Simple Summary
The tumor microenvironment (TME) exhibits a pivotal function in the progression of cancer through the bidirectional communication between cellular and noncellular components. During the exchange and interplay of biological signals between immune cells and tumor cells in TME, exosomal lncRNAs are viewed as one of the vital factors, which can interfere with innate and adaptive immune responses to affect the therapeutic efficiency. This review aims to summarize the advance in the roles of exosomal lncRNAs on tumor-immune regulation and propose potential treatment strategies.
Abstract
Exosomes are nanovesicles secreted into biofluids by various cell types and have been implicated in different physiological and pathological processes. Interestingly, a plethora of studies emphasized the mediating role of exosomes in the bidirectional communication between donor and recipient cells. Among the various cargoes of exosomes, long non-coding RNAs (lncRNAs) have been identified as crucial regulators between cancer cells and immune cells in the tumor microenvironment (TME) that can interfere with innate and adaptive immune responses to affect the therapeutic efficiency. Recently, a few major studies have focused on the exosomal lncRNA-mediated interaction between cancer cells and immune cells infiltrated into TME. Nevertheless, a dearth of studies pertains to the immune regulating role of exosomal lncRNAs in cancer and is still in the early stages. Comprehensive mechanisms of exosomal lncRNAs in tumor immunity are not well understood. Herein, we provide an overview of the immunomodulatory function of exosomal lncRNAs in cancer and treatment resistance. In addition, we also summarize the potential therapeutic strategies toward exosomal lncRNAs in TME.
1. Background
Cancer is still life-threatening, with sustained growth in incidence and mortality worldwide. In tumor occurrence and progression, the tumor microenvironment (TME) has been recognized as a complex and diverse multicellular ecosystem shaped by neoplastic cells [1]. Constituted by cellular agents (tumor cells, immune cells, endothelial cells, fibroblasts, etc.) and other soluble mediators (cytokines, chemokines, extracellular vesicles, etc.) [2,3], TME exhibits a pivotal function in the progression of cancer through the bidirectional communication between cellular and noncellular components. Noteworthy, the transmission of stimulatory signals between tumor cells and immune cells in TME contributes to tumor progression by evading immune surveillance [4,5]. Accordingly, disruption of the signal delivery has drawn much research interest and could prove a potential intervention target in cancer. A growing body of studies has shown that cargo-carrying extracellular vesicles (EVs) are critical messengers in the interaction between tumor cells and the microenvironment [6,7]. In addition, non-coding RNAs (ncRNAs), such as long-noncoding RNAs (lncRNAs), have been implicated in abnormal immune responses and the subsequent tumor progression and therapy resistance [8,9]. Moreover, lncRNAs can be encapsulated by EVs and delivered to recipient cells in TME [10]. Therefore, understanding the mechanism of the dysregulated immune response involved in TME, which is mediated by the transportation of lncRNAs through EVs, is critical for identifying the possible therapeutic approach to cancer.
EVs are lipid bilayer-bounded vesicles originating from various cells, such as tumor cells, stromal cells, and immune cells, and diffusely present in a wide range of biofluids such as blood, sweat, urine, and ascites [11,12]. EVs are further classified into exosomes, micro-vesicles, and apoptotic bodies based on their sizes and biogenesis mechanisms [13]. In exosome production and secretion, cellular membranes tend to invaginate to form the early endosomes. Thereafter, the early endosomes are transformed into late endosomes by the Golgi complex. The late endosomal compartments limiting membranes further invaginate to form intraluminal vesicles (ILVs) while encapsulating cargoes from donor cells. Plenty of ILVs aggregate into multivesicular bodies (MVBs). Some MVBs are degraded by intracellular lysosomes, and a few can merge with the cytoplasmic membrane, thus releasing ILVs to the extracellular environment, ultimately forming exosomes [14]. According to the newly-revised ISEV2018 guideline, the naming standard of EVs is based on the physical characteristics (size or density), biochemical composition, or cell of origin [15]. In this review, all cited studies used exosomes sedimented at 100.000 g or defined as exosomes based on their protein composition and identified by transmission electron microscope.
Since exosomes can be loaded with biological information, including proteins and nuclear acids (e.g., messenger RNAs (mRNAs), microRNAs (miRNAs), and lncRNAs) [16], they are widely known as shuttles for cell-cell communication, usually along with the transmission of biological signals [17]. Such exosome-mediated signal exchanges between cells exist in physiological conditions, such as embryonic development and material metabolism [18], and also participate in tumor progression. An increasing number of studies have indicated that exosomes may exert a key function in immune regulation and thus contribute to establishing an immunosuppressive TME [19]. For example, CD73 expression of exosomes in the serum of melanoma patients can inhibit the immune response of T cells, thus enhancing the resistance to immunotherapy [20]. Melanoma-derived exosomes can inhibit the infiltration of CD8+T cells into tumors [21]. Pancreatic cancer-secreted exosomes can boost the conversion of tumor-associated macrophages to M2 phenotype to favor tumor metastasis [22]. Colorectal cancer-derived exosomes induced the expansion of regulatory T cells (Tregs) to construct an immunosuppressive TME that promotes tumor growth and chemoresistance [23]. Taken together, these studies have demonstrated that exosomes may regulate the innate and adaptive immune cells in TME to affect tumor progression and treatment. Accordingly, systematically clarifying the mechanism of exosomes targeting the immune response of cancer may be beneficial to explore potential therapeutic options.
In the human genome, no more than 2% of genes can be transcribed into mRNAs and thus translated into proteins [24]. However, most genes are transcribed into RNAs without the protein-coding ability known as ncRNAs [24]. According to the length of nucleotides, ncRNAs are mainly classified into (1) short ncRNAs, including miRNAs, small interfering RNAs (siRNAs), and small nucleolar RNAs (snoRNAs), and (2) lncRNAs [25]. With the development of sequencing technology, much more quantities of lncRNAs have been found in the human genome. Furthermore, lncRNAs can affect gene expression through more complex mechanisms by interplaying with DNA, RNA, and protein [26]. Therefore, lncRNAs have rapidly become the research focus in recent years.
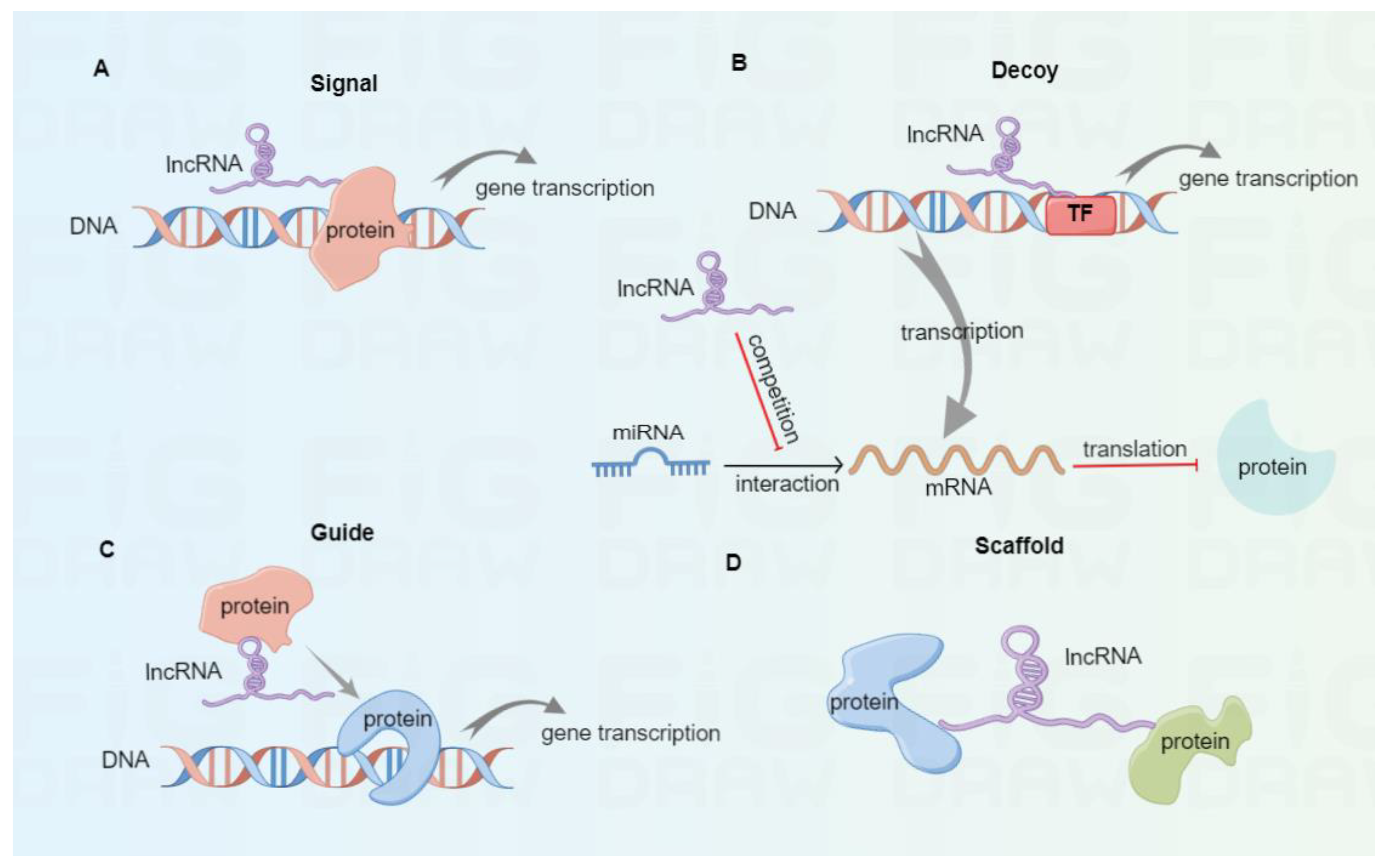
As a class of ncRNAs, lncRNAs involve more than 200 nucleotides and exist in the nucleus and cytoplasm [27]. Based on the transcription onset and direction relative to neighboring protein-coding genes, lncRNAs are classified into sense, antisense, intronic, bidirectional, and intergenic [28]. According to their molecular mechanisms (Figure 1), lncRNAs are divided into signal, decoy, guide, and scaffold [29,30]. Once subjected to external stimulation, signal lncRNAs can modulate the downstream gene transcription independently or interact with proteins like transcription factors, thus affecting specific signaling pathways. For instance, the p53-induced lncRNA-p21 was found to interplay with the heterogeneous nuclear ribonucleoprotein-K, thus suppressing the expression of subsequent genes related to the p53 signaling pathway [31]. Decoy lncRNAs function as decoys of transcription factors or competing endogenous RNAs (ceRNAs) to trap miRNAs and reverse their inhibitory effect in mRNA translation. In tongue squamous cell carcinoma, lncKRT16P6 acts as a trap for binding miR-3180 to promote the expression of GATA zinc finger domain containing 2A (GATAD2A), thus leading to tumorigenesis and metastasis [32]. Highly expressed lncRNA MALAT1 promotes tumor cell migration and invasion by sinking miR-384 in glioma [33]. Guide lncRNAs can bring ribonucleoprotein complexes, such as chromatin modification enzymes, to target positions for various biological functions. Xiao et al. reported that LINC00839 boosted the formation of RUVBL1/Tip60 complexes and guided them to the promoter of NRF1 to induce NRF1 expression, therefore affecting colorectal cancer progression [34]. Scaffold lncRNAs can serve as platforms to interplay with multiple protein complexes, thus facilitating the fusion and incorporation of messages among diverse signaling pathways. For instance, lncRNA ENSG00000274093.1 was found to promote the combination of histone deacetylase 2 (HDAC2) with histone methyltransferase EZH2 to induce epithelial-mesenchymal transition, thus promoting the migration and invasion of colorectal cancer cells [35].
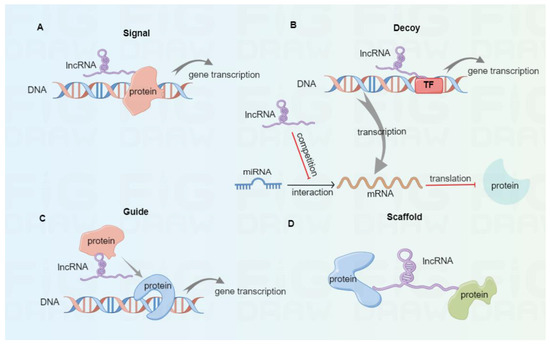
Figure 1.
The classification of lncRNAs based on molecular mechanisms. (A) Signal lncRNAs can modulate downstream gene transcription independently or by interacting with proteins. (B) Decoy lncRNAs serve as decoys of transcription factors or ceRNAs to bind with miRNAs, thus eliminating their inhibitory effect in mRNA translation. (C) Guide lncRNAs can bring protein complexes to target sites to promote gene transcription. (D) Scaffold lncRNAs serve as scaffold molecules to interplay with multiple protein complexes.
Owing to their transcriptional modulation to DNA, post-transcriptional modulation to target miRNA, and epigenetic regulation to proteins, lncRNAs are involved in many physiological processes, such as cell survival, growth [36,37], and maintenance of stemness [38]. Accumulating studies have demonstrated that dysregulated expression of lncRNAs is closely associated with tumor progression and metastasis [39]. Notably, lncRNAs can modulate tumor immunity between tumor cells and immune cells in TME. For instance, Fang et al. demonstrated that lncRNA GAS5 overexpression in NK cells increases their cytotoxicity to liver cancer [40]. Upregulated LncRNA HCG18 in tumor tissues was reported to inhibit CD8+T cells, thus increasing the chemoresistance of colorectal cancer [41]. LncRNA-PACERR was found to promote the polarization of M2 macrophages to regulate the progression of pancreatic ductal adenocarcinoma [42]. These results suggest that lncRNAs can participate in tumor progression by regulating the function of immune cells. Therefore, further understanding the role of lncRNAs in regulatory tumor immunity can provide insights for tumor immunotherapy.
During the exchange and interplay of biological signals between non-tumor cells and tumor cells in TME, exosomal lncRNAs are viewed as one of the vital factors affecting tumorigenesis and progression. Since many kinds of cells infiltrated into TME, several stromal cells, such as CAFs and endothelial cells, also communicate with tumor cells based on exosomal lncRNAs. CAFs can promote immune suppression by inhibiting the function of immune cells or secreting inflammatory factors in TME [43]. Inflammatory cytokines, such as IL-6, can take part in the regulation of tumor activity [44]. For instance, LncRNA POU3F3 can be packaged into exosomes derived from esophageal squamous cell carcinoma cells (ESCC) to promote the transformation from normal fibroblasts (NFs) into CAFs. The increased CAFs can enhance the cisplatin resistance of ESCC by producing IL-6 [45]. Ding et al. discovered an uncharacterized lncRNA FLJ22447, which can be transported to fibroblasts via tumor-derived exosomes. High expression of lncRNA FLJ22447 was found to promote the transformation of NFs to CAFs by upregulating IL-33 expression, thus facilitating OSCC progression [46]. In addition, CD90+ liver cancer cell-secreted exosomes containing lncRNA H19 can be transferred to endothelial cells to promote angiogenesis and increase their adhesive characteristic, thereby leading to tumor metastasis [47]. These results indicate that exosomal lncRNAs also mediate the communication between stromal cells and tumor cells to affect tumor progression. Accordingly, searching for potential therapeutics to target exosomal lncRNAs may offer value in cancer treatment.
In addition, immune cells infiltrating into TME mainly include tumor-associated macrophages (TAMs), neutrophils, NK cells, and T and B cell subsets [48]. These immune cells could interact with tumor cells to exert pro- or anti-tumor effects, which may be attributed to exosomal lncRNAs mediated immune regulation in TME [49,50]. Although the effects of exosomes or lncRNAs alone in cancer immunity are still under investigation, the immunomodulation effects of exosome-derived lncRNAs in cancer also need clarification. In this review, we used the combined terms “exosomes,” “lncRNAs,” and “cancer immune” to search the related literature in the PubMed database and summarized the newest advance in the roles of exosomal lncRNAs on tumor-immune regulation and potential treatment strategies.
2. Exosomal lncRNA-Mediated Immune Regulation
The immune responses of the host are classically divided into innate and adaptive immunity against foreign substances, which are known as antigens. Innate immunity (also called natural immunity) tends to occur early and is the first barrier against microorganism invasion. It primarily comprises physical barriers like epithelial surface, cells (neutrophils, macrophages, natural killer (NK) cells, dendritic cells, etc.), and noncellular factors (complements and mediators of inflammation) [51,52]. Intact epithelial barriers construct the first line of defense to impede microbial entry into the host. The macrophages/neutrophils-mediated phagocytosis or the NK cells-mediated killing activity could eliminate the microbes and infected cells inside the host [53]. In addition, innate immunity could initiate adaptive immunity through the secretion of cytokines or complement fragments. Adaptive immunity (acquired immunity) is often divided into B-cell-mediated humoral immunity and T-cell-mediated cellular immunity [54]. In humoral immunity, activated B cells secrete antibodies to eliminate extracellular microbes. After recognizing the antigens presented by dendritic cells, naive CD4+T cells can be activated and differentiated into effector CD4+T cells. Moreover, naive CD8+T cells can be activated and developed into CD8+ cytotoxic T lymphocytes (CTLs) targeting phagocytosed and intracellular microbes in adaptive immunity. As one of the classical immune response mechanisms, effector CD4+T cells secrete cytokines to activate macrophages to eliminate microbes. Meanwhile, effector CD8+T cell-mediated cytotoxicity produces granzymes and perforins to cause apoptotic death of the infected cells [55]. In addition, the naive T and B cells could also differentiate into long-lived memory T and B cells during the microorganism stimulation. Once exposed to the same microbial stimulation, the immune system can mobilize memory T and B cells to respond more rapidly and vigorously, known as immunological memory [56,57]. Based on the current study results, we discussed the regulation role of exosomal lncRNAs secreted from tumor cells or immune cells in innate or adaptive immunity.
2.1. Tumor Cell-Derived Exosomal lncRNAs
2.1.1. The Role of Exosomal lncRNAs in Macrophages
Macrophages in TME are also called tumor-associated macrophages (TAMs) and are a crucial population among innate immune cells [58]. TAMs play pro-tumor or anti-tumor roles depending on their activation status. After exposure to diverse stimuli, TAMs can be transformed into classical M1 or alternative M2 macrophages. M1 macrophages are more likely to produce pro-inflammatory cytokines and possess anti-tumor immunity, whereas M2 macrophages are responsible for anti-inflammation and pro-tumor progression [59]. The conversion of TAMs to M1 or M2 macrophages often occurs in the tumor progression from the early to the advanced stage [60]. Therefore, the interaction between TAM and cancer cells through exosomes needs investigation.
Brain cancer. Microglia are the unique lineage of resident macrophages in the brain [61]. Once activated, microglia can be polarized to M1 or M2 phenotypes. Xing et al. found that the downregulation of lncRNA XIST promoted the production of exosomes containing miRNA-503 in brain metastasis tumors of breast cancer, converting microglia to M2 to exert tumor-promoting effects [62]. In addition, exosomal lncRNA TALC secreted by glioblastoma can be transmitted to microglia, producing complement C5/C5a to activate the ENO1/p38 MAPK axis, thus manipulating the M2 polarization to achieve temozolomide resistance [63].
Colorectal cancer. Transcriptomics analysis has revealed the upregulation of 13 lncRNAs in BRAF mutant colorectal cancer (CRC) cells. The 13 lncRNAs can be secreted to promote the CD163+M2 macrophage polarization through exosomes. The increased IL-6 production from M2 and the elevated TGF-β generation from cancer-associated fibroblasts (CAFs) constitute the pro-tumor immunosuppressive microenvironment [64]. Furthermore, exosomal lncRNA RPPH1 can be transported to macrophages to provoke M2 polarization, thus promoting CRC cell metastasis and proliferation indicated by the increased levels of vimentin and Ki-67 [65]. Furthermore, exosomal lnc-HOXB8-1:2 secreted from neuroendocrine differentiation CRC cells is also found to promote M2 polarization, thus facilitating the progression and metastasis of CRC cells [66].
Esophageal and gastric cancer. Esophageal cancer (EC) cell-secreted exosomal lncRNA RP11-465B22.8 could be transferred to macrophages to induce M2 polarization, resulting in tumor cell migration and invasion [67]. Gastric cancer (GC) cell-secreted exosomal lncRNA HCG18 can promote the polarization of M2 macrophages by lessening miR-875-3p to enhance the expression of Kruppel-like factor 4 [68].
Hepatocellular carcinoma. The upregulated exosomal lncRNA HMMR-AS1 could boost M2 polarization via the miR-147a/ARID3A axis in hepatocellular carcinoma cells (HCC), thus leading to disease progression and poor prognosis [69]. In addition, the overexpression of lncRNA TUC399 can be encapsulated by HCC-derived exosomes and then taken up by a monocytic cell line THP-1 cells. This process benefits the polarization of THP-1 cells towards the M2 phenotype and diminishes its phagocytosis and pro-inflammatory cytokine production [70].
Lung cancer. Exosomal lncRNA SNHG7 secreted from lung adenocarcinoma (LUAD) cells is transferred to macrophages, which activates the phosphatidylinositol 3-kinase (PI3K)/AKT pathway and induces the phosphatase and tensin homolog (PTEN) degradation by recruiting cullin 4A, thus accelerating M2 polarization. The polarization of M2 macrophage can increase the docetaxel resistance in LUAD [71]. In non-small cell lung cancer (NSCLC), exosomal lncRNA SOX2 overlapping transcript (SOX2-OT) secreted from NSCLC cells boosts M2 polarization, thus increasing EGFR-TKI resistance by targeting the miR-627-3p/SMADs pathway [72]. In addition, NSCLC cell-secreted exosomal lncRNA PCAT6 has been found to induce M2 polarization, favoring tumor growth by promoting tumor cell invasion and migration [73]. Exosomal LINC00313 derived from NSCLC cells has also been reported to induce M2 polarization by activating STAT6 and benefit tumor progression [74]. Silencing of exosomal lncRNA FGD5-AS1 secreted from NSCLC cells can suppress M2 polarization to reduce migration and invasion of lung cancer cells [75].
Other tumors. Exosome-carrying lncRNA TP73-AS1 derived from nasopharyngeal carcinoma (NPC) can be transported to macrophages, which leads to M2 polarization and increased macrophage migration and tube formation [76]. In addition, LncRNA BCRT1 can be incorporated into exosomes to facilitate M2 polarization by targeting the miR-1303/PTBP3 axis in breast cancer, thus enhancing tumor cell migration and angiogenesis [77]. The overexpressed LncRNA ELFN1-AS1 in osteosarcoma (OS) cells could be transported via exosomes to macrophage, thereby boosting M2 polarization and OS progression [78]. Pancreatic cancer (PC) cell-derived exosomal FGD5-AS1 has been found to induce M2 polarization, thus accelerating PC cell growth and invasion [79]. Exosomes containing lncARSR secreted from renal cell carcinoma (RCC) were found to promote M2 polarization by activating the STAT3 pathway, thus favoring RCC progression [80]. In clear cell renal cell carcinoma (ccRCC), exosomal AP000439.2 derived from the tumor cells can promote their migration and growth by inducing M2 polarization via activating the STAT3/NF-κB signaling pathway [81].
To sum up, the above results indicate that exosomal lncRNAs derived from tumors usually promote M2 polarization or the production of inflammatory cytokines to support tumor progression and drug resistance.
2.1.2. The Role of Exosomal lncRNAs in NK Cells
NK cells are an important innate immune cell population, functionally killing tumor cells or producing cytokines to strengthen immune response [82]. Upregulated expression of lncRNA GAS5 in NK cells is found to enhance their cytotoxicity to different cancer cells [40,83]. CRC-cell secreted exosomal lncRNA SNHG10 can be transferred into NK cells to activate the inhibin subunit beta C (INHBC)-dependent TGF-β signaling pathway, thus inhibiting NK cell-mediated cytotoxicity [84]. These results indicate that exosomal lncRNAs can regulate the cytotoxic activity of NK cells, therefore, participate in the immune response to affect tumor progression.
2.1.3. The Role of Exosomal lncRNAs in CD4+ and CD8+T Cells
The adaptive immune response is mediated by T and B effector lymphocytes. Based on the different surface receptors and antigen specificity, the effector T cells could be classified into CD4+T helper (Th) and CD8+cytotoxic T lymphocytes (CTL) cells [85]. Due to the different biological roles, CD4+T cells can be further divided into T helper (Th) 1 cells, Th2, Th9, Th22, Th17 cells, follicular T helper (Tfh), and regulatory T (Treg) cells [86].
Th17 cells have been reported to benefit tumor progression by producing interleukin 17 (IL-17) [87]. Sun et al. revealed that CRC-secreted exosomes containing lncRNA CRNDE-h could be transmitted to CD4+T cells to increase RORγt transcription and further suppress its interaction with E3 ubiquitin ligase Itch to facilitate Th17 cells differentiation [88]. CRC-derived exosomes loaded with lncRNA KCNQ1OT1 can restrain the anticancer effect of CD8+T cells, thus facilitating CRC progression [89]. Collectively, tumor-secreted exosomal lncRNAs can promote Th17 cell differentiation or inhibit the function of normal CD8+T cells, thus contributing to tumorigenesis.
2.1.4. The Role of Exosomal lncRNAs in Tregs
Tregs, a specialized subset of T cells, can be recruited to TME to construct an immunosuppressive environment [90]. Ni et al. found that CD73+γδ1 Tregs were the primary ones exerting the immunosuppressive function in the progression of breast cancer. Mechanistically, breast tumor cell-secreted lncRNA SNHG16 shuttles through exosomes to boost CD73 expression in γδ1 Tregs and activate the TGF-β1/SMAD5 pathway [91]. Furthermore, Wang et al. reported that exosomal lncRNA RP11-323N12.5 could be delivered to tumor-infiltrating leukocytes, enhancing Treg differentiation, and thus contributing to immunosuppression and tumor growth of GC through the YAP1/c-MYC axis [92]. To sum up, tumor-derived exosomal lncRNA can trigger Tregs differentiation to induce immunosuppression in TME.
2.1.5. The Role of Exosomal lncRNAs in Bregs
Regulatory B cells (Bregs) are a special subpopulation of B lymphocytes that can inhibit the inflammatory response and hold immunological tolerance [93]. A few studies have demonstrated that Bregs can secrete IL-10 and TGF-β to restrain the immune response against tumors, thus beneficial to carcinogenesis [94,95,96]. A previous study has shown that ESCC-derived exosomes can restrict the expansion of B cells and promote the differentiation of Interleukin-10+ Bregs conversely. Further bioinformatic analysis has confirmed the significantly differentially expressed lncRNA profiles in ESCC-Exo compared with normal exosomes [97]. This result reveals that tumor-derived exosomal lncRNAs may promote Breg generation to escape immune surveillance.
2.2. Immune Cell-Derived Exosomal lncRNAs
In addition, exosomal lncRNAs from immune cells could be delivered into tumor cells to affect tumor progression and treatment efficacy. The underlying roles of macrophages- or CD8+T cell-derived exosomal lncRNAs in immune regulation and cancer pathogenesis are discussed as follows.
Macrophage-secreted exosomes. TAMs are one of the most plentiful immune cells infiltrated into TME [98]. TAMs can secrete exosomes containing lncRNAs to regulate tumor cell metabolism and survival. For example, TAM-secreted exosomes containing HIF-1α-stabilizing long noncoding RNA (HISLA) enhanced the aerobic glycolysis of breast cancer cells, further inhibiting cell apoptosis and promoting chemoresistance [99]. Furthermore, exosomal lncMMPA derived from TAMs is also found to facilitate M2 polarization and augment tumor’ aerobic glycolysis to promote HCC proliferation [100]. TAM-secreted exosomes with highly-expressed LIFR-AS1 can sink miR-29a to upregulate NFIA, thus supporting cell proliferation and invasion and suppressing apoptosis of OS cells [101]. However, the overexpression of recombination signal binding protein for immunoglobulin kappa J region in macrophage can secrete lncRNA LBX1-AS1-loaded exosomes to repress the proliferation and invasion of oral squamous cell carcinoma (OSCC) cells [102]. These studies indicated that the beneficial or harmful effects on tumors depend on the characteristics of macrophages.
M2-secreted exosomes. The lncRNAs encapsulated by M2-secreted exosomes have been proven to participate in tumor growth regulation and immune response by secreting cytokines like IL-10 and TGF-β. For instance, M2 macrophage-secreted exosomes (M2-Exo) transferred lncRNA AFAP1-AS1 at a high expression level to EC cells, which boosted the lung metastasis of cancer cells [103]. Hepatitis B virus (HBV)-related HCC can produce HBeAg to facilitate the expression of lncRNA MAPKAPK5_AS1 (MAAS) in M2 macrophages, thus transmitted by M2-Exo to HBV+HCC cells to promote their proliferation [104]. Upregulated lncRNA CRNDE in M2-Exo can be delivered to cisplatin-treated GC cells to promote their proliferation and enhance the resistance to cisplatin [105]. M2-Exo-loaded lncRNA AGAP2 antisense RNA 1 (AGAP2-AS1) can promote the radio resistance of lung cancer cells by negatively regulating miR-296 [106]. LINC00273-containing exosomes secreted from the M2 subtype are found to improve the invasion, migration, and metastasis of LUAD cells by regulating the Hippo/Yes-associated transcriptional regulator pathway [107]. The M2-Exo containing the highly expressed lncRNA SBF2-AS1 can significantly upregulate the expression of X-linked inhibitor of apoptosis protein by competitively binding to miR-122-5p, thus contributing to PC progression [108]. M2-secreted exosomal lncRNA H19 can facilitate the autophagy process of bladder cancer (BC) through stabilizing Unc-51-like autophagy activating kinase 1 (ULK1) [109]. Taken together, the delivery of aberrant lncRNA in M2-secreted exosomes often benefits tumor progression or enhances the chemoresistance or radio resistance of tumor cells.
M1-secreted exosomes. M1 macrophages have been reported to transfer exosomal lncRNA to tumor cells to affect tumor progression. M1-derived exosomes (M1-Exo) loaded with lncRNA HOTTIP can promote the apoptosis of head and neck squamous cell carcinoma (HNSCC) cells and repress the progression. The mechanistic investigation has unveiled that lncRNA HOTTIP upregulates the TLR5/NF-κB signaling pathway by binding to miR-19a-3p and miR-19b-3p [110]. Therefore, certain lncRNAs in M1-Exo may inhibit the progression of tumors and serve as a potential therapeutic target.
CD8+T cell-secreted exosomes. As previously mentioned, tumor-secreted exosomal lncRNAs can interfere with the cytotoxic ability of CD8+T cells to tumor cells. Furthermore, normal CD8+T cells could be inhibited by their exhausted population. Accompanied by low cytotoxic ability, the exhausted CD8+T cells have been shown to deliver self-generated exosomes to normal CD8+T cells, thus impairing their proliferation and activity [111].
3. Exosomal lncRNA-Based Cancer Treatment
Emerging studies have demonstrated the significant linkage between exosomal lncRNAs and immune regulation in cancers. Aberrant exosome-associated lncRNAs could affect the sensitivity to therapeutic strategies by regulating the infiltration and function of immune cells in cancers (Table 1). Based on the immune-regulatory mechanism of exosomal lncRNAs in cancers, increasing studies tend to explore the corresponding therapeutic strategies targeting exosomal lncRNAs.

Table 1.
Summary of immunoregulation role of exosomal lncRNAs in cancer.
Given the side-effect and resistance of traditional therapy, such as chemotherapy and radiotherapy, immunotherapy is considered a promising option to fight cancer [112,113]. The application of immunotherapy can effectively identify and eliminate tumor cells by enhancing the natural anticancer capacity of the host’s immune cells. With the advance in research of immunotherapy in past decades, vaccines, chimeric antigen receptor T-cell (CAR-T) therapy, immune checkpoint inhibitors, and combinatorial therapies have emerged as potential immunotherapies against cancer [114,115,116,117]. One of the most prospective immunotherapy strategies is the immune checkpoint inhibitors. In physiological conditions, immune checkpoints, such as PD-1 and CTLA-4, are expressed on the surface of immune cells, maintaining immunological tolerance and avoiding autoimmunity appearance [118]. Tumor cells exploit their surface ligands to combine with corresponding immune checkpoint receptors, thus suppressing the anti-tumor immune response of immune cells [119].
Emerging evidence has revealed that exosomal lncRNAs can regulate the ligand-receptor interplay of PD-L1/PD-1 to escape the immune attack of immune cells in various cancers. For instance, Xing et al. reported that the downregulation of lncRNA XIST boosted the production of tumor-secreted exosomal miR-503, which enhanced the expression of PD-L1 on M2 macrophages to suppress T proliferation in breast cancer [62]. Exosomes secreted by ESCC cells can induce the high expression of PD-1 on Bregs by activating the TLR4/MAPK signaling pathways, thus promoting B cell-mediated immunosuppression [97]. CRC-secreted exosomes can deliver lncRNA KCNQ1OT1 to themselves, thus inhibiting the ubiquitination of PD-L1 from escaping the CD8+T cell-mediated immune surveillance [89]. Exosomal lncRNA PCED1B-AS1 derived from HCC can enhance the expression of PD-L1 and PD-L2 on HCC cells and function as a suppressor to T cells and macrophages [120]. These results demonstrate that exosomal lncRNAs can promote the tumor’s escape from immune surveillance by enhancing the immunosuppression of M2 and Bregs or upregulating the expression of PD-L1 and PD-L2 on tumor cells. Therefore, the application of immune checkpoint inhibitors may block the exosomal lncRNA-mediated receptor-ligand interaction between tumor and immune cells, thus enhancing anti-tumor immunity [121].
A recent study also revealed that lncRNA NORAD knockdown induces the secretion of exosomes containing miR-199a-5p to suppress the ubiquitination of PD-L1 in an ESCC mouse model, resulting in enhanced efficacy of anti-PD-1 in combination with radiation [122]. In addition, Li et al. also identified that B7-H3 and VSIR might be novel checkpoints for their upregulated expression in GC patients, affecting the efficacy of immunotherapy [123]. These results indicate that the application of immune checkpoint inhibitors or antibodies aiming to interfere with exosomal lncRNA-mediated checkpoint interaction benefits the normal function of immune cells, thereby improving the efficacy of cancer treatment.
Given that exosomes carrying lncRNAs play the role of immune regulation in cancer, interventions of exosomal lncRNAs, such as siRNA, are also one of the potential therapeutic modalities. Characterized by double-stranded RNAs with 20 to 24 nucleotides, siRNA can inhibit mRNA transcription by inducing heterochromatin assembly and suppress mRNA translation or facilitate the degradation of mRNA or pre-mRNA to regulate gene expression [124]. Furthermore, siRNAs can be delivered and integrated into the genome of target cells, thus leading to post-transcriptional regulation of lncRNAs. Certain siRNAs complementarily combine with target lncRNAs to construct an RNA-induced silencing complex, thereby splicing and degrading the target lncRNA [125]. Accordingly, siRNAs can interfere with aberrant lncRNA expression, thus affecting tumor progression [126]. Chen et al. revealed that using siRNAs to knock down PCAT6 in NSCLC cells can suppress M2 polarization, thus inhibiting tumor growth [73]. Due to the good biocompatibility of exosomes, they are used as delivery mediators to protect siRNAs from endosome-mediated degradation. For instance, upregulated lncRNA DARS-AS1 was found to facilitate the tumorigenesis and metastasis of triple-negative breast cancer (TNBC), whereas DARS-AS1 siRNA-loaded exosomes inhibited the growth and metastasis of TNBC [127]. Guo et al. found that upregulated lncRNA H19 in M2-secreted exosomes promoted BC cell growth, whereas the application of M2-Exo-siRNA H19 inhibited BC progression [109]. These results indicate that exosomal siRNAs targeting exosomal lncRNAs may offer potential value in cancer therapy.
4. Conclusions
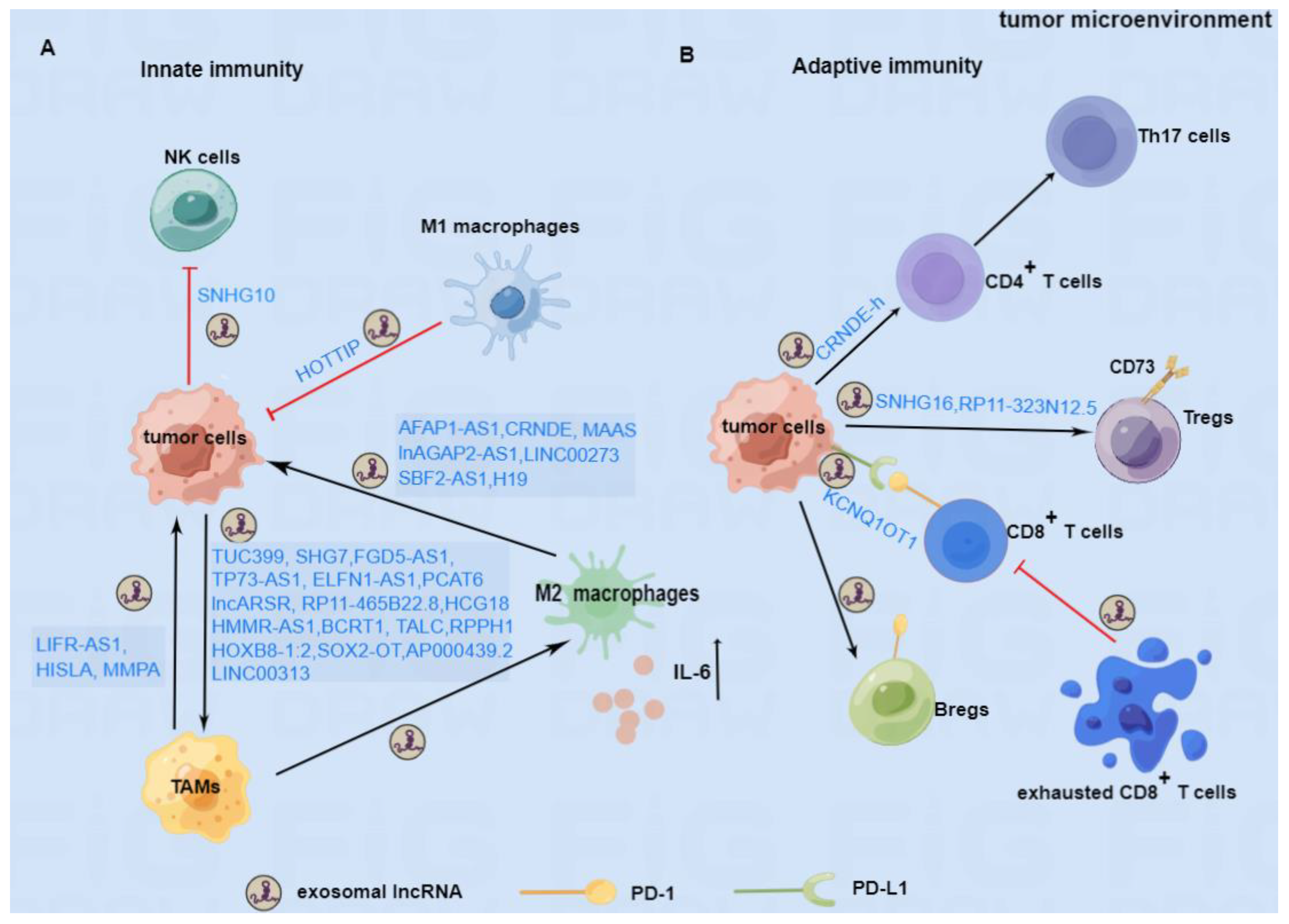
In this review, we summarized the emerging studies about the role of exosomal lncRNAs in the immune response of cancer and proposed possible therapeutic strategies targeting exosomal lncRNAs. As shown in Figure 2, exosomal lncRNAs can regulate macrophage polarization, inhibit the function of NK cells, and promote the secretion of inflammatory factors to participate in the tumor’s innate immune responses. Furthermore, exosomal lncRNAs can inhibit the activity of CD8+ T cells and facilitate the differentiation of Th17 cells, Tregs, and Bregs to regulate the adaptive immune response in cancer. Based on the immunomodulatory mechanism of exosomal lncRNAs, blockade of immune checkpoints represented by the PD-1/PD-L1 antibodies or inhibitors to stimulate the immune response, and exosomes carrying siRNAs to interfere with exosomal lncRNAs have made promising progress in cancer therapy. However, most results are still at the stage of basic investigation or preclinical tests. To date, effectively targeting exosomal lncRNAs to achieve better outcomes in cancer remains a challenge. The research needs to be accelerated to solve the existing problems and obtain clinical benefits.
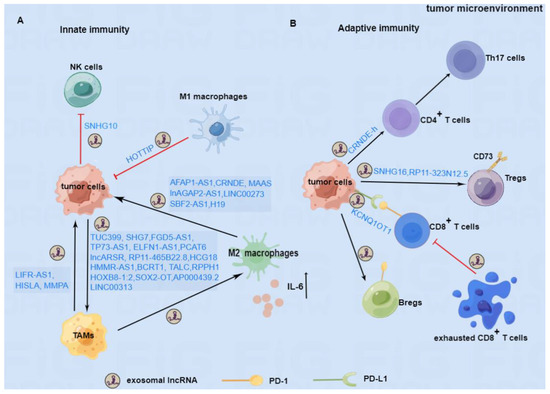
Figure 2.
The immunomodulatory role of exosomal lncRNAs in TME. (A) In innate immunity, tumor cells can secret exosomal lncRNAs to boost the polarization of M2 macrophages or inhibit the killing function of NK cells. TAMs and M2 macrophage-secreted exosomal lncRNAs can support the progression of tumor cells, whereas M1 macrophage-derived exosomal lncRNA may inhibit the progression of tumors. (B) In adaptive immunity, tumor cells can secrete exosomal lncRNAs to promote the differentiation of Th17 cells, Tregs, and Bregs and the PD-1 expression on the surface of Bregs, which are beneficial to the progression of tumors. In addition, tumor cells can also secrete exosomes containing lncRNAs to maintain the PD-L1 expression on their surface, thus restraining the biological function of CD8+T cells. Furthermore, exhausted CD8+T cells can also produce certain exosomal lncRNA to impair the ability of normal CD8+T cells. LncRNAs involved in the mechanism of immune regulation are depicted above.
Author Contributions
W.Z., K.Y. and Z.X. contributed to the conception and design of the study. J.P., K.Y. and A.T. contributed to the writing, review, and revision of the manuscript. Y.Y. and N.B. performed the administrative, technical and material support. All authors have read and agreed to the published version of the manuscript.
Funding
This study is supported by grants from the Science and Technology Innovation Program of Hunan Province (Yan, Y. 2021RC3029) and the horizontal project (Peng, J. 2021-021, 1 43010100). The Figures were created using Figdraw.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. CB 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kanada, M.; Ye, J.; Deng, Y.; He, Q.; Lei, Z.; Chen, Y.; Li, Y.; Qin, P.; Zhang, J.; et al. Exosome-mediated remodeling of the tumor microenvironment: From local to distant intercellular communication. Cancer Lett. 2022, 543, 215796. [Google Scholar] [CrossRef] [PubMed]
- Rajput, P.K.; Sharma, J.R.; Yadav, U.C.S. Cellular and molecular insights into the roles of visfatin in breast cancer cells plasticity programs. Life Sci. 2022, 304, 120706. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wasielewski, L.J.; Yang, J.C.; Cai, D.; Evans, C.P.; Murphy, W.J.; Liu, C. The Immunotherapy and Immunosuppressive Signaling in Therapy-Resistant Prostate Cancer. Biomedicines 2022, 10, 1778. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kannan, B.; Lim, B.Y.; Li, Z.; Lim, A.H.; Loh, J.W.; Ko, T.K.; Ng, C.C.; Chan, J.Y. The Multi-Dimensional Biomarker Landscape in Cancer Immunotherapy. Int. J. Mol. Sci. 2022, 23, 7839. [Google Scholar] [CrossRef]
- Glass, S.E.; Coffey, R.J. Recent Advances in the Study of Extracellular Vesicles in Colorectal Cancer. Gastroenterology 2022, 163, 1188–1197. [Google Scholar] [CrossRef]
- Li, Q. Role of exosomes in cellular communication between tumor cells and the tumor microenvironment. Oncol. Lett. 2022, 24, 240. [Google Scholar] [CrossRef]
- Peltier, D.C.; Roberts, A.; Reddy, P. LNCing RNA to immunity. Trends Immunol. 2022, 43, 478–495. [Google Scholar] [CrossRef]
- Yang, P.; Ding, J.; Bian, Y.; Ma, Z.; Wang, K.; Li, J. Long non-coding RNAs and cancer mechanisms: Immune cells and inflammatory cytokines in the tumor microenvironment. Med. Oncol. 2022, 39, 108. [Google Scholar] [CrossRef]
- Entezari, M.; Ghanbarirad, M.; Taheriazam, A.; Sadrkhanloo, M.; Zabolian, A.; Goharrizi, M.; Hushmandi, K.; Aref, A.R.; Ashrafizadeh, M.; Zarrabi, A.; et al. Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed. Pharmacother. Biomed. Pharmacother. 2022, 150, 112963. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zeng, S.; Gong, Z.; Yan, Y. Exosome-based immunotherapy: A promising approach for cancer treatment. Mol. Cancer 2020, 19, 160. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Daßler-Plenker, J.; Küttner, V.; Egeblad, M. Communication in tiny packages: Exosomes as means of tumor-stroma communication. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188340. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Sung, B.H.; Parent, C.A.; Weaver, A.M. Extracellular vesicles: Critical players during cell migration. Dev. Cell 2021, 56, 1861–1874. [Google Scholar] [CrossRef]
- Yi, Y.; Wu, M.; Zeng, H.; Hu, W.; Zhao, C.; Xiong, M.; Lv, W.; Deng, P.; Zhang, Q.; Wu, Y. Tumor-Derived Exosomal Non-Coding RNAs: The Emerging Mechanisms and Potential Clinical Applications in Breast Cancer. Front. Oncol. 2021, 11, 738945. [Google Scholar] [CrossRef]
- Turiello, R.; Capone, M.; Morretta, E.; Monti, M.C.; Madonna, G.; Azzaro, R.; Del Gaudio, P.; Simeone, E.; Sorrentino, A.; Ascierto, P.A.; et al. Exosomal CD73 from serum of patients with melanoma suppresses lymphocyte functions and is associated with therapy resistance to anti-PD-1 agents. J. Immunother. Cancer 2022, 10, e004043. [Google Scholar] [CrossRef]
- Guan, L.; Wu, B.; Li, T.; Beer, L.A.; Sharma, G.; Li, M.; Lee, C.N.; Liu, S.; Yang, C.; Huang, L.; et al. HRS phosphorylation drives immunosuppressive exosome secretion and restricts CD8(+) T-cell infiltration into tumors. Nat. Commun. 2022, 13, 4078. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed]
- Ning, T.; Li, J.; He, Y.; Zhang, H.; Wang, X.; Deng, T.; Liu, R.; Li, H.; Bai, M.; Fan, Q.; et al. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 2723–2736. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- McDonel, P.; Guttman, M. Approaches for Understanding the Mechanisms of Long Noncoding RNA Regulation of Gene Expression. Cold Spring Harb. Perspect. Biol. 2019, 11, a032151. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Ning, S.; Zhang, J.; Wang, P.; Zhi, H.; Wang, J.; Liu, Y.; Gao, Y.; Guo, M.; Yue, M.; Wang, L.; et al. Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016, 44, D980–D985. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, L.; Wang, X.; Chen, J. lncKRT16P6 promotes tongue squamous cell carcinoma progression by sponging miR-3180 and regulating GATAD2A expression. Int. J. Oncol. 2022, 61, 111. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zhang, B.W.; Zhang, Z.B.; Deng, Q.J. LncRNA MALAT1 knockdown inhibits cell migration and invasion by suppressing autophagy through miR-384/GOLM1 axis in glioma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2601–2615. [Google Scholar] [CrossRef]
- Liu, X.; Chen, J.; Zhang, S.; Liu, X.; Long, X.; Lan, J.; Zhou, M.; Zheng, L.; Zhou, J. LINC00839 promotes colorectal cancer progression by recruiting RUVBL1/Tip60 complexes to activate NRF1. EMBO Rep. 2022, 23, e54128. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.P.; Yalikong, A.; Zhang, J.W.; Cai, S.L.; Li, B.; Di, S.; Lv, Z.T.; Xu, E.P.; Zhong, Y.S.; Zhou, P.H. HDAC2 promotes the EMT of colorectal cancer cells and via the modular scaffold function of ENSG00000274093.1. J. Cell. Mol. Med. 2021, 25, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.T.; Ni, Y.R.; Zeng, F.J. The roles of long noncoding RNAs in the regulation of OCT4 expression. Stem Cell Res. Ther. 2022, 13, 383. [Google Scholar] [CrossRef] [PubMed]
- Léveillé, N.; Melo, C.A.; Rooijers, K.; Díaz-Lagares, A.; Melo, S.A.; Korkmaz, G.; Lopes, R.; Moqadam, F.A.; Maia, A.R.; Wijchers, P.J.; et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015, 6, 6520. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Fang, P.; Xiang, L.; Chen, W.; Li, S.; Huang, S.; Li, J.; Zhuge, L.; Jin, L.; Feng, W.; Chen, Y.; et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019, 25, 99–109. [Google Scholar] [CrossRef]
- Xu, Y.J.; Zhao, J.M.; Ni, X.F.; Wang, W.; Hu, W.W.; Wu, C.P. LncRNA HCG18 suppresses CD8(+) T cells to confer resistance to cetuximab in colorectal cancer via miR-20b-5p/PD-L1 axis. Epigenomics 2021, 13, 1281–1297. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, M.; He, X.; Cao, Y.; Liu, P.; Li, F.; Zou, S.; Wen, C.; Zhan, Q.; Xu, Z.; et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2022, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Gok Yavuz, B.; Gunaydin, G.; Gedik, M.E.; Kosemehmetoglu, K.; Karakoc, D.; Ozgur, F.; Guc, D. Cancer associated fibroblasts sculpt tumour microenvironment by recruiting monocytes and inducing immunosuppressive PD-1(+) TAMs. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Liu, D.; Liu, S.; Li, D.; Horecny, I.; Zhang, X.; Li, P.; Chen, L.; Miller, M.; Chowdhury, R.; et al. SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis. Sci. Rep. 2022, 12, 8579. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Yang, L.; Yu, C.; Zhu, W.; Zhou, X.; Xiong, Y.; Wang, W.; Ji, F.; He, D.; Cao, X. Tumor-Secreted Exosomal lncRNA POU3F3 Promotes Cisplatin Resistance in ESCC by Inducing Fibroblast Differentiation into CAFs. Mol. Ther. Oncolytics 2020, 18, 1–13. [Google Scholar] [CrossRef]
- Ding, L.; Ren, J.; Zhang, D.; Li, Y.; Huang, X.; Hu, Q.; Wang, H.; Song, Y.; Ni, Y.; Hou, Y. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis 2018, 39, 397–406. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155–165. [Google Scholar] [CrossRef]
- Wen, Y.; Zhu, Y.; Zhang, C.; Yang, X.; Gao, Y.; Li, M.; Yang, H.; Liu, T.; Tang, H. Chronic inflammation, cancer development and immunotherapy. Front. Pharmacol. 2022, 13, 1040163. [Google Scholar] [CrossRef]
- Zhou, H.; Wan, H.; Feng, Y.; Zhu, L.; Mi, Y. The diagnostic role and mechanistic functions of exosomal lncRNAs in prostate cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2022. [Google Scholar] [CrossRef]
- Jia, Z.; Jia, J.; Yao, L.; Li, Z. Crosstalk of Exosomal Non-Coding RNAs in The Tumor Microenvironment: Novel Frontiers. Front. Immunol. 2022, 13, 900155. [Google Scholar] [CrossRef]
- Garg, A.D.; Agostinis, P. Cell death and immunity in cancer: From danger signals to mimicry of pathogen defense responses. Immunol. Rev. 2017, 280, 126–148. [Google Scholar] [CrossRef]
- Hewitt, R.J.; Lloyd, C.M. Regulation of immune responses by the airway epithelial cell landscape. Nat. Rev. Immunol. 2021, 21, 347–362. [Google Scholar] [CrossRef]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Masopust, D.; Schenkel, J.M. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 2013, 13, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.N.; Mackay, L.K. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 2016, 16, 79–89. [Google Scholar] [CrossRef]
- Dijkgraaf, F.E.; Matos, T.R.; Hoogenboezem, M.; Toebes, M.; Vredevoogd, D.W.; Mertz, M.; van den Broek, B.; Song, J.Y.; Teunissen, M.B.M.; Luiten, R.M.; et al. Tissue patrol by resident memory CD8(+) T cells in human skin. Nat. Immunol. 2019, 20, 756–764. [Google Scholar] [CrossRef]
- Belgiovine, C.; D’Incalci, M.; Allavena, P.; Frapolli, R. Tumor-associated macrophages and anti-tumor therapies: Complex links. Cell. Mol. Life Sci. CMLS 2016, 73, 2411–2424. [Google Scholar] [CrossRef]
- Wang, J.; Mi, S.; Ding, M.; Li, X.; Yuan, S. Metabolism and polarization regulation of macrophages in the tumor microenvironment. Cancer Lett. 2022, 543, 215766. [Google Scholar] [CrossRef]
- Schweer, D.; McAtee, A.; Neupane, K.; Richards, C.; Ueland, F.; Kolesar, J. Tumor-Associated Macrophages and Ovarian Cancer: Implications for Therapy. Cancers 2022, 14, 2220. [Google Scholar] [CrossRef]
- Konishi, H.; Koizumi, S.; Kiyama, H. Phagocytic astrocytes: Emerging from the shadows of microglia. Glia 2022, 70, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Liu, Y.; Wu, S.Y.; Wu, K.; Sharma, S.; Mo, Y.Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018, 78, 4316–4330. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Meng, X.; Wu, P.; Zha, C.; Han, B.; Li, L.; Sun, N.; Qi, T.; Qin, J.; Zhang, Y.; et al. Glioblastoma Cell-Derived lncRNA-Containing Exosomes Induce Microglia to Produce Complement C5, Promoting Chemotherapy Resistance. Cancer Immunol. Res. 2021, 9, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Zhi, J.; Jia, X.J.; Yan, J.; Wang, H.C.; Feng, B.; Xing, H.Y.; Jia, Y.T. BRAF(V600E) mutant colorectal cancer cells mediate local immunosuppressive microenvironment through exosomal long noncoding RNAs. World J. Gastrointest. Oncol. 2021, 13, 2129–2148. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.X.; Liu, H.S.; Wang, F.W.; Xiong, L.; Zhou, C.; Hu, T.; He, X.W.; Wu, X.J.; Xie, D.; Wu, X.R.; et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lan, Q.; Lai, W.; Wu, H.; Xu, H.; Fang, K.; Chu, Z.; Zeng, Y. Exosome-derived lnc-HOXB8-1:2 induces tumor-associated macrophage infiltration to promote neuroendocrine differentiated colorectal cancer progression by sponging hsa-miR-6825-5p. BMC Cancer 2022, 22, 928. [Google Scholar] [CrossRef]
- Hu, R.; Bi, R.; Jiang, L.; Xiao, H.; Xie, X.; Liu, H.; Hu, F. LncRNA RP11-465B22.8 triggers esophageal cancer progression by targeting miR-765/KLK4 axis. Cell Death Discov. 2021, 7, 262. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Wu, Y.; Liu, C.; Zeng, F.; Wang, J.L.; Wu, D.Z.; Wu, J.P.; Yue, Z.Q.; Gan, J.H.; Lu, H.; et al. Exosome-mediated transfer of lncRNA HCG18 promotes M2 macrophage polarization in gastric cancer. Mol. Immunol. 2021, 140, 196–205. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Y.; Dong, K.; Zhang, H.; Gong, J.; Wang, S. Exosomal lncRNA HMMR-AS1 mediates macrophage polarization through miR-147a/ARID3A axis under hypoxia and affects the progression of hepatocellular carcinoma. Environ. Toxicol. 2022, 37, 1357–1372. [Google Scholar] [CrossRef]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, J.; Li, C.; Yuan, Y.; Fang, S.; Liu, W.; Qian, Y.; Ma, J.; Chang, L.; Chen, F.; et al. Exosome-mediated transfer of SNHG7 enhances docetaxel resistance in lung adenocarcinoma. Cancer Lett. 2022, 526, 142–154. [Google Scholar] [CrossRef]
- Zhou, D.; Xia, Z.; Xie, M.; Gao, Y.; Yu, Q.; He, B. Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975. Hum. Cell 2021, 34, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hong, C.; Qu, J.; Chen, J.; Qin, Z. Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered 2022, 13, 12834–12846. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, L.; Chen, Y.; Yu, Z.; Zhao, Z. Cancer cell-derived exosomal LINC00313 induces M2 macrophage differentiation in non-small cell lung cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2022, 24, 2395–2408. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Li, Q.; Ma, R.; Wang, Z.; Yu, Y.; Liu, H.; Miao, Y.; Jiang, S. Long Noncoding RNA FGD5-AS1 Knockdown Decrease Viability, Migration, and Invasion of Non-Small Cell Lung Cancer (NSCLC) Cells by Regulating the MicroRNA-944/MACC1 Axis. Technol. Cancer Res. Treat. 2021, 20, 1533033821990090. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Tian, L.; Yan, B.; Yang, L.; Li, Y. LncRNA TP73-AS1 promotes nasopharyngeal carcinoma progression through targeting miR-342-3p and M2 polarization via exosomes. Cancer Cell Int. 2022, 22, 16. [Google Scholar] [CrossRef]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.; Li, P.; Niu, X.; Liang, X.; Liu, G.; Liu, Z.; Ge, H. Osteosarcoma Cell-Derived Exosomal ELFN1-AS1 Mediates Macrophage M2 Polarization via Sponging miR-138-5p and miR-1291 to Promote the Tumorgenesis of Osteosarcoma. Front. Oncol. 2022, 12, 881022. [Google Scholar] [CrossRef]
- He, Z.; Wang, J.; Zhu, C.; Xu, J.; Chen, P.; Jiang, X.; Chen, Y.; Jiang, J.; Sun, C. Exosome-derived FGD5-AS1 promotes tumor-associated macrophage M2 polarization-mediated pancreatic cancer cell proliferation and metastasis. Cancer Lett. 2022, 548, 215751. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, X.; Yu, Y.; Zheng, L.; Lan, J.; Wu, Y.; Liu, H.; Zhao, A.; Huang, H.; Chen, W. Renal cell carcinoma-derived exosomes deliver lncARSR to induce macrophage polarization and promote tumor progression via STAT3 pathway. Int. J. Biol. Sci. 2022, 18, 3209–3222. [Google Scholar] [CrossRef]
- Shen, T.; Miao, S.; Zhou, Y.; Yi, X.; Xue, S.; Du, B.; Tang, C.; Qu, L.; Fu, D.; Jia, R.; et al. Exosomal AP000439.2 from clear cell renal cell carcinoma induces M2 macrophage polarization to promote tumor progression through activation of STAT3. Cell Commun. Signal. CCS 2022, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.R.; Felices, M.; Miller, J.S. Challenges to the broad application of allogeneic natural killer cell immunotherapy of cancer. Stem Cell Res. Ther. 2022, 13, 165. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.F.; Gu, Z.S.; Zheng, L.L.; Zhao, M.X.; Wang, X.J. Long non-coding RNA GAS5 promotes natural killer cell cytotoxicity against gastric cancer by regulating miR-18a. Neoplasma 2020, 67, 1085–1093. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, Y.; Ou, W.; Wang, Y.; Dong, D.; Peng, X.; Luo, Y. Exosomal lncRNA SNHG10 derived from colorectal cancer cells suppresses natural killer cell cytotoxicity by upregulating INHBC. Cancer Cell Int. 2021, 21, 528. [Google Scholar] [CrossRef]
- Brummelman, J.; Pilipow, K.; Lugli, E. The Single-Cell Phenotypic Identity of Human CD8(+) and CD4(+) T Cells. Int. Rev. Cell Mol. Biol. 2018, 341, 63–124. [Google Scholar] [CrossRef] [PubMed]
- Sungnak, W.; Wang, C.; Kuchroo, V.K. Multilayer regulation of CD4 T cell subset differentiation in the era of single cell genomics. Adv. Immunol. 2019, 141, 1–31. [Google Scholar] [CrossRef]
- Bilska, M.; Pawłowska, A.; Zakrzewska, E.; Chudzik, A.; Suszczyk, D.; Gogacz, M.; Wertel, I. Th17 Cells and IL-17 As Novel Immune Targets in Ovarian Cancer Therapy. J. Oncol. 2020, 2020, 8797683. [Google Scholar] [CrossRef]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 123. [Google Scholar] [CrossRef]
- Xian, D.; Niu, L.; Zeng, J.; Wang, L. LncRNA KCNQ1OT1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating PD-L1 Ubiquitination via MiR-30a-5p/USP22. Front. Cell Dev. Biol. 2021, 9, 653808. [Google Scholar] [CrossRef]
- Goswami, T.K.; Singh, M.; Dhawan, M.; Mitra, S.; Emran, T.B.; Rabaan, A.A.; Mutair, A.A.; Alawi, Z.A.; Alhumaid, S.; Dhama, K. Regulatory T cells (Tregs) and their therapeutic potential against autoimmune disorders—Advances and challenges. Hum. Vaccines Immunother. 2022, 18, 2035117. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, F.; Shi, Y.; Zhang, Q.; Xu, S.; Yao, Y.; Jiang, R. RP11-323N12.5 promotes the malignancy and immunosuppression of human gastric cancer by increasing YAP1 transcription. Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2021, 24, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Flores-Borja, F.; Blair, P. Mechanisms of induction of regulatory B cells in the tumour microenvironment and their contribution to immunosuppression and pro-tumour responses. Clin. Exp. Immunol. 2022, 209, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, L.; Wei, W. Regulatory B cells in inflammatory diseases and tumor. Int. Immunopharmacol. 2019, 67, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Leitner, W.W.; Golding, B.; Scott, D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006, 66, 7741–7747. [Google Scholar] [CrossRef]
- Liu, R.; Lu, Z.; Gu, J.; Liu, J.; Huang, E.; Liu, X.; Wang, L.; Yang, J.; Deng, Y.; Qian, J.; et al. MicroRNAs 15A and 16-1 Activate Signaling Pathways That Mediate Chemotaxis of Immune Regulatory B cells to Colorectal Tumors. Gastroenterology 2018, 154, 637–651.e637. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, Y.; Dong, L.; Zhang, Q.; Wang, C.; Zhang, Y.; Li, X.; Fu, Z. Circulating exosomes from esophageal squamous cell carcinoma mediate the generation of B10 and PD-1(high) Breg cells. Cancer Sci. 2019, 110, 2700–2710. [Google Scholar] [CrossRef]
- Ruffell, B.; Affara, N.I.; Coussens, L.M. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012, 33, 119–126. [Google Scholar] [CrossRef]
- Chen, F.; Chen, J.; Yang, L.; Liu, J.; Zhang, X.; Zhang, Y.; Tu, Q.; Yin, D.; Lin, D.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510. [Google Scholar] [CrossRef]
- Xu, M.; Zhou, C.; Weng, J.; Chen, Z.; Zhou, Q.; Gao, J.; Shi, G.; Ke, A.; Ren, N.; Sun, H.; et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J. Exp. Clin. Cancer Res. CR 2022, 41, 253. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Y.; Wang, J.; Han, Y.; Ren, T.; Huang, Y.; Chen, C.; Huang, Q.; Wang, W.; Niu, J.; et al. Macrophages-derived exosomal lncRNA LIFR-AS1 promotes osteosarcoma cell progression via miR-29a/NFIA axis. Cancer Cell Int. 2021, 21, 192. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Wei, H.; Wu, S.; Tang, Z.; Li, X.; Zou, C. Exosomal LncRNA LBX1-AS1 Derived From RBPJ Overexpressed-Macrophages Inhibits Oral Squamous Cell Carcinoma Progress via miR-182-5p/FOXO3. Front. Oncol. 2021, 11, 605884. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Xu, R.; Hong, S.; Xu, T.; Zhang, W.; Liu, M. M2 Macrophage-Derived Exosomal lncRNA AFAP1-AS1 and MicroRNA-26a Affect Cell Migration and Metastasis in Esophageal Cancer. Mol. Therapy Nucleic Acids 2020, 22, 779–790. [Google Scholar] [CrossRef]
- Tao, L.; Li, D.; Mu, S.; Tian, G.; Yan, G. LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab. Investig. 2022, 102, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhou, L.Q.; Liu, C.; Zeng, F.; Yuan, Y.W.; Zhou, Q.; Li, S.H.; Wu, Y.; Wang, J.L.; Wu, D.Z.; et al. Transfer of LncRNA CRNDE in TAM-derived exosomes is linked with cisplatin resistance in gastric cancer. EMBO Rep. 2021, 22, e52124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sang, Y.; Chen, D.; Wu, X.; Wang, X.; Yang, W.; Chen, Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021, 12, 467. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, K.; Zhi, Y.; Wu, Y.; Chen, B.; Bai, J.; Wang, X. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin. Transl. Med. 2021, 11, e478. [Google Scholar] [CrossRef]
- Yin, Z.; Zhou, Y.; Ma, T.; Chen, S.; Shi, N.; Zou, Y.; Hou, B.; Zhang, C. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J. Cell. Mol. Med. 2020, 24, 5028–5038. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, W.; Gao, W.; Li, L.; Liang, Y.; Mei, Z.; Liu, B.; Wang, R. Long Noncoding RNA H19 Derived from M2 Tumor-Associated Macrophages Promotes Bladder Cell Autophagy via Stabilizing ULK1. J. Oncol. 2022, 2022, 3465459. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, L.; Shen, N.; Ning, X.; Wu, D.; Jiang, K.; Huang, X. M1 macrophage-derived exosomes and their key molecule lncRNA HOTTIP suppress head and neck squamous cell carcinoma progression by upregulating the TLR5/NF-kappaB pathway. Cell Death Dis. 2022, 13, 183. [Google Scholar] [CrossRef]
- Wang, X.; Shen, H.; He, Q.; Tian, W.; Xia, A.; Lu, X.J. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J. Med. Genet. 2019, 56, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Cheng, J.; Zhang, L.Y.; Zhang, J.G. Self-assembling peptides-based nano-cargos for targeted chemotherapy and immunotherapy of tumors: Recent developments, challenges, and future perspectives. Drug Deliv. 2022, 29, 1184–1200. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chan, T.A. Solving the puzzle of what makes immunotherapies work. Trends Cancer 2022, 8, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Lasvergnas, J.; Naigeon, M.; Chouahnia, K.; Zelek, L.; Chaput, N.; Duchemann, B. Adoptive cell therapies in thoracic malignancies. Cancer Immunol. Immunother. CII 2022, 71, 2077–2098. [Google Scholar] [CrossRef]
- Luginbuehl, V.; Abraham, E.; Kovar, K.; Flaaten, R.; Müller, A.M.S. Better by design: What to expect from novel CAR-engineered cell therapies? Biotechnol. Adv. 2022, 58, 107917. [Google Scholar] [CrossRef]
- Filippone, A.; Lanza, M.; Mannino, D.; Raciti, G.; Colarossi, C.; Sciacca, D.; Cuzzocrea, S.; Paterniti, I. PD1/PD-L1 immune checkpoint as a potential target for preventing brain tumor progression. Cancer Immunol. Immunother. CII 2022, 71, 2067–2075. [Google Scholar] [CrossRef]
- Yuan, B.; Wang, G.; Tang, X.; Tong, A.; Zhou, L. Immunotherapy of glioblastoma: Recent advances and future prospects. Hum. Vaccines Immunother. 2022, 18, 2055417. [Google Scholar] [CrossRef]
- Fife, B.T.; Bluestone, J.A. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol. Rev. 2008, 224, 166–182. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Koukourakis, I.M.; Balaska, K.; Mitrakas, A.G.; Harris, A.L.; Koukourakis, M.I. Programmed death-1 receptor (PD-1) and PD-ligand-1 (PD-L1) expression in non-small cell lung cancer and the immune-suppressive effect of anaerobic glycolysis. Med. Oncol. 2019, 36, 76. [Google Scholar] [CrossRef]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, J.; Ma, Y.; Li, J.; Sun, X.; Zhao, X.; Shi, X.; Hu, Y.; Qu, F.; Zhang, X. Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J. Exp. Clin. Cancer Res. 2021, 40, 306. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Z.; Peng, E.; Peng, J. Role of an Exosomes-Related lncRNAs Signature in Tumor Immune Microenvironment of Gastric Cancer. Front. Cell Dev. Biol. 2022, 10, 873319. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010, 20, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhao, W.; Guo, D.; Li, Z. LncRNA E2F-Mediated Cell Proliferation Enhancing lncRNA Regulates Cancer Cell Behaviors and Affects Prognosis of Gastric Cancer. Dig. Dis. Sci. 2020, 65, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, G.; Yu, T.; He, J.; Liu, J.; Chai, X.; Zhao, G.; Yin, D.; Zhang, C. Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis. Cancer Lett. 2022, 543, 215781. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).