartoszewska, S.; Cabaj, A.; Dabrowski, M.; Collawn, J.F.; Bartoszewski, R. MiR-34c-5p modulates X-box-binding protein 1 (XBP1) expression during the adaptive phase of the unfolded protein response. FASEB J. 2019, 33, 11541–11554. [Google Scholar] [CrossRef]
- Zhao, Z.; Lin, C.Y.; Cheng, K. SiRNA- and miRNA-based therapeutics for liver fibrosis. Transl. Res. J. Lab. Clin. Med. 2019, 214, 17–29. [Google Scholar] [CrossRef]
- Miroshnichenko, S.; Patutina, O. Enhanced inhibition of tumorigenesis using combinations of miRNA-targeted therapeutics. Front. Pharmacol. 2019, 10, 488. [Google Scholar] [CrossRef]
- Bansal, P.; Kumar, A.; Chandna, S.; Arora, M.; Bansal, R. Targeting miRNA for therapeutics using a micronome based method for identification of miRNA-mRNA pairs and validation of key regulator miRNA. Methods Mol. Biol. 2018, 1823, 185–195. [Google Scholar] [PubMed]
- Li, D.J.; Sun, C.C. Editorial: Towards miRNA based therapeutics for lung cancer. Curr. Pharm. Des. 2018, 23, 5971–5972. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, G.; Tian, W.; Deng, Y.; Xu, Y. MiRNA-based therapeutics for lung cancer. Curr. Pharm. Des. 2018, 23, 5989–5996. [Google Scholar] [CrossRef]
- Wen, M.M. Getting miRNA Therapeutics into the Target Cells for Neurodegenerative Diseases: A Mini-Review. Front. Mol. Neurosci. 2016, 9, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Liu, D.; Lai, H.; Li, J.; Wang, C. Developing miRNA therapeutics for cardiac repair in ischemic heart disease. J. Thorac. Dis. 2016, 8, E918–E927. [Google Scholar] [CrossRef]
- Kouri, F.M.; Ritner, C.; Stegh, A.H. miRNA-182 and the regulation of the glioblastoma phenotype—Toward miRNA-based precision therapeutics. Cell Cycle 2015, 14, 3794–3800. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Yadav, T.; Rani, V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. 2016, 98, 12–23. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Ooi, J.Y.Y.; Lin, R.C.Y.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Nouraee, N.; Mowla, S.J. miRNA therapeutics in cardiovascular diseases: Promises and problems. Front. Genet. 2015, 6, 232. [Google Scholar] [CrossRef]
- Kwekkeboom, R.F.J.; Lei, Z.; Doevendans, P.A.; Musters, R.J.P.; Sluijter, J.P.G. Targeted delivery of miRNA therapeutics for cardiovascular diseases: Opportunities and challenges. Clin. Sci. 2014, 127, 351–365. [Google Scholar] [CrossRef]
- Pereira, D.; Rodrigues, P.; Borralho, P.; Rodrigues, C. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today 2013, 18, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Ford, L.P. Using synthetic miRNA mimics for diverting cell fate: A possibility of miRNA-based therapeutics? Leuk. Res. 2006, 30, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewska, S.; Kamysz, W.; Jakiela, B.; Sanak, M.; Króliczewski, J.; Bebok, Z.; Bartoszewski, R.; Collawn, J.F. miR-200b downregulates CFTR during hypoxia in human lung epithelial cells. Cell. Mol. Biol. Lett. 2017, 22, 23. [Google Scholar] [CrossRef]
- Kalinowski, L.; Janaszak-Jasiecka, A.; Siekierzycka, A.; Bartoszewska, S.; Woźniak, M.; Lejnowski, D.; Collawn, J.F.; Bartoszewski, R. Posttranscriptional and transcriptional regulation of endothelial nitric-oxide synthase during hypoxia: The role of microRNAs. Cell. Mol. Biol. Lett. 2016, 21, 16. [Google Scholar] [CrossRef]
- Kleinman, M.E.; Yamada, K.; Takeda, A.; Chandrasekaran, V.; Nozaki, M.; Baffi, J.Z.; Albuquerque, R.J.C.; Yamasaki, S.; Itaya, M.; Pan, Y.; et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 2008, 452, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.; Vaishnaw, A.; Cehelsky, J.; Meyers, R.; Nochur, S.; Harrison, L.; Meeking, P.; Mann, A.; Moane, E.; Oxford, J.; et al. Viral Load Drives Disease in Humans Experimentally Infected with Respiratory Syncytial Virus. Am. J. Respir. Crit. Care Med. 2010, 182, 1305–1314. [Google Scholar] [CrossRef]
- DeVincenzo, J.; Lambkin-Williams, R.; Wilkinson, T.; Cehelsky, J.; Nochur, S.; Walsh, E.; Meyers, R.; Gollob, J.; Vaishnaw, A. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 2010, 107, 8800–8805. [Google Scholar] [CrossRef]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Davis, M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nature reviews. Drug Discov. 2015, 14, 843–856. [Google Scholar] [CrossRef]
- Ackley, K.L. Are we there yet? An update on oligonucleotide drug development. Chim. Oggi. 2016, 34, Xxxv–Xxxviii. [Google Scholar]
- Setten, R.L.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Gurtan, A.M.; Sharp, P.A. The Role of miRNAs in Regulating Gene Expression Networks. J. Mol. Biol. 2013, 425, 3582–3600. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, Y.; Xie, S.J.; Xu, S.J.; Zhou, H.; Qu, L.H. Argonaute hits-clip decodes microRNA-mRNA interaction maps during heart development. Cardiology 2013, 126, 62. [Google Scholar]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute hits-clip decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell. Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of Stress-Dependent Cardiac Growth and Gene Expression by a MicroRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef]
- Leung, A.; Sharp, P.A. MicroRNA Functions in Stress Responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef]
- Mendell, J.T.; Olson, E.N. MicroRNAs in Stress Signaling and Human Disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Ge, X.-L.; Wang, J.-L.; Liu, X.; Zhang, J.; Liu, C.; Guo, L. Inhibition of miR-19a protects neurons against ischemic stroke through modulating glucose metabolism and neuronal apoptosis. Cell. Mol. Biol. Lett. 2019, 24, 37. [Google Scholar] [CrossRef]
- Lu, G.; Zhang, Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell. Mol. Biol. Lett. 2019, 24, 34. [Google Scholar] [CrossRef]
- Fu, Y.; Lin, L.; Xia, L. MiR-107 function as a tumor suppressor gene in colorectal cancer by targeting transferrin receptor 1. Cell. Mol. Biol. Lett. 2019, 24, 31. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zuo, Y.; Xu, Y.; Zhang, Z.; Li, Y.; Pang, J. MiR-613 inhibits proliferation and invasion and induces apoptosis of rheumatoid arthritis synovial fibroblasts by direct down-regulation of DKK1. Cell. Mol. Biol. Lett. 2019, 24, 8. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y. Upregulation of miR-29b-3p protects cardiomyocytes from hypoxia-induced apoptosis by targeting TRAF5. Cell. Mol. Biol. Lett. 2019, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Z.; Chen, T.; Pan, J.; Shen, Y.; Chen, X.; Zhou, X.; Cheng, R.; Yang, Y. The role of miR-431-5p in regulating pulmonary surfactant expression in vitro. Cell. Mol. Biol. Lett. 2019, 24, 25. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lian, J.X.; Meng, S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell. Mol. Biol. Lett. 2019, 24, 23. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wang, Y.; Gao, J.; Yan, Z.; Li, Z.; Zou, X.; Li, Y.; Wang, J.; Guo, Y. miR-29b-3p regulated osteoblast differentiation via regulating IGF-1 secretion of mechanically stimulated osteocytes. Cell. Mol. Biol. Lett. 2019, 24, 11. [Google Scholar] [CrossRef]
- Yang, Y.; Bao, Y.; Yang, G.-K.; Wan, J.; Du, L.-J.; Ma, Z.-H. MiR-214 sensitizes human colon cancer cells to 5-FU by targeting Hsp27. Cell. Mol. Biol. Lett. 2019, 24, 22. [Google Scholar] [CrossRef]
- Ma, F.; Lin, P.; Chen, Q.; Lu, X.; Zhang, Y.E.; Wu, C.-I. Direct measurement of pervasive weak repression by microRNAs and their role at the network level. BMC Genom. 2018, 19, 362. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- Moszynska, A.; Jaskiewicz, M.; Serocki, M.; Cabaj, A.; Crossman, D.K.; Bartoszewska, S.; Gebert, M.; Dabrowski, M.; Collawn, J.F.; Bartoszewski, R. The hypoxia-induced changes in miRNA-mRNA in RNA-induced silencing complexes and HIF-2 induced miRNAs in human endothelial cells. FASEB J. 2022, 36, e22412. [Google Scholar] [CrossRef]
- Serocki, M.; Bartoszewska, S.; Janaszak-Jasiecka, A.; Ochocka, R.J.; Collawn, J.F.; Bartoszewski, R. miRNAs regulate the HIF switch during hypoxia: A novel therapeutic target. Angiogenesis 2018, 21, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.M.; Liu, S.Q.; Sun, M.Z. miR-429 as biomarker for diagnosis, treatment and prognosis of cancers and its potential action mechanisms: A systematic literature review. Neoplasma 2020, 67, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, I.; Ciccarese, F.; Sharova, E.; Urso, L.; Raimondi, V.; Silic-Benussi, M.; D’agostino, D.M.; Ciminale, V. The miR-200 Family of microRNAs: Fine Tuners of Epithelial-Mesenchymal Transition and Circulating Cancer Biomarkers. Cancers 2021, 13, 5874. [Google Scholar] [CrossRef]
- Klicka, K.; Grzywa, T.M.; Mielniczuk, A.; Klinke, A.; Włodarski, P.K. The role of miR-200 family in the regulation of hallmarks of cancer. Front. Oncol. 2022, 12, 965231. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Wang, R.; Chen, L.-B. Review of MiR-200b and cancer chemosensitivity. Biomed. Pharmacother. 2012, 66, 397–402. [Google Scholar] [CrossRef]
- Huang, G.-L.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G.A. MiR-200 family and cancer: From a meta-analysis view. Mol. Asp. Med. 2019, 70, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Riascos, Z.V.; Ginesta, M.M.; Fabregat, J.; Serrano, T.; Busquets, J.; Buscail, L.; Cordelier, P.; Capellá, G. Expression and Role of MicroRNAs from the miR-200 Family in the Tumor Formation and Metastatic Propensity of Pancreatic Cancer. Mol. Ther.—Nucleic Acids 2019, 17, 491–503. [Google Scholar] [CrossRef]
- Senfter, D.; Madlener, S.; Krupitza, G.; Mader, R.M. The microRNA-200 family: Still much to discover. Biomol. Concepts 2016, 7, 311–319. [Google Scholar] [CrossRef]
- Hauschild, A.-C.; Pastrello, C.; Ekaputeri, G.K.A.; Bethune-Waddell, D.; Abovsky, M.; Ahmed, Z.; Kotlyar, M.; Lu, R.; Jurisica, I. MirDIP 5.2: Tissue context annotation and novel microRNA curation. Nucleic Acids Res. 2023, 51, D217–D225. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. Mirdip 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. Mirtarbase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Antes, G.; von Elm, E. The prisma statement—What should be reported about systematic reviews? Dtsch. Med. Wochenschr. 2009, 134, 1619. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. J. Clin. Epidemiol. 2009, 62, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, F. Comment on: “MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19”. Drugs 2021, 81, 1691–1692. [Google Scholar] [CrossRef]
- Bartoszewski, R.; Sikorski, A.F. Editorial focus: Understanding off-target effects as the key to successful RNAi therapy. Cell. Mol. Biol. Lett. 2019, 24, 69. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, Y.; Peng, H.; Chen, Y.; Zhu, P.; Huang, Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015, 81, 142–160. [Google Scholar] [CrossRef]
- Terasawa, K.; Shimizu, K.; Tsujimoto, G. Synthetic pre-miRNA-based shRNA as potent RNAi triggers. J. Nucleic Acids 2011, 2011, 131579. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef]
- Bartoszewski, R.; Sikorski, A.F. Editorial focus: Entering into the non-coding RNA era. Cell. Mol. Biol. Lett. 2018, 23, 45. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol. Biol. 2013, 936, 117–127. [Google Scholar]
- Diener, C.; Hart, M.; Fecher-Trost, C.; Knittel, J.; Rheinheimer, S.; Meyer, M.R.; Mayer, J.; Flockerzi, V.; Keller, A.; Meese, E. Outside the limit: Questioning the distance restrictions for cooperative miRNA binding sites. Cell. Mol. Biol. Lett. 2023, 28, 8. [Google Scholar] [CrossRef] [PubMed]
- Gebert, M.; Bartoszewska, S.; Janaszak-Jasiecka, A.; Moszyńska, A.; Cabaj, A.; Króliczewski, J.; Madanecki, P.; Ochocka, R.J.; Crossman, D.K.; Collawn, J.F.; et al. PIWI proteins contribute to apoptosis during the UPR in human airway epithelial cells. Sci. Rep. 2018, 8, 16431. [Google Scholar] [CrossRef] [PubMed]
- Gebert, M.; Sobolewska, A.; Bartoszewska, S.; Cabaj, A.; Crossman, D.K.; Króliczewski, J.; Madanecki, P.; Dąbrowski, M.; Collawn, J.F.; Bartoszewski, R. Genome-wide mRNA profiling identifies X-box-binding protein 1 (XBP1) as an IRE1 and PUMA repressor. Cell. Mol. Life Sci. 2021, 78, 7061–7080. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewski, R.; Gebert, M.; Janaszak-Jasiecka, A.; Cabaj, A.; Króliczewski, J.; Bartoszewska, S.; Sobolewska, A.; Crossman, D.K.; Ochocka, R.; Kamysz, W.; et al. Genome-wide mRNA profiling identifies RCAN1 and GADD45A as regulators of the transitional switch from survival to apoptosis during ER stress. FEBS J. 2020, 287, 2923–2947. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef]
- Esau, C.C. Inhibition of microRNA with antisense oligonucleotides. Methods 2008, 44, 55–60. [Google Scholar] [CrossRef]
- Davis, S.; Propp, S.; Freier, S.M.; Jones, L.E.; Serra, M.J.; Kinberger, G.; Bhat, B.; Swayze, E.E.; Bennett, C.F.; Esau, C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009, 37, 70–77. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef]
- Torres, A.G.; Fabani, M.M.; Vigorito, E.; Gait, M.J. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA 2011, 17, 933–943. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Kuwajima, S.; Braich, R.; Rajeev, K.G.; Pena, J.; Tuschl, T.; Manoharan, M.; Stoffel, M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007, 35, 2885–2892. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Moszynska, A.; Gebert, M.; Collawn, J.F.; Bartoszewski, R. EPAS1 resistance to miRNA-based regulation contributes to prolonged expression of hif-2 during hypoxia in human endothelial cells. Gene 2023, 868, 147376. [Google Scholar] [CrossRef] [PubMed]
- Staton, A.A.; Giraldez, A.J. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat. Protoc. 2011, 6, 2035–2049. [Google Scholar] [CrossRef] [PubMed]
- Summerton, J.E. Morpholino, siRNA, and S-DNA Compared: Impact of Structure and Mechanism of Action on Off-Target Effects and Sequence Specificity. Curr. Top. Med. Chem. 2007, 7, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Hudziak, R.M.; Barofsky, E.; Barofsky, D.F.; Weller, D.L.; Huang, S.-B.; Weller, D.D. Resistance of Morpholino Phosphorodiamidate Oligomers to Enzymatic Degradation. Antisense Nucleic Acid Drug Dev. 1996, 6, 267–272. [Google Scholar] [CrossRef]
- Summerton, J. Morpholino antisense oligomers: The case for an RNase H-independent structural type. Biochim. Biophys. Acta 1999, 1489, 141–158. [Google Scholar] [CrossRef]
- Gagliardi, M.; Matarazzo, M.R. RIP: RNA Immunoprecipitation. Methods Mol. Biol. 2016, 1480, 73–86. [Google Scholar] [CrossRef]
- Jayaseelan, S.; Doyle, F.; Tenenbaum, S.A. Profiling post-transcriptionally networked mRNA subsets using RIP-Chip and RIP-Seq. Methods 2014, 67, 13–19. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Y.; Wu, S.; Shi, X.; Li, L.; Zhao, J.; Xu, H. Tumor-Suppressing Effects of miR-429 on Human Osteosarcoma. Cell Biochem. Biophys. 2014, 70, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Chen, X.; Li, H.; Zheng, L. Delta-tocotrienol suppresses the migration and angiogenesis of trophoblasts in preeclampsia and promotes their apoptosis via miR-429/ ZEB1 axis. Bioengineered 2021, 12, 1861–1873. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ouyang, X.; Wang, Y.; Fan, Q. MAPKAPK5-AS1 drives the progression of hepatocellular carcinoma via regulating mir-429/zeb1 axis. BMC Mol. Cell. Biol. 2022, 23, 21. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, X.; Hu, B.; Wang, X.J.; Wang, Q.; Wang, W.L. The effects of microRNA-429 on inhibition of cervical cancer cells through targeting zeb1 and crkl. Biomed. Pharmacother. 2016, 80, 311–321. [Google Scholar] [CrossRef]
- Tian, Y.; Pan, Q.; Shang, Y.; Zhu, R.; Ye, J.; Liu, Y.; Zhong, X.; Li, S.; He, Y.; Chen, L.; et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (ASCL2): Impact on the epithelial-mesenchymal transition in colon cancer cells. J. Biol. Chem. 2014, 289, 36101–36115. [Google Scholar] [CrossRef]
- Shen, J.; Hong, L.; Yu, D.; Cao, T.; Zhou, Z.; He, S. LncRNA XIST promotes pancreatic cancer migration, invasion and EMT by sponging miR-429 to modulate ZEB1 expression. Int. J. Biochem. Cell Biol. 2019, 113, 17–26. [Google Scholar] [CrossRef]
- Sun, B.; Zheng, X.; Ye, W.; Zhao, P.; Ma, G. LncRNA linc01303 promotes the progression of oral squamous cell carcinomas via the mir-429/ZEB1/EMT axis. J. Oncol. 2021, 2021, 7974012. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, W.; Wang, X. Inhibition of long non-coding RNA MALAT1 elevates microRNA-429 to suppress the progression of hypopharyngeal squamous cell carcinoma by reducing ZEB1. Life Sci. 2020, 262, 118480. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, M.; Song, Y.; Yang, N. RNF185 antisense RNA 1 (RNF185-AS1) promotes proliferation, migration, and invasion in papillary thyroid carcinoma. Anti-Cancer Drugs 2022, 33, 595–606. [Google Scholar] [CrossRef]
- Bartoszewska, S.; Kochan, K.; Piotrowski, A.; Kamysz, W.; Ochocka, R.J.; Collawn, J.F.; Bartoszewski, R. The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1alpha expression in human endothelial cells through a negative feedback loop. FASEB J. 2015, 29, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Wang, Y.; Cao, Y.; Li, G.; Sun, R.; Teng, P.; Wang, Y.; Bi, Y.; Guo, Z.; Yuan, Y.; et al. MiR-429 improved the hypoxia tolerance of human amniotic cells by targeting HIF-1α. Biotechnol. Lett. 2018, 40, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Janaszak-Jasiecka, A.; Bartoszewska, S.; Kochan, K.; Piotrowski, A.; Kalinowski, L.; Kamysz, W.; Ochocka, R.J.; Bartoszewski, R.; Collawn, J.F. miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells. Sci. Rep. 2016, 6, 22775. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, L.; Luan, S.; Jiang, Y.; Wang, Q. MiR-429 inhibits osteosarcoma progression by targeting HOXA9 through suppressing WNT/beta-catenin signaling pathway. Oncol. Lett. 2020, 20, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Tian, G.-Y. MiR-429 regulates the metastasis and EMT of HCC cells through targeting RAB23. Arch. Biochem. Biophys. 2018, 637, 48–55. [Google Scholar] [CrossRef]
- Chen, L.; Xue, Y.; Zheng, J.; Liu, X.; Liu, J.; Chen, J.; Li, Z.; Xi, Z.; Teng, H.; Wang, P.; et al. MiR-429 Regulated by Endothelial Monocyte Activating Polypeptide-II (EMAP-II) Influences Blood-Tumor Barrier Permeability by Inhibiting the Expressions of ZO-1, Occludin and Claudin-5. Front. Mol. Neurosci. 2018, 11, 35. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Wang, S.; Dai, J.; Yan, L.; Wang, J.; Sun, Y. Tacrolimus induces fibroblasts apoptosis and reduces epidural fibrosis by regulating miR-429 and its target of RhoE. Biochem. Biophys. Res. Commun. 2017, 490, 1197–1204. [Google Scholar] [CrossRef]
- Su, Z.; Jiang, G.; Chen, J.; Liu, X.; Zhao, H.; Fang, Z.; He, Y.; Jiang, X.; Xu, G. Erratum: MicroRNA-429 inhibits cancer cell proliferation and migration by targeting AKT1 in renal cell carcinoma. Mol. Clin. Oncol. 2020, 13, 92. [Google Scholar]
- Tian, X.; Wei, Z.; Wang, J.; Liu, P.; Qin, Y.; Zhong, M. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting pak6/cofilin signaling. Oncol. Rep. 2015, 34, 707–714. [Google Scholar] [CrossRef]
- Qiu, M.; Liang, Z.; Chen, L.; Tan, G.; Wang, K.; Liu, L.; Liu, J.; Chen, H. MicroRNA-429 suppresses cell proliferation, epithelial-mesenchymal transition, and metastasis by direct targeting of BMI1 and E2F3 in renal cell carcinoma. Urol. Oncol. 2015, 33, 332.e9–332.e18. [Google Scholar] [CrossRef]
- Xu, H.; Jin, L.; Chen, Y.; Li, J. Downregulation of microRNA-429 protects cardiomyocytes against hypoxia-induced apoptosis by increasing Notch1 expression. Int. J. Mol. Med. 2016, 37, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Liu, F.; Zhang, T.; Li, Y.; Ye, L.; Zhang, X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis 2013, 34, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Guo, T.; Qu, X.; Che, X.; Li, C.; Wang, S.; Gong, J.; Wu, P.; Liu, Y.; Liu, Y.; et al. PD-L1 Under Regulation of miR-429 Influences the Sensitivity of Gastric Cancer Cells to TRAIL by Binding of EGFR. Front. Oncol. 2020, 10, 1067. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Yao, Q.; Wang, Y.; Liu, Z.; Zhang, L. LncRNA SNHG6 promotes wilms’ tumor progression through regulating mir-429/frs2 axis. Cancer Biother. Radiopharm. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tong, X.; Zhou, Z.; Wang, S.; Lei, Z.; Zhang, T.; Liu, Z.; Zeng, Y.; Li, C.; Zhao, J.; et al. Circular RNA hsa_circ_0008305 (circptk2) inhibits tgf-beta-induced epithelial-mesenchymal transition and metastasis by controlling tif1gamma in non-small cell lung cancer. Mol. Cancer 2018, 17, 140. [Google Scholar] [CrossRef]
- Wang, J.; Lai, X.; Peng, X. CircLIFR Inhibits Non-small Cell Lung Cancer Progression by Acting as a miR-429 Sponge to Enhance CELF2 Expression. Biochem. Genet. 2023, 61, 725–741. [Google Scholar] [CrossRef]
- Shen, F.; Zheng, H.; Zhou, L.; Li, W.; Xu, X. Overexpression of MALAT1 contributes to cervical cancer progression by acting as a sponge of miR-429. J. Cell. Physiol. 2019, 234, 11219–11226. [Google Scholar] [CrossRef]
- Bi, M.; Zheng, L.; Chen, L.; He, J.; Yuan, C.; Ma, P.; Zhao, Y.; Hu, F.; Tang, W.; Sheng, M. Ln RNA linc01234 promotes triple-negative breast cancer progression through regulating the miR-429/SYNJ1 axis. Am. J. Transl. Res. 2021, 13, 11399–11412. [Google Scholar]
- Luo, N. LncRNA MSC-AS1/miRNA-429 axis mediates growth and metastasis of nasopharyngeal carcinoma via JAK1/STAT3 signaling pathway. Comput. Math. Methods Med. 2022, 2022, 1447207. [Google Scholar] [CrossRef]
- Cao, L.; Zhou, X.; Ding, X.; Gao, D. Knockdown of circ-pvt1 inhibits the progression of lung adenocarcinoma and enhances the sensitivity to cisplatin via the miR-429/FOXK1 signaling axis. Mol. Med. Rep. 2021, 24, 684. [Google Scholar] [CrossRef]
- Meng, D.-F.; Shao, H.; Feng, C.-B. LINC00894 Enhances the Progression of Breast Cancer by Sponging miR-429 to Regulate ZEB1 Expression. OncoTargets Ther. 2021, 14, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, J.; Wa, Q.; He, M.; Wang, X.; Zhou, J.; Cen, Y. Knockdown of circ_0084043 suppresses the development of human melanoma cells through mir-429/tribbles homolog 2 axis and Wnt/beta-catenin pathway. Life Sci. 2020, 243, 117323. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hou, Z.; Luo, B.; Li, C.; Liu, J.; Liu, J.; Tang, J.; Yao, G. Circular RNA circRNA_0082835 promotes progression and lymphatic metastasis of primary melanoma by sponging microRNA miRNA-429. Bioengineered 2021, 12, 4159–4173. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lu, H.; Li, F.; Hao, X.; Han, L.; Dong, Q.; Chen, X. MicroRNA-429 inhibits neuroblastoma cell proliferation, migration and invasion via the NF-κB pathway. Cell. Mol. Biol. Lett. 2020, 25, 5. [Google Scholar] [CrossRef]
- Kawasaki, H.; Amano, H. Anti-inflammatory role of microRNA-429 in human gingival epithelial cells-inhibition of Il-8 production through direct binding to ikkbeta mRNA. Mol. Med. Rep. 2021, 24, 581. [Google Scholar] [CrossRef]
- Zhang, Z.; Wen, H.; Peng, B.; Weng, J.; Zeng, F. Hfd-induced TRAF6 upregulation promotes liver cholesterol accumulation and fatty liver development via ezh2-mediated miR-429/pparalpha axis. Molecular therapy. Nucleic Acids 2021, 24, 711–727. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, Z.; Ma, K.; Li, X.; Tian, N.; Duan, J.; Xiao, X.; Wang, Y. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J. Cancer 2017, 8, 4106–4116. [Google Scholar] [CrossRef]
- Lang, Y.; Xu, S.; Ma, J.; Wu, J.; Jin, S.; Cao, S.; Yu, Y. MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem. Biophys. Res. Commun. 2014, 450, 154–159. [Google Scholar] [CrossRef]
- Tang, J.; Li, L.; Huang, W.; Sui, C.; Yang, Y.; Lin, X.; Hou, G.; Chen, X.; Fu, J.; Yuan, S.; et al. MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial–mesenchymal transition. Cancer Lett. 2015, 364, 33–43. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Li, Q.; Guo, C.; Sun, W.; Zhao, D.; Jiang, S.; Hao, L.; Tian, Y.; Liu, S.; et al. MiR-429-CRKL axis regulates clear cell renal cell carcinoma malignant progression through sOS1/MEK/ERK/MMP2/MMP9 pathway. Biomed. Pharmacother. 2020, 127, 110215. [Google Scholar] [CrossRef]
- Guo, C.; Zhao, D.; Zhang, Q.; Liu, S.; Sun, M.-Z. miR-429 suppresses tumor migration and invasion by targeting CRKL in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and epithelial-mesenchymal transition. Sci. Rep. 2018, 8, 2375. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tang, J.; Zhang, B.; Yang, W.; LiuGao, M.; Wang, R.; Tan, Y.; Fan, J.; Chang, Y.; Fu, J.; et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut 2015, 64, 156–167. [Google Scholar] [CrossRef]
- Colquhoun, D. An investigation of the false discovery rate and the misinterpretation of p-values. R Soc. Open Sci. 2014, 1, 140216. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A. Why Most Published Research Findings Are False. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-Mesenchymal Transition in Cancer: Parallels Between Normal Development and Tumor Progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A Double-Negative Feedback Loop between ZEB1-SIP1 and the microRNA-200 Family Regulates Epithelial-Mesenchymal Transition. Cancer Res 2008, 68, 7846–7854. [Google Scholar] [CrossRef]
- Bartoszewski, R.; Serocki, M.; Janaszak-Jasiecka, A.; Bartoszewska, S.; Kochan-Jamrozy, K.; Piotrowski, A.; Króliczewski, J.; Collawn, J.F. miR-200b downregulates Kruppel Like Factor 2 (KLF2) during acute hypoxia in human endothelial cells. Eur. J. Cell Biol. 2017, 96, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, H.; Zhao, J.; Vieth, E.; Nephew, K.P.; Matei, D. EZH2 inhibition promotes epithelial-to-mesenchymal transition in ovarian cancer cells. Oncotarget 2016, 7, 84453–84467. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.D.; Lv, Z.; Zhu, W.F. RBBP4 promotes colon cancer malignant progression via regulating Wnt/beta-catenin pathway. World J. Gastroenterol. 2020, 26, 5328–5342. [Google Scholar] [CrossRef]
- Shao, S.; Zhao, X.; Zhang, X.; Luo, M.; Zuo, X.; Huang, S.; Wang, Y.; Gu, S.; Zhao, X. Notch1 signaling regulates the epithelial–mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer 2015, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-W.; Xia, W.; Lim, S.-O.; Hsu, J.L.; Huo, L.; Wu, Y.; Li, L.-Y.; Lai, C.-C.; Chang, S.-S.; Hsu, Y.-H.; et al. AKT1 Inhibits Epithelial-to-Mesenchymal Transition in Breast Cancer through Phosphorylation-Dependent Twist1 Degradation. Cancer Res 2016, 76, 1451–1462. [Google Scholar] [CrossRef]
- Kyuno, D.; Yamaguchi, H.; Ito, T.; Kono, T.; Kimura, Y.; Imamura, M.; Konno, T.; Hirata, K.; Sawada, N.; Kojima, T. Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J. Gastroenterol. 2014, 20, 10813–10824. [Google Scholar] [CrossRef]
- Lee, S.H.; Paek, A.R.; Yoon, K.; Kim, S.H.; Lee, S.Y.; You, H.J. Tight junction protein 1 is regulated by transforming growth factor-beta and contributes to cell motility in nsclc cells. BMB Rep. 2015, 48, 115–120. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, Z.; Zhang, M.; Wang, W.; Liang, H.; Zeng, J.; Wu, K.; Wang, X.; Hsieh, J.; Guo, P.; et al. HIF-1α promotes ZEB1 expression and EMT in a human bladder cancer lung metastasis animal model. Oncol. Lett. 2018, 15, 3482–3489. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Tanaka, F.; Morita, H.; Hiraki, A.; Hashimoto, S. Hypoxia-induced HIF-1alpha and ZEB1 are critical for the malignant transformation of ameloblastoma via TGF-beta-dependent emt. Cancer Med. 2019, 8, 7822–7832. [Google Scholar] [CrossRef]
- Shih, C.-H.; Chuang, L.-L.; Tsai, M.-H.; Chen, L.-H.; Chuang, E.Y.; Lu, T.-P.; Lai, L.-C. Hypoxia-Induced MALAT1 Promotes the Proliferation and Migration of Breast Cancer Cells by Sponging MiR-3064-5p. Front. Oncol. 2021, 11, 658151. [Google Scholar] [CrossRef]
- Zheng, X.; Linke, S.; Dias, J.M.; Zheng, X.; Gradin, K.; Wallis, T.P.; Hamilton, B.R.; Gustafsson, M.; Ruas, J.L.; Wilkins, S.; et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 3368–3373. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-L.; Park, J.-S.; Manzanero, S.; Choi, Y.; Baik, S.-H.; Okun, E.; Gelderblom, M.; Fann, D.Y.-W.; Magnus, T.; Launikonis, B.S.; et al. Evidence that collaboration between HIF-1α and Notch-1 promotes neuronal cell death in ischemic stroke. Neurobiol. Dis. 2014, 62, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-L.; Wan, L.; Xu, Q.-R.; Zhao, Y.; Liu, J.-C. Notch signaling activation contributes to cardioprotection provided by ischemic preconditioning and postconditioning. J. Transl. Med. 2013, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Sallé-Lefort, S.; Miard, S.; Nolin, M.-A.; Boivin, L.; Paré, M.-È.; Debigaré, R.; Picard, F. Hypoxia upregulates Malat1 expression through a CaMKK/AMPK/HIF-1α axis. Int. J. Oncol. 2016, 49, 1731–1736. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Gao, J.; Liao, Q. Circular RNA circptk2 regulates oxygen-glucose deprivation-activated microglia-induced hippocampal neuronal apoptosis via miR-29b-sOCS-1-JAK2/STAT3-IL-1beta signaling. Int. J. Biol. Macromol. 2019, 129, 488–496. [Google Scholar] [CrossRef]
- Zhang, M.-X.; Zhang, L.-Z.; Fu, L.-M.; Yao, H.-H.; Tan, L.; Feng, Z.-H.; Li, J.-Y.; Lu, J.; Pan, Y.-H.; Shu, G.-N.; et al. Positive feedback regulation of lncRNA PVT1 and HIF2α contributes to clear cell renal cell carcinoma tumorigenesis and metastasis. Oncogene 2021, 40, 5639–5650. [Google Scholar] [CrossRef]
- Wang, J.; Dong, Z.; Sheng, Z.; Cai, Y. Hypoxia-induced PVT1 promotes lung cancer chemoresistance to cisplatin by autophagy via PVT1/miR-140-3p/ATG5 axis. Cell Death Discov. 2022, 8, 104. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, Y.; Chen, Y.; Bai, Y.; Zhang, Y. Long noncoding RNA LINC01234 promotes serine hydroxymethyltransferase 2 expression and proliferation by competitively binding miR-642a-5p in colon cancer. Cell Death Dis. 2019, 10, 137. [Google Scholar] [CrossRef]
- Wang, L.; Sun, L.; Liu, R.; Mo, H.; Niu, Y.; Chen, T.; Wang, Y.; Han, S.; Tu, K.; Liu, Q. Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1alpha signaling loop promotes hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 2021, 40, 72. [Google Scholar] [CrossRef]
- Hu, C.; Bai, X.; Liu, C.; Hu, Z. Long noncoding RNA XIST participates hypoxia-induced angiogenesis in human brain microvascular endothelial cells through regulating miR-485/SOX7 axis. Am. J. Transl. Res. 2019, 11, 6487–6497. [Google Scholar]
- Xie, J. Long Noncoding RNA XIST Regulates Myocardial Infarction via miR-486-5p/SIRT1 Axis. Appl. Biochem. Biotechnol. 2023, 195, 725–734. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Li, X.-F. Hypoxia and the Tumor Microenvironment. Technol. Cancer Res. Treat. 2021, 20, 15330338211036304. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int. J. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Nejad, A.E.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Javanmard, S.H.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Jacob, S.T. Emerging Role of MicroRNAs in Drug-Resistant Breast Cancer. Gene Expr. 2011, 15, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol. Chem. 2017, 398, 929–938. [Google Scholar] [CrossRef]
- Baxter, D.E.; Allinson, L.M.; Al Amri, W.S.; Poulter, J.A.; Pramanik, A.; Thorne, J.L.; Verghese, E.T.; Hughes, T.A. MiR-195 and Its Target SEMA6D Regulate Chemoresponse in Breast Cancer. Cancers 2021, 13, 5979. [Google Scholar] [CrossRef]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to s-1 in colon cancer. Cancer Genom. Proteom. 2006, 3, 317–324. [Google Scholar]
- Bogutz, A.B.; Oh-McGinnis, R.; Jacob, K.J.; Ho-Lau, R.; Gu, T.; Gertsenstein, M.; Nagy, A.; Lefebvre, L. Transcription factor ASCL2 is required for development of the glycogen trophoblast cell lineage. PLoS Genet. 2018, 14, e1007587. [Google Scholar] [CrossRef]
- Ziello, J.E.; Jovin, I.S.; Huang, Y. Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway and its Potential for Therapeutic Intervention in Malignancy and Ischemia. Yale J. Biol. Med. 2007, 80, 51–60. [Google Scholar]
- Semenza, G.L. Hypoxia-Inducible Factor 1 (HIF-1) Pathway. Sci. STKE 2007, 2007, cm8. [Google Scholar] [CrossRef] [PubMed]
- Salceda, S.; Caro, J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions: Its stabilization by hypoxia depends on redox-induced changes*. J. Biol. Chem. 1997, 272, 22642–22647. [Google Scholar] [CrossRef] [PubMed]
- Jaśkiewicz, M.; Moszyńska, A.; Króliczewski, J.; Cabaj, A.; Bartoszewska, S.; Charzyńska, A.; Gebert, M.; Dąbrowski, M.; Collawn, J.F.; Bartoszewski, R. The transition from HIF-1 to HIF-2 during prolonged hypoxia results from reactivation of PHDs and HIF1A mRNA instability. Cell. Mol. Biol. Lett. 2022, 27, 109. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-Inducible Factors in Physiology and Medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef]
- Li, C.; Jackson, R.M. Reactive species mechanisms of cellular hypoxia-reoxygenation injury. Am. J. Physiol. Physiol. 2002, 282, C227–C241. [Google Scholar] [CrossRef]
- Talks, K.L.; Turley, H.; Gatter, K.C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J.; Harris, A.L. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 2000, 157, 411–421. [Google Scholar] [CrossRef]
- Tian, H.; McKnight, S.L.; Russell, D.W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997, 11, 72–82. [Google Scholar] [CrossRef]
- Flamme, I.; Fröhlich, T.; von Reutern, M.; Kappel, A.; Damert, A.; Risau, W. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1α and developmentally expressed in blood vessels. Mech. Dev. 1997, 63, 51–60. [Google Scholar] [CrossRef]
- Koh, M.Y.; Powis, G. Passing the baton: The HIF switch. Trends Biochem. Sci. 2012, 37, 364–372. [Google Scholar] [CrossRef]
- Bartoszewski, R.; Moszynska, A.; Serocki, M.; Cabaj, A.; Polten, A.; Ochocka, R.; Dell’ Italia, L.; Bartoszewska, S.; Kroliczewski, J.; Dabrowski, M.; et al. Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. FASEB J. 2019, 33, 7929–7941. [Google Scholar] [CrossRef]
- Carmeliet, P.; Dor, Y.; Herbert, J.M.; Fukumura, D.; Brusselmans, K.; Dewerchin, M.; Neeman, M.; Bono, F.; Abramovitch, R.; Maxwell, P.; et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998, 394, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Shibasaki, F. Hypoxia-Inducible Factor as an Angiogenic Master Switch. Front. Pediatr. 2015, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Ryan, H.E.; Lo, J.; Johnson, R.S. HIF-1alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998, 17, 3005–3015. [Google Scholar] [CrossRef] [PubMed]
- Packer, M. Mutual Antagonism of Hypoxia-Inducible Factor Isoforms in Cardiac, Vascular, and Renal Disorders. JACC Basic Transl. Sci. 2020, 5, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, J.-P.; Heikkilä, M.; Malinen, M.; Lee, H.-M.; Palvimo, J.J.; Wei, G.-H.; Myllyharju, J. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell. Mol. Life Sci. 2020, 77, 3627–3642. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Moszynska, A.; Serocki, M.; Kroliczewski, J.; Bartoszewska, S.; Collawn, J.F.; Bartoszewski, R. Hypoxia-inducible factor (HIF)-3a2 serves as an endothelial cell fate executor during chronic hypoxia. EXCLI J. 2022, 21, 454–469. [Google Scholar]
- Ravenna, L.; Salvatori, L.; Russo, M.A. HIF3α: The little we know. FEBS J. 2016, 283, 993–1003. [Google Scholar] [CrossRef]
- Pasanen, A.; Heikkilä, M.; Rautavuoma, K.; Hirsilä, M.; Kivirikko, K.I.; Myllyharju, J. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int. J. Biochem. Cell Biol. 2010, 42, 1189–1200. [Google Scholar] [CrossRef]
- Maynard, M.A.; Evans, A.J.; Hosomi, T.; Hara, S.; Jewett, M.A.; Ohh, M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005, 19, 1396–1406. [Google Scholar] [CrossRef]
- Bartoszewska, S.; Collawn, J.F.; Bartoszewski, R. The Role of the Hypoxia-Related Unfolded Protein Response (UPR) in the Tumor Microenvironment. Cancers 2022, 14, 4870. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia--a key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Pore, N.; Jiang, Z.; Shu, H.-K.; Bernhard, E.; Kao, G.D.; Maity, A. Akt1 Activation Can Augment Hypoxia-Inducible Factor-1α Expression by Increasing Protein Translation through a Mammalian Target of Rapamycin–Independent Pathway. Mol. Cancer Res. 2006, 4, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Emerling, B.M.; Weinberg, F.; Liu, J.-L.; Mak, T.W.; Chandel, N.S. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a). Proc. Natl. Acad. Sci. USA 2008, 105, 2622–2627. [Google Scholar] [CrossRef] [PubMed]
- Zundel, W.; Schindler, C.; Haas-Kogan, D.; Koong, A.; Kaper, F.; Chen, E.; Gottschalk, A.R.; Ryan, H.E.; Johnson, R.S.; Jefferson, A.B.; et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000, 14, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Narravula, S.; Colgan, S.P. Hypoxia-Inducible Factor 1-Mediated Inhibition of Peroxisome Proliferator-Activated Receptor α Expression During Hypoxia. J. Immunol. 2001, 166, 7543–7548. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.A.; Abd Jamil, A.H.; Heather, L.C.; Murray, A.J.; Sutton, E.R.; Slingo, M.; Sebag-Montefiore, L.; Tan, S.C.; Aksentijevic, D.; Gildea, O.S.; et al. On the pivotal role of pparalpha in adaptation of the heart to hypoxia and why fat in the diet increases hypoxic injury. FASEB J. 2016, 30, 2684–2697. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Anand, V.; Roy, S. Vascular Endothelial Growth Factor Signaling in Hypoxia and Inflammation. J. Neuroimmune Pharmacol. 2014, 9, 142–160. [Google Scholar] [CrossRef]
- Park, T. Crk and CrkL as Therapeutic Targets for Cancer Treatment. Cells 2021, 10, 739. [Google Scholar] [CrossRef]
- Chen, P.Y.; Qin, L.; Zhuang, Z.W.; Tellides, G.; Lax, I.; Schlessinger, J.; Simons, M. The docking protein Frs2alpha is a critical regulator of Vegf receptors signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 5514–5519. [Google Scholar] [CrossRef]
- Stetler-Stevenson, W.G.; Seo, D.-W. TIMP-2: An endogenous inhibitor of angiogenesis. Trends Mol. Med. 2005, 11, 97–103. [Google Scholar] [CrossRef]
- Xue, C.; Xie, J.; Zhao, D.; Lin, S.; Zhou, T.; Shi, S.; Shao, X.; Lin, Y.; Zhu, B.; Cai, X. The JAK/STAT3 signalling pathway regulated angiogenesis in an endothelial cell/adipose-derived stromal cell co-culture, 3D gel model. Cell Prolif. 2017, 50, e12307. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Ganini, C.; Candi, E.; Melino, G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, L.; Zhou, J.; Chen, Y.; Xie, D.; Yao, Y.; Cui, D. Novel Insights into MALAT1 Function as a MicroRNA Sponge in NSCLC. Front. Oncol. 2021, 11, 758653. [Google Scholar] [CrossRef]
- Choi, P.S.; Zakhary, L.; Choi, W.-Y.; Caron, S.; Alvarez-Saavedra, E.; Miska, E.A.; McManus, M.; Harfe, B.; Giraldez, A.J.; Horvitz, R.H.; et al. Members of the miRNA-200 Family Regulate Olfactory Neurogenesis. Neuron 2008, 57, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ding, S.; Fan, Y.; Shen, F.; Dong, Q.; Zhao, B.; Pan, Y.; Wan, J. MiR-429 inhibits the angiogenesis of human brain microvascular endothelial cells through Snai2-mediated Gsk-3beta/beta-catenin pathway. Comput. Math. Methods Med. 2021, 2021, 6753926. [Google Scholar] [CrossRef]
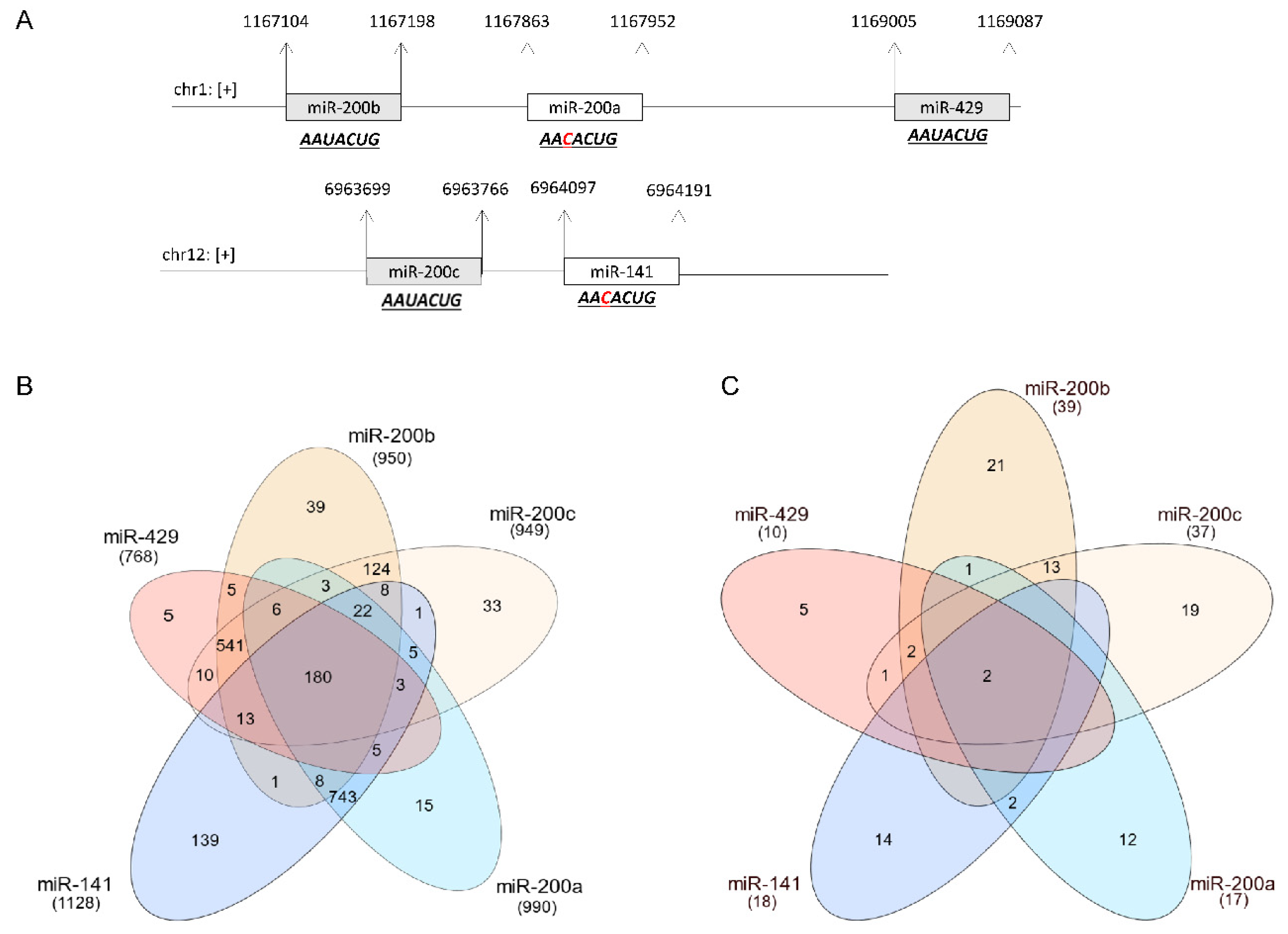

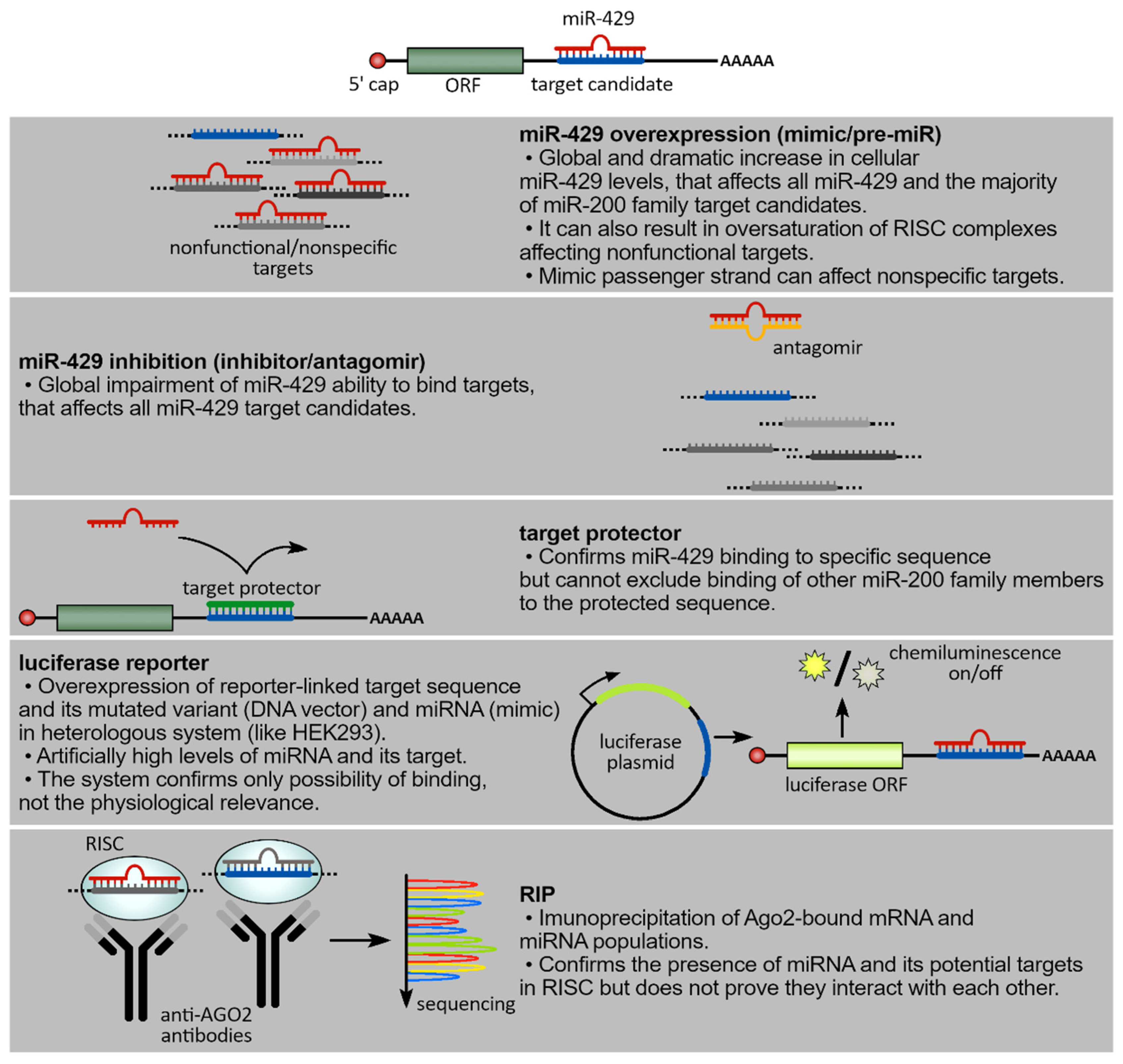
| DOI | Targets | Sponge (*) | Transcriptional Regulator | Mimic | Inhibitor | Luciferase Reporter | Target Protector | RIP | qPCR | WB | Animal Model | miR-200 Family Verification | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ttAV10.1038/ncb1722 | ZEB1 ZEB2 | − | ZEB1 | + | + | + | − | − | + | + | + | + | [91] |
| 10.1007/s12013-014-9885-8 | ZEB1 | − | − | + | + | + | − | − | + | + | + | + | [92] |
| 10.1080/21655979.2021.1923238 | ZEB1 | − | − | + | + | + | − | − | + | + | + | − | [93] |
| 10.1186/s12860-022-00420-x | ZEB1 MAPKAPK5-AS1 * | + | − | + | + | + | − | + | − | + | + | − | [94] |
| Q31016/j.biopha.2016.03.035 | ZEB1 CRKL | − | − | + | + | + | − | − | + | + | + | − | [95] |
| 10.1074/jbc.M114.598383 | ZEB1 ZEB2 | − | ASCL2 | + | + | + | − | + | − | − | − | + | [96] |
| 10.1016/j.biocel.2019.05.021 | ZEB1 XIST * | + | − | + | + | + | − | − | + | + | − | − | [97] |
| 10.1155/2021/7974012 | ZEB1 LINC01303 * | + | − | + | + | + | − | + | + | + | + | − | [98] |
| 10.1016/j.lfs.2020.118480 | ZEB1 MALAT1 * | + | − | + | + | + | − | + | + | + | − | − | [99] |
| 10.1097/CAD.0000000000001295 | LRP4 RNF185-AS1 * | + | − | + | + | + | − | + | + | + | + | − | [100] |
| 10.1096/fj.14-267054 | HIF1A | − | HIF-1 | + | + | − | + | − | + | + | − | + | [101] |
| 10.1007/s10529-018-2604-6 | HIF1A | − | − | + | + | + | − | − | + | + | − | − | [102] |
| 10.1038/srep22775 | HIF3A | − | HIF-1 | + | + | − | + | − | + | + | − | + | [103] |
| 10.3892/ol.2020.11766 | HOXA9 | − | − | + | + | + | − | − | + | + | + | − | [104] |
| 10.1016/j.abb.2017.11.011 | RAB23 | − | − | + | + | + | − | − | + | + | + | − | [105] |
| 10.3389/fnmol.2018.00035 | TJP1 OCLN | − | − | + | + | + | − | − | + | + | − | − | [106] |
| 10.1016/j.bbrc.2017.06.181 | RohE | − | − | + | + | + | − | − | + | + | + | − | [107] |
| 10.3892/mco.2019.1940 | AKT1 | − | − | + | + | + | − | − | + | + | + | − | [108] |
| 10.3892/or.2015.4039 | PAK6 | − | − | + | + | + | − | − | + | + | − | − | [109] |
| 10.1016/j.urolonc.2015.03.016 | BMI1 E2F3 | − | − | + | + | + | − | − | + | + | − | − | [110] |
| 10.3892/ijmm.2016.2558 | NOTCH1 | − | − | + | + | + | − | − | + | + | − | − | [111] |
| 10.1093/carcin/bgt089 | RAB18 | − | HBx | + | + | + | − | − | + | + | − | + | [112] |
| 10.3389/fonc.2020.01067 | CD274 | − | − | + | + | + | − | − | + | + | − | − | [113] |
| 10.1089/cbr.2020.3705 | FRS2 SNHG6 * | + | − | + | + | + | − | + | + | + | + | − | [114] |
| 10.1186/s12943-018-0889-7 | TRIM33 circPTK2 * | + | − | + | + | + | − | + | + | + | − | + | [115] |
| 10.1007/s10528-022-10285-6 | CELF2 circLIFR * | + | − | + | + | + | − | + | + | + | + | − | [116] |
| 10.1002/jcp.27772 | MALAT1 * | + | − | + | + | + | − | + | − | + | + | − | [117] |
| PMID: 34786067 | SYNJ1 LINC01234 * | + | − | + | + | + | − | + | + | + | + | − | [118] |
| 10.1155/2022/1447207 | JAK1 MSC-AS1 * | + | − | + | + | + | − | + | + | + | − | − | [119] |
| 10.3892/mmr.2021.12323 | FOXK1 circPVT1 * | + | − | + | + | + | − | − | + | + | − | − | [120] |
| 10.2147/OTT.S277284 | SCAMP1 | + | − | + | + | + | − | − | + | + | + | − | [121] |
| 10.1016/j.lfs.2020.117323 | TRIB2 circ_0084043 * | + | − | + | + | + | − | + | + | + | + | − | [122] |
| 10.1080/21655979.2021.1953822 | EZH2 circRNA_0082835 * | + | − | + | + | + | − | + | + | + | − | − | [123] |
| 10.1186/s11658-020-0202-9 | IKKB | − | − | + | + | + | − | − | + | + | + | − | [124] |
| 10.3892/mmr.2021.12220 | IKKB | − | − | + | + | + | − | − | + | + | − | − | [125] |
| 10.1016/j.omtn.2021.01.026 | PPARA | − | EZH2 | + | + | + | − | − | + | + | + | − | [126] |
| 10.7150/jca.21024 | XIST * | + | − | + | + | + | − | − | + | + | + | − | [127] |
| 10.1016/j.bbrc.2014.05.084 | PTEN RASSF8 TIMP2 | − | − | + | + | + | − | − | + | + | − | − | [128] |
| 10.1016/j.canlet.2015.04.023 | PTEN | − | − | + | + | + | − | − | + | + | + | + | [129] |
| 10.1016/j.biopha.2020.110215 | CRKL | − | − | + | + | + | − | − | + | + | − | − | [130] |
| 10.1038/s41598-018-20258-8 | CRKL | − | − | + | + | + | − | + | − | + | − | − | [131] |
| 10.1136/gutjnl-2013-305715 | RBBP4 | − | − | + | + | + | − | − | + | + | − | + | [132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartoszewska, S.; Sławski, J.; Collawn, J.F.; Bartoszewski, R. HIF-1-Induced hsa-miR-429: Understanding Its Direct Targets as the Key to Developing Cancer Diagnostics and Therapies. Cancers 2023, 15, 2903. https://doi.org/10.3390/cancers15112903
Bartoszewska S, Sławski J, Collawn JF, Bartoszewski R. HIF-1-Induced hsa-miR-429: Understanding Its Direct Targets as the Key to Developing Cancer Diagnostics and Therapies. Cancers. 2023; 15(11):2903. https://doi.org/10.3390/cancers15112903
Chicago/Turabian StyleBartoszewska, Sylwia, Jakub Sławski, James F. Collawn, and Rafal Bartoszewski. 2023. "HIF-1-Induced hsa-miR-429: Understanding Its Direct Targets as the Key to Developing Cancer Diagnostics and Therapies" Cancers 15, no. 11: 2903. https://doi.org/10.3390/cancers15112903
APA StyleBartoszewska, S., Sławski, J., Collawn, J. F., & Bartoszewski, R. (2023). HIF-1-Induced hsa-miR-429: Understanding Its Direct Targets as the Key to Developing Cancer Diagnostics and Therapies. Cancers, 15(11), 2903. https://doi.org/10.3390/cancers15112903







