Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis
Abstract
Simple Summary
Abstract
1. Background
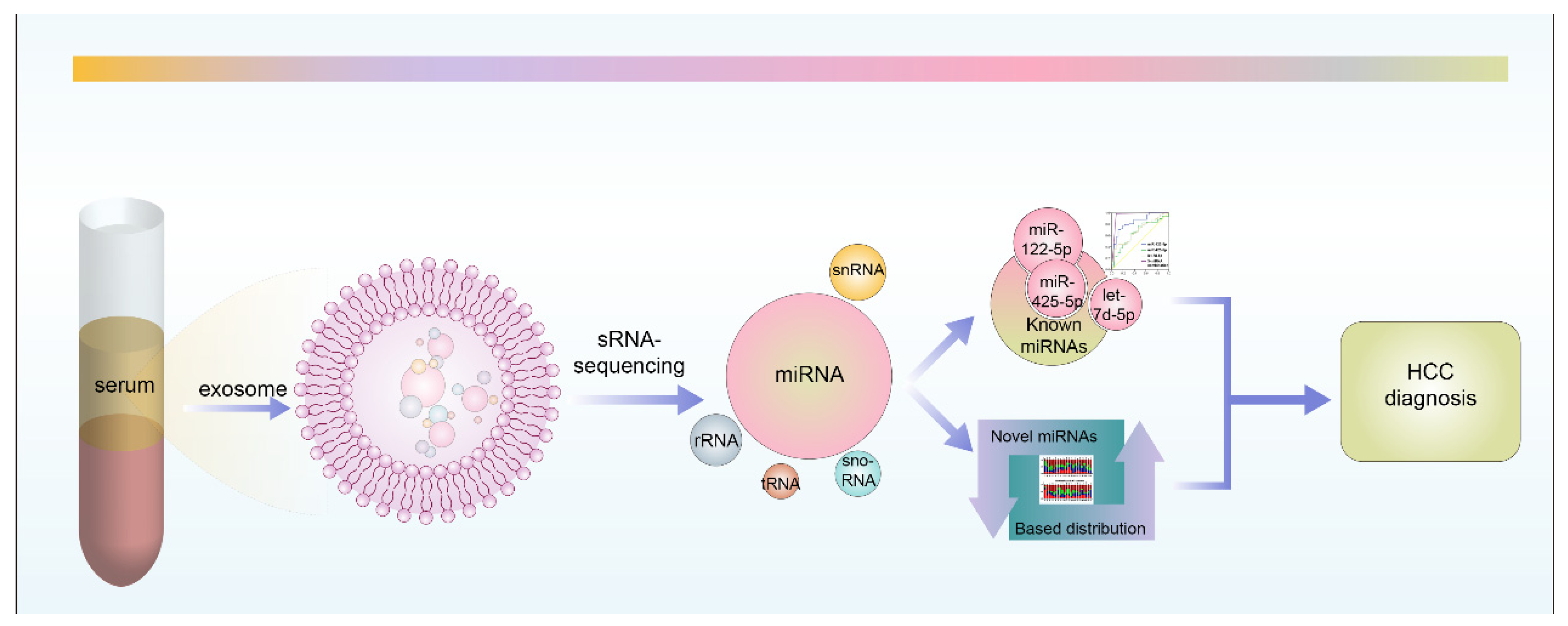
2. Materials and Methods
2.1. Patients and Specimens
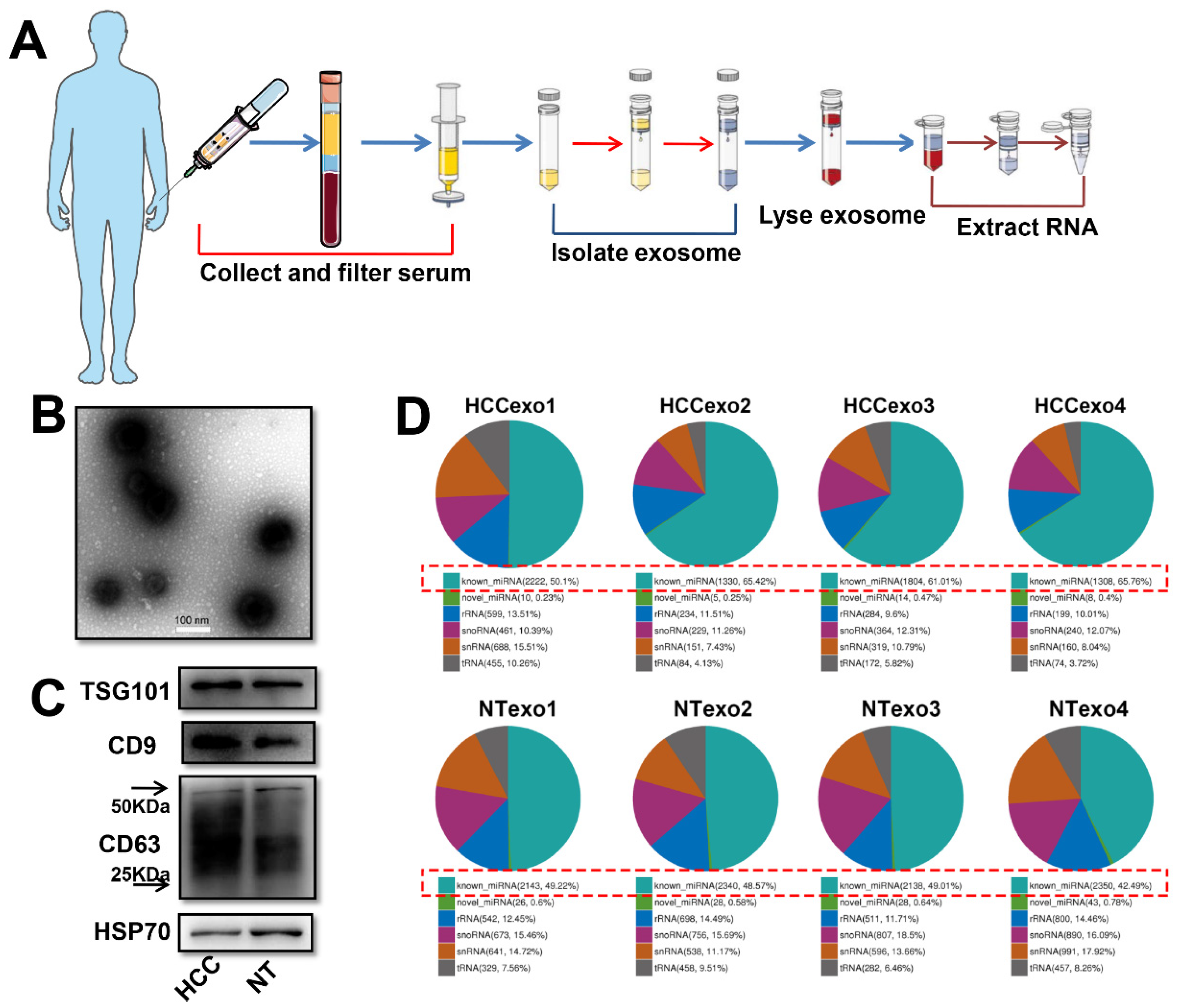
2.2. Serum Exosome Purification and Exosome RNA Extraction
2.3. sRNA-Sequencing Analysis
2.4. Exosome Identification
2.5. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.6. Western Blot Analysis
2.7. Statistical Analysis
3. Results
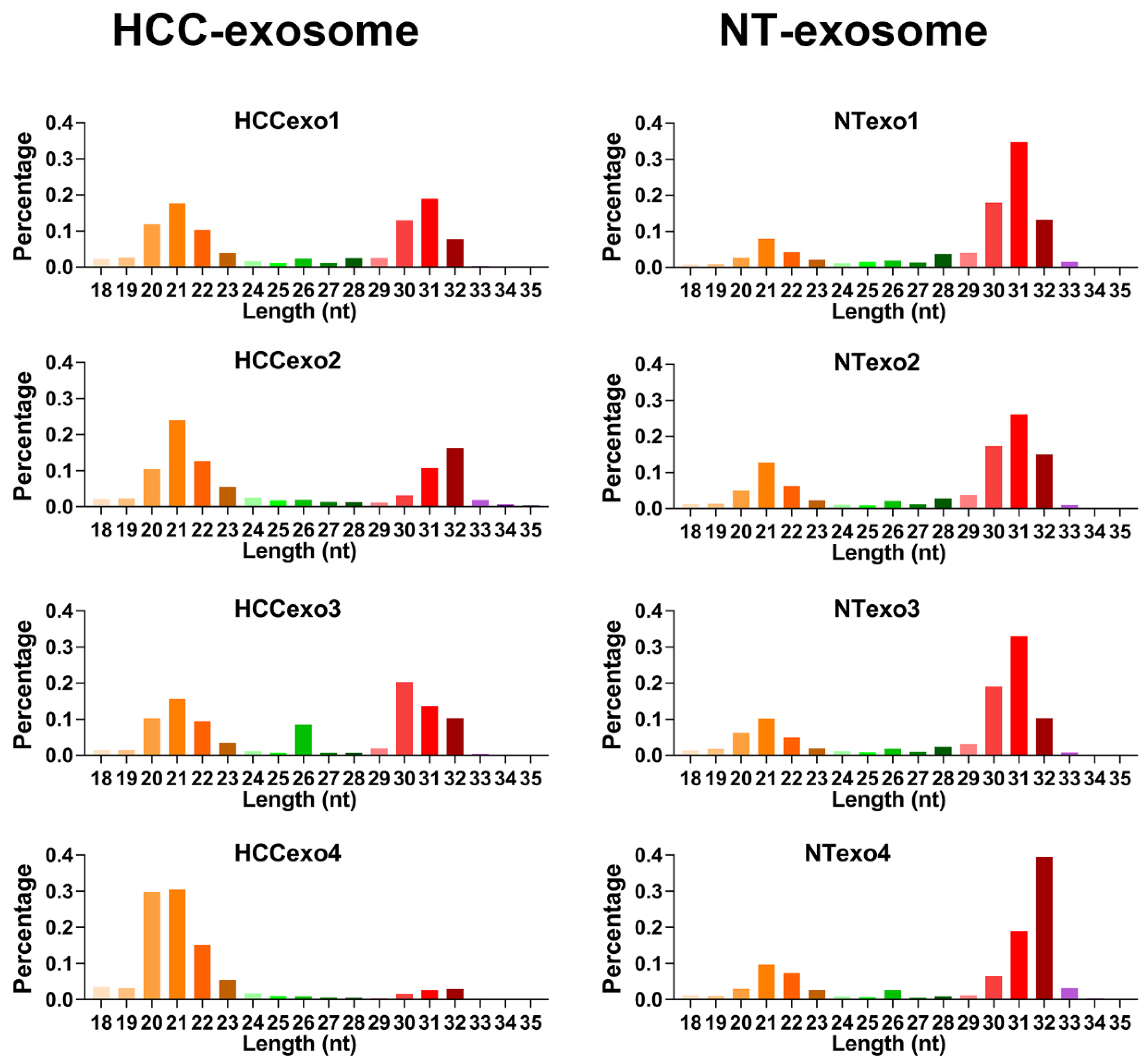
3.1. Profiling and Analyzing of Exosome-Derived Small RNAs in the Serum of HCC Patients
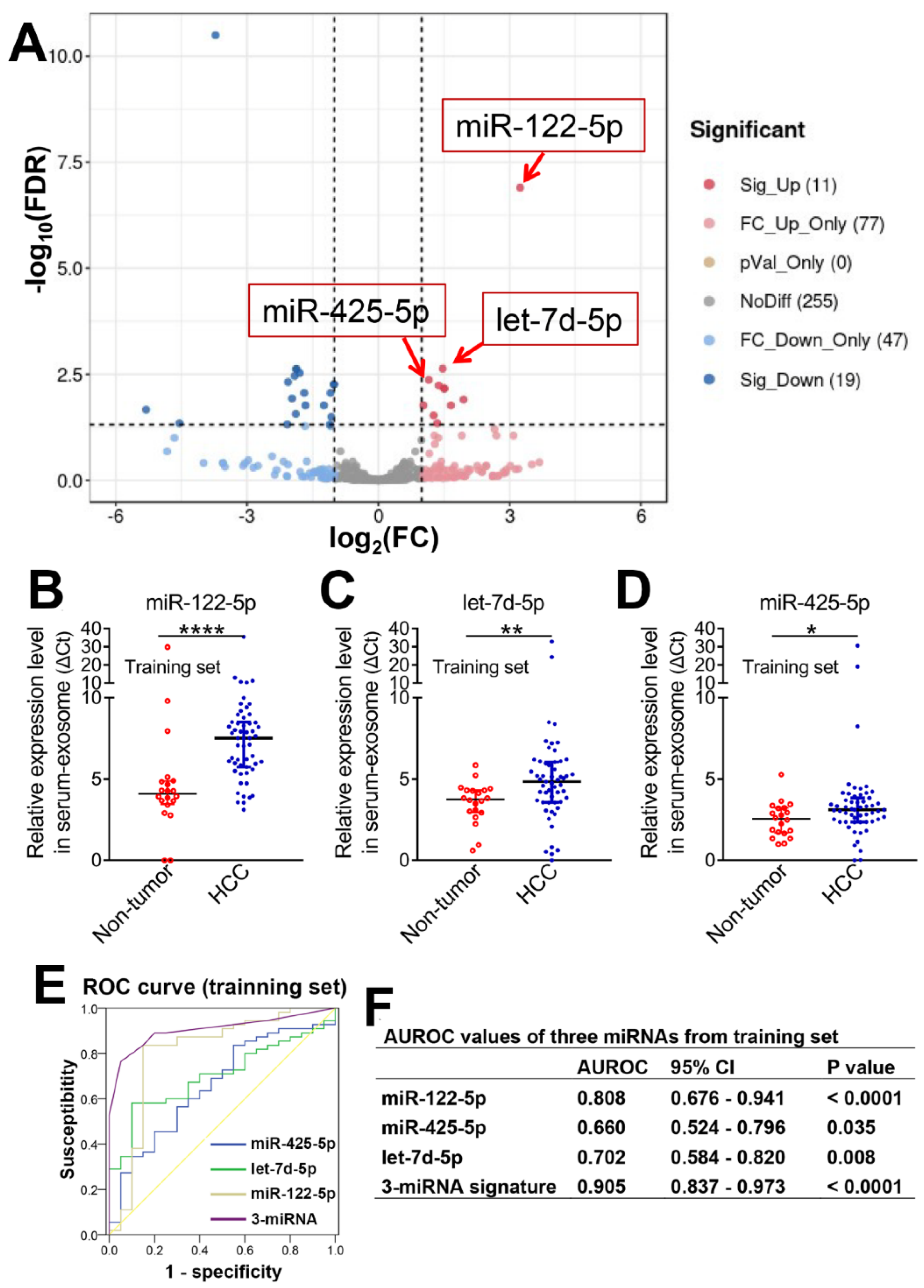
3.2. Three Serum-Exosome-Derived miRNAs Acted as Accurate Biomarkers for the Diagnosis of HCC
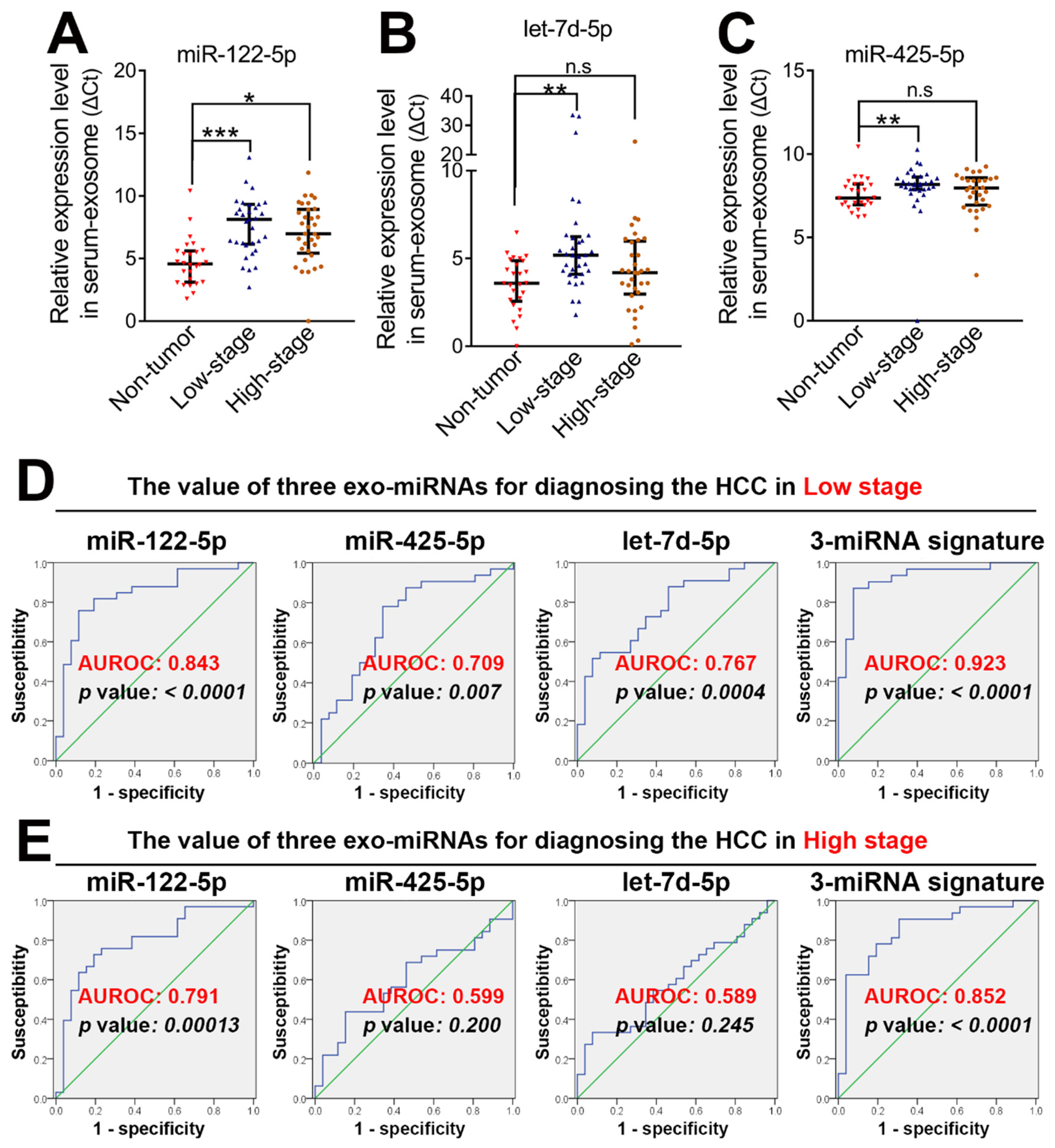
3.3. Three Serum-Exosome-Derived miRNAs Were Preferred for the Early Diagnosis of HCC
3.4. Novel HCC Serum-exosome-derived miRNAs’ Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Gupta, S.; Bent, S.; Kohlwes, J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann. Intern. Med. 2003, 139, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Wei, W.; Ma, G. Exosomes: The Indispensable Messenger in Tumor Pathogenesis and the Rising Star in Antitumor Applications. Adv. Biosyst. 2019, 3, e1900008. [Google Scholar] [CrossRef] [PubMed]
- Słomka, A.; Kornek, M.; Cho, W.C. Small Extracellular Vesicles and Their Involvement in Cancer Resistance: An Up-to-Date Review. Cells 2022, 11, 2913. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Fu, Y.; Liu, G.; Shu, B.; Davis, J.; Tofaris, G.K. Multiplexed Profiling of Extracellular Vesicles for Biomarker Development. Nano-Micro Lett. 2021, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2018, 19, 11–34. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, H.T.; Wang, S.; Huang, S.L.; Zheng, Y.; Wang, C.Q.; Hu, B.Y.; Qin, W.; Zou, T.T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152. [Google Scholar] [CrossRef]
- Fan, Z.; Yu, J.; Lin, J.; Liu, Y.; Liao, Y. Exosome-specific tumor diagnosis via biomedical analysis of exosome-containing microRNA biomarkers. Analyst 2019, 144, 5856–5865. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef]
- Rayamajhi, S.; Nguyen, T.; Marasini, R.; Aryal, S. Macrophage-derived exosome-mimetic hybrid vesicles for tumor targeted drug delivery. Acta Biomater. 2019, 94, 482–494. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef]
- Ohno, S.; Drummen, G.P.; Kuroda, M. Focus on Extracellular Vesicles: Development of Extracellular Vesicle-Based Therapeutic Systems. Int. J. Mol. Sci. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Shen, J.; Wang, R.; Wang, L.; Dai, Y.; Zhang, Y.; Huang, X. Role of exosomal small RNA in prostate cancer metastasis. Cancer Manag. Res. 2018, 10, 4029–4038. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Zhao, J.; Li, M.; Liu, X.; Xu, Y.; Li, W.; Wu, S.; Su, Z. Exosomal miRNAs are potential diagnostic biomarkers between malignant and benign thyroid nodules based on next-generation sequencing. Carcinogenesis 2020, 41, 18–24. [Google Scholar] [CrossRef]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef]
- Chun, Y.S.; Pawlik, T.M.; Vauthey, J.N. 8th Edition of the AJCC Cancer Staging Manual: Pancreas and Hepatobiliary Cancers. Ann. Surg. Oncol. 2018, 25, 845–847. [Google Scholar] [CrossRef]
- Rui, T.; Zhang, X.; Feng, S.; Huang, H.; Zhan, S.; Xie, H.; Zhou, L.; Zheng, S.; Ling, Q. MiR-516a-3p is a Novel Mediator of Hepatocellular Carcinoma Oncogenic Activity and Cellular Metabolism. Engineering 2021, 16, 162–175. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Rui, T.; Zhang, X.; Feng, S.; Huang, H.; Zhan, S.; Xie, H.; Zhou, L.; Ling, Q.; Zheng, S. The Similar Effects of miR-512-3p and miR-519a-2-5p on the Promotion of Hepatocellular Carcinoma: Different Tunes Sung with Equal Skill. Front. Oncol. 2020, 10, 1244. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Matsui, T.; Nishikawa, K.; Yukimoto, H.; Katsuta, K.; Nakamura, Y.; Tanaka, S.; Oiwa, M.; Nakahashi, H.; Shomi, Y.; Haruki, Y.; et al. Needle tract seeding following endoscopic ultrasound-guided fine-needle aspiration for pancreatic cancer: A report of two cases. World J. Surg. Oncol. 2019, 17, 134. [Google Scholar] [CrossRef]
- Durand, F.; Belghiti, J.; Paradis, V. Liver transplantation for hepatocellular carcinoma: Role of biopsy. Liver Transpl. 2007, 13, S17–S23. [Google Scholar] [CrossRef]
- Ahmadi, S.A.; Moinfar, M.; Gohari Moghaddam, K.; Bahadori, M. Practical application of angiogenesis and vasculogenic mimicry in prostatic adenocarcinoma. Arch. Iran. Med. 2010, 13, 498–503. [Google Scholar]
- Maspes, A.; Pizzetti, F.; Rossetti, A.; Makvandi, P.; Sitia, G.; Rossi, F. Advances in Bio-Based Polymers for Colorectal CancerTreatment: Hydrogels and Nanoplatforms. Gels 2021, 7, 6. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Wang, S.; Fletcher, S.; Niu, D.; Mitter, N.; Birch, P.; Jin, H. Message in a Bubble: Shuttling Small RNAs and Proteins Between Cells and Interacting Organisms Using Extracellular Vesicles. Annu. Rev. Plant Biol. 2021, 72, 497–524. [Google Scholar] [CrossRef]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef]
- Gorji-Bahri, G.; Hashemi, A.; Moghimi, H.R. ExomiRs: A Novel Strategy in Cancer Diagnosis and Therapy. Curr. Gene Ther. 2018, 18, 336–350. [Google Scholar] [CrossRef]
- Li, S.; Yi, M.; Dong, B.; Tan, X.; Luo, S.; Wu, K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int. J. Cancer 2021, 148, 2640–2651. [Google Scholar] [CrossRef]
- Jalalian, S.H.; Ramezani, M.; Jalalian, S.A.; Abnous, K.; Taghdisi, S.M. Exosomes, new biomarkers in early cancer detection. Anal. Biochem. 2019, 571, 1–13. [Google Scholar] [CrossRef]
- Sorop, A.; Constantinescu, D.; Cojocaru, F.; Dinischiotu, A.; Cucu, D.; Dima, S.O. Exosomal microRNAs as Biomarkers and Therapeutic Targets for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 4997. [Google Scholar] [CrossRef]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef]
- Xue, X.; Zhao, Y.; Wang, X.; Qin, L.; Hu, R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J. Cell. Biochem. 2019, 120, 135–142. [Google Scholar] [CrossRef]
- Yang, X.; Sun, L.; Wang, L.; Yao, B.; Mo, H.; Yang, W. LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed. Pharmacother. 2019, 118, 109386. [Google Scholar] [CrossRef]
- Rao, D.; Guan, S.; Huang, J.; Chang, Q.; Duan, S. miR-425-5p Acts as a Molecular Marker and Promoted Proliferation, Migration by Targeting RNF11 in Hepatocellular Carcinoma. Biomed. Res. Int. 2020, 2020, 6530973. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Shang, J.; Zhan, W.; Liu, J.; Ning, H.; Chen, N. miR-425-5p promotes cell proliferation, migration and invasion by directly targeting FOXD3 in hepatocellular carcinoma cells. Mol. Med. Rep. 2019, 20, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Ren, C.C.; Yang, L.; Nai, M.M.; Xu, Y.M.; Zhang, F.; Liu, Y. MicroRNA let-7d-5p rescues ovarian cancer cell apoptosis and restores chemosensitivity by regulating the p53 signaling pathway via HMGA1. Int. J. Oncol. 2019, 54, 1771–1784. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ju, L.; Zhai, J.; Song, Y.; Song, J.; Ma, C. miRLocator: A Python Implementation and Web Server for Predicting miRNAs from Pre-miRNA Sequences. Methods Mol. Biol. 2019, 1932, 89–97. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rui, T.; Zhang, X.; Guo, J.; Xiang, A.; Tang, N.; Liu, J.; Mao, Z. Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis. Cancers 2023, 15, 205. https://doi.org/10.3390/cancers15010205
Rui T, Zhang X, Guo J, Xiang A, Tang N, Liu J, Mao Z. Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis. Cancers. 2023; 15(1):205. https://doi.org/10.3390/cancers15010205
Chicago/Turabian StyleRui, Tao, Xiaobing Zhang, Jufeng Guo, Aizhai Xiang, Ning Tang, Jian Liu, and Zonglei Mao. 2023. "Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis" Cancers 15, no. 1: 205. https://doi.org/10.3390/cancers15010205
APA StyleRui, T., Zhang, X., Guo, J., Xiang, A., Tang, N., Liu, J., & Mao, Z. (2023). Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis. Cancers, 15(1), 205. https://doi.org/10.3390/cancers15010205




