Long Non-Coding RNAs as Epigenetic Regulators of Immune Checkpoints in Cancer Immunity
Abstract
Simple Summary
Abstract
1. Introduction
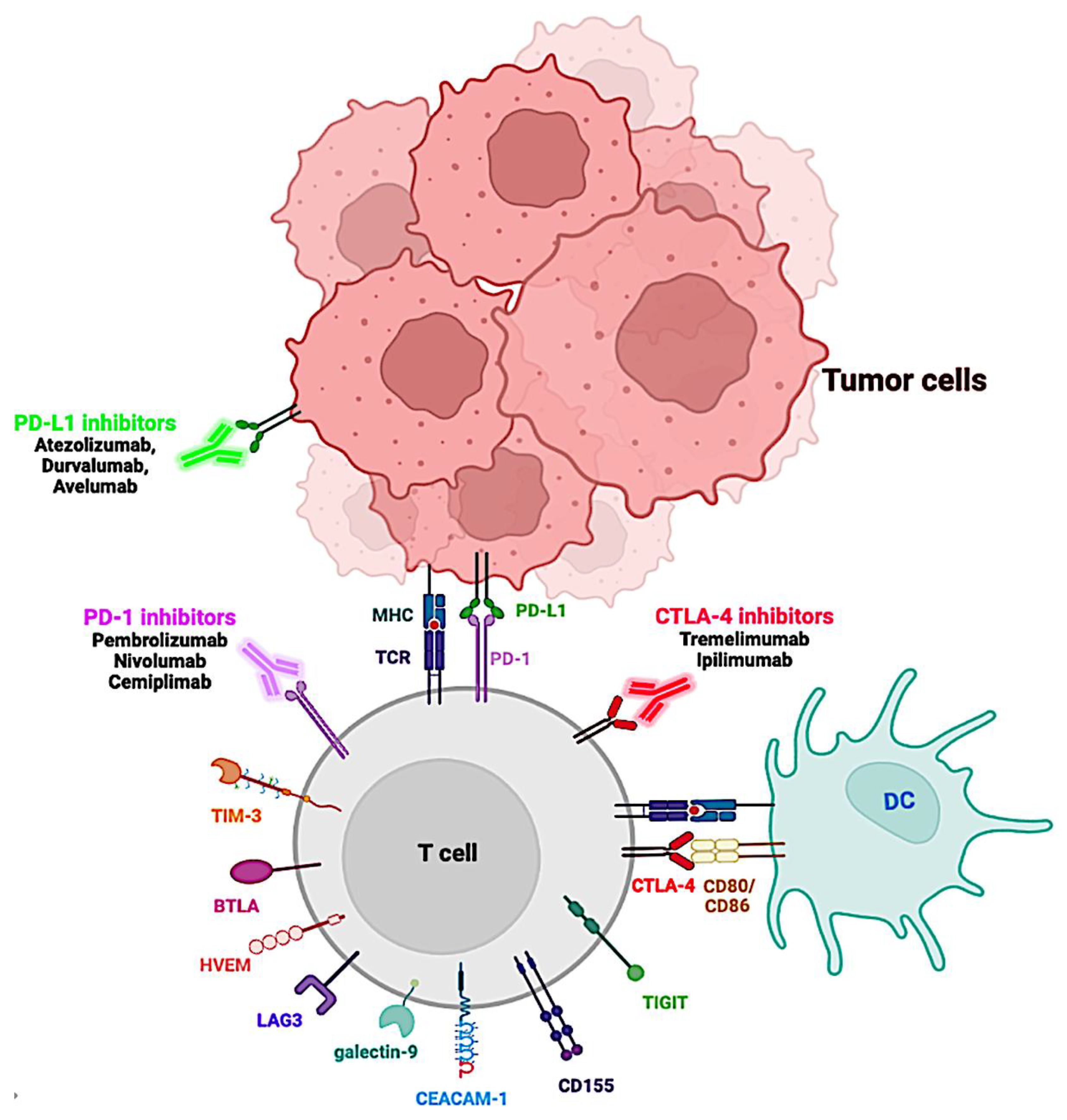
2. Immune Checkpoint Molecules and Cancer Progression
3. Long Non-Coding RNAs: An Overview
4. LncRNAs Involved in Epigenetic Regulation of Immune Checkpoints in Malignancies
5. Super-Enhancer lncRNAs
6. Circular RNAs Mediating Immune Checkpoint Regulation
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer; Lyon, France, 2020; Available online: http://gco.iarc.fr/today/home (accessed on 12 July 2022).
- Dawson, M.A.; Kouzarides, T. Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Devaskar, S.U.; Raychaudhuri, S. Epigenetics—A Science of Heritable Biological Adaptation. Pediatr. Res. 2007, 61, 1–4. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.; Tsai, H.-C.; Baylin, S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef]
- Peschansky, V.J.; Wahlestedt, C. Non-Coding RNAs as Direct and Indirect Modulators of Epigenetic Regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef]
- Ghorbaninejad, M.; Khademi-Shirvan, M.; Hosseini, S.; Baghaban Eslaminejad, M. Epidrugs: Novel Epigenetic Regulators That Open a New Window for Targeting Osteoblast Differentiation. Stem Cell Res. Ther. 2020, 11, 456. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-Coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Patnaik, S. Anupriya Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front. Pharmacol. 2019, 10, 588. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Teng, M.W.; Smyth, M.J. Cancer Immunoediting and Resistance to T Cell-Based Immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef]
- Pardoll, D.M. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Hemmings, B.A. AGC Kinases in Cancer Metastasis, Immune Checkpoint Regulation and Drug Resistance. Semin. Cancer Biol. 2018, 48, iii–iv. [Google Scholar] [CrossRef] [PubMed]
- Ohta, A. Oxygen-Dependent Regulation of Immune Checkpoint Mechanisms. Int. Immunol. 2018, 30, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Cao, W.; Wang, Z.; Zhang, B.; Liu, J. Regulation of Cancer Immune Escape: The Roles of MiRNAs in Immune Checkpoint Proteins. Cancer Lett. 2018, 431, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Robert, C. A Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus Docetaxel for Previously Treated, PD-L1-Positive, Advanced Non-Small-Cell Lung Cancer (KEYNOTE-010): A Randomised Controlled Trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Lacchetti, C.; Schneider, B.J.; Atkins, M.B.; Brassil, K.J.; Caterino, J.M.; Chau, I.; Ernstoff, M.S.; Gardner, J.M.; Ginex, P.; et al. Management of Immune-Related Adverse Events in Patients Treated With Immune Checkpoint Inhibitor Therapy: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2018, 36, 1714–1768. [Google Scholar] [CrossRef]
- Héninger, E.; Krueger, T.E.; Lang, J.M. Augmenting Antitumor Immune Responses with Epigenetic Modifying Agents. Front. Immunol. 2015, 6, 29. [Google Scholar] [CrossRef]
- Dobosz, P.; Stempor, P.A.; Ramírez Moreno, M.; Bulgakova, N.A. Transcriptional and Post-Transcriptional Regulation of Checkpoint Genes on the Tumour Side of the Immunological Synapse. Heredity 2022, 129, 64–74. [Google Scholar] [CrossRef]
- Barragan, I. Epigenetics Modulates the Complexity of the Response to Immune Checkpoint Blockade. EBioMedicine 2020, 60, 103005. [Google Scholar] [CrossRef]
- Fife, B.T.; Pauken, K.E. The Role of the PD-1 Pathway in Autoimmunity and Peripheral Tolerance. Ann. N. Y. Acad. Sci. 2011, 1217, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Krause, R.M. Paul Ehrlich and O.T. Avery: Pathfinders in the Search for Immunity. Vaccine 1999, 17 (Suppl. S3), S64–S67. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Kunimasa, K.; Goto, T. Immunosurveillance and Immunoediting of Lung Cancer: Current Perspectives and Challenges. Int. J. Mol. Sci. 2020, 21, 597. [Google Scholar] [CrossRef]
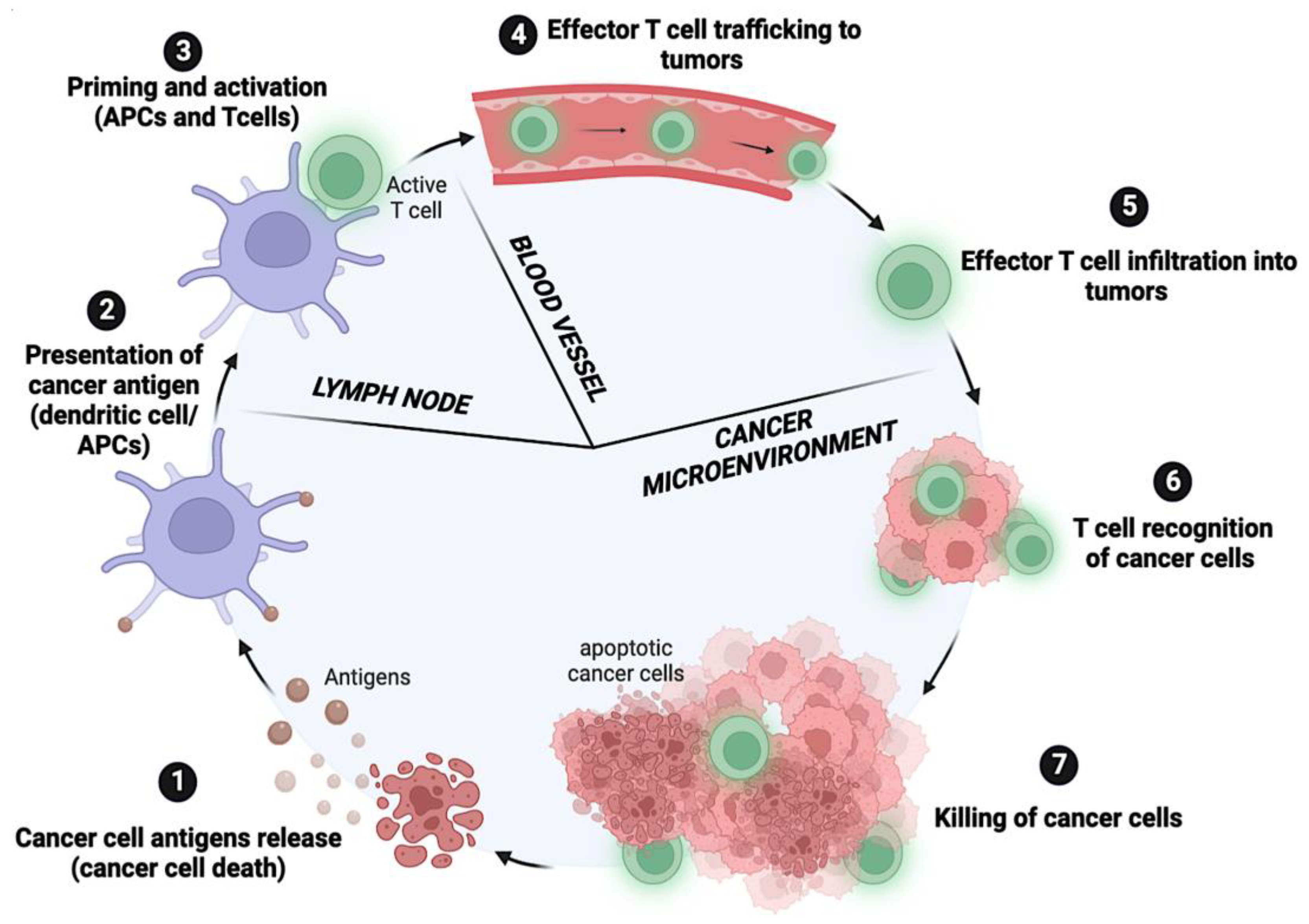
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Lou, H.; Le, F.; Hu, M.; Yang, X.; Li, L.; Wang, L.; Wang, N.; Gao, H.; Jin, F. Aberrant DNA Methylation of IGF2-H19 Locus in Human Fetus and in Spermatozoa From Assisted Reproductive Technologies. Reprod. Sci. 2018, 26, 1004–1997. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The Future of Immune Checkpoint Therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- O’Day, S.J.; Hamid, O.; Urba, W.J. Targeting Cytotoxic T-Lymphocyte Antigen-4 (CTLA-4): A Novel Strategy for the Treatment of Melanoma and Other Malignancies. Cancer 2007, 110, 2614–2627. [Google Scholar] [CrossRef]
- Bardhan, K.; Anagnostou, T.; Boussiotis, V.A. The PD1:PD-L1/2 Pathway from Discovery to Clinical Implementation. Front. Immunol. 2016, 7, 550. [Google Scholar] [CrossRef]
- Long, L.; Zhang, X.; Chen, F.; Pan, Q.; Phiphatwatchara, P.; Zeng, Y.; Chen, H. The Promising Immune Checkpoint LAG-3: From Tumor Microenvironment to Cancer Immunotherapy. Genes Cancer 2018, 9, 176. [Google Scholar] [CrossRef]
- Shan, C.; Li, X.; Zhang, J. Progress of Immune Checkpoint LAG-3 in Immunotherapy. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Ha, S.-J.; Kim, H.R. Clinical Insights into Novel Immune Checkpoint Inhibitors. Front. Pharmacol. 2021, 12, 1074. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Gey, A.; Dariane, C.; Mejean, A.; Timsit, M.-O.; Blanc, C.; Verkarre, V.; Radulescu, C.; Fabre, E.; Vano, Y. Tim-3-Biomarqueur et Cible Thérapeutique En Cancérologie. Médecine/Sciences 2018, 34, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Peppelenbosch, M.P.; Sprengers, D.; Kwekkeboom, J. TIGIT, the next Step towards Successful Combination Immune Checkpoint Therapy in Cancer. Front. Immunol. 2021, 12, 699895. [Google Scholar] [CrossRef] [PubMed]
- Raphael, I.; Kumar, R.; McCarl, L.H.; Shoger, K.; Wang, L.; Sandlesh, P.; Sneiderman, C.T.; Allen, J.; Zhai, S.; Campagna, M.L. TIGIT and PD-1 Immune Checkpoint Pathways Are Associated with Patient Outcome and Anti-Tumor Immunity in Glioblastoma. Front. Immunol. 2021, 12, 637146. [Google Scholar] [CrossRef] [PubMed]
- Radwan, S.M.; Elleboudy, N.S.; Nabih, N.A.; El-kholy, A.; Kamal, A.M. The Prospective Prognostic Value of the Immune Checkpoint BTLA Expression in Adult Acute Myeloid Leukemia Patients. Egypt. J. Med. Hum. Genet. 2021, 22, 1–8. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Lin, H.-W.; Chien, C.-L.; Lai, Y.-L.; Sun, W.-Z.; Chen, C.-A.; Cheng, W.-F. BTLA Blockade Enhances Cancer Therapy by Inhibiting IL-6/IL-10-Induced CD19high B Lymphocytes. J. Immunother. Cancer 2019, 7, 313. [Google Scholar] [CrossRef]
- Meireson, A.; Devos, M.; Brochez, L. IDO Expression in Cancer: Different Compartment, Different Functionality? Front. Immunol. 2020, 11, 531491. [Google Scholar] [CrossRef]
- Ward, F.J.; Dahal, L.N.; Wijesekera, S.K.; Abdul-Jawad, S.K.; Kaewarpai, T.; Xu, H.; Vickers, M.A.; Barker, R.N. The Soluble Isoform of CTLA-4 as a Regulator of T-Cell Responses. Eur. J. Immunol. 2013, 43, 1274–1285. [Google Scholar] [CrossRef]
- Elhag, O.A.O.; Hu, X.-J.; Wen-Ying, Z.; Li, X.; Yuan, Y.-Z.; Deng, L.-F.; Liu, D.-L.; Liu, Y.-L.; Hui, G. Reconstructed Adeno-Associated Virus with the Extracellular Domain of Murine PD-1 Induces Antitumor Immunity. Asian Pac. J. Cancer Prev. 2012, 13, 4031–4036. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, Y.; Su, M.; Zhang, R.; Ding, J.; Hao, X.; Ma, Y. Role of Soluble Programmed Death-1 (SPD-1) and SPD-Ligand 1 in Patients with Cystic Echinococcosis. Exp. Ther. Med. 2016, 11, 251–256. [Google Scholar] [CrossRef]
- Jalali, S.; Price-Troska, T.; Paludo, J.; Villasboas, J.; Kim, H.-J.; Yang, Z.-Z.; Novak, A.J.; Ansell, S.M. Soluble PD-1 Ligands Regulate T-Cell Function in Waldenstrom Macroglobulinemia. Blood Adv. 2018, 2, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Dowell, A.C.; Munford, H.; Goel, A.; Gordon, N.S.; James, N.D.; Cheng, K.; Zeegers, M.P.; Ward, D.G.; Bryan, R.T. PD-L2 Is Constitutively Expressed in Normal and Malignant Urothelium. Front. Oncol. 2021, 11, 626748. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Arooj, S.; Wang, H. Soluble B7-CD28 Family Inhibitory Immune Checkpoint Proteins and Anti-Cancer Immunotherapy. Front. Immunol. 2021, 12, 651634. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, D.; Chen, Q.; Yang, C.; Wang, B.; Wu, H. Soluble B7-H3 Promotes the Invasion and Metastasis of Pancreatic Carcinoma Cells through the TLR4/NF-ΚB Pathway. Sci. Rep. 2016, 6, 27528. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, J.; Shi, J.; Yu, G.; Lu, B.; Zhang, X. Soluble CD276 (B7-H3) Is Released from Monocytes, Dendritic Cells and Activated T Cells and Is Detectable in Normal Human Serum. Immunology 2008, 123, 538–546. [Google Scholar] [CrossRef]
- Bartkowiak, T.; Curran, M.A. 4-1BB Agonists: Multi-Potent Potentiators of Tumor Immunity. Front. Oncol. 2015, 5, 117. [Google Scholar] [CrossRef]
- Kamal, A.M.; Wasfey, E.F.; Elghamry, W.R.; Sabry, O.M.; Elghobary, H.A.; Radwan, S.M. Genetic Signature of CTLA-4, BTLA, TIM-3 and LAG-3 Molecular Expression in Colorectal Cancer Patients: Implications in Diagnosis and Survival Outcomes. Clin. Biochem. 2021, 96, 13–18. [Google Scholar] [CrossRef]
- Marin-Acevedo, J.A.; Kimbrough, E.O.; Lou, Y. Next Generation of Immune Checkpoint Inhibitors and Beyond. J. Hematol. Oncol. 2021, 14, 45. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Pauken, K.E. The Diverse Functions of the PD1 Inhibitory Pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef]
- Ward, J.P.; Gubin, M.M.; Schreiber, R.D. The Role of Neoantigens in Naturally Occurring and Therapeutically Induced Immune Responses to Cancer. Adv. Immunol. 2016, 130, 25–74. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Wojtukiewicz, M.Z.; Rek, M.M.; Karpowicz, K.; Górska, M.; Polityńska, B.; Wojtukiewicz, A.M.; Moniuszko, M.; Radziwon, P.; Tucker, S.C.; Honn, K.V. Inhibitors of Immune Checkpoints-PD-1, PD-L1, CTLA-4-New Opportunities for Cancer Patients and a New Challenge for Internists and General Practitioners. Cancer Metastasis Rev. 2021, 40, 949–982. [Google Scholar] [CrossRef]
- Boussiotis, V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016, 375, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel Immune Checkpoint Targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef] [PubMed]
- Toor, S.M.; Murshed, K.; Al-Dhaheri, M.; Khawar, M.; Abu Nada, M.; Elkord, E. Immune Checkpoints in Circulating and Tumor-Infiltrating CD4+ T Cell Subsets in Colorectal Cancer Patients. Front. Immunol. 2019, 10, 2936. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, T.; Xuan, Q.; Zhao, H.; Qin, L.; Zhang, Q. Lymphocyte-Activation Gene-3 Expression and Prognostic Value in Neoadjuvant-Treated Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 124–133. [Google Scholar] [CrossRef]
- Zhou, E.; Huang, Q.; Wang, J.; Fang, C.; Yang, L.; Zhu, M.; Chen, J.; Chen, L.; Dong, M. Up-Regulation of Tim-3 Is Associated with Poor Prognosis of Patients with Colon Cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 8018. [Google Scholar]
- Yan, W.; Liu, X.; Ma, H.; Zhang, H.; Song, X.; Gao, L.; Liang, X.; Ma, C. Tim-3 Fosters HCC Development by Enhancing TGF-β-Mediated Alternative Activation of Macrophages. Gut 2015, 64, 1593–1604. [Google Scholar] [CrossRef]
- Li, H.; Wu, K.; Tao, K.; Chen, L.; Zheng, Q.; Lu, X.; Liu, J.; Shi, L.; Liu, C.; Wang, G. Tim-3/Galectin-9 Signaling Pathway Mediates T-cell Dysfunction and Predicts Poor Prognosis in Patients with Hepatitis B Virus-associated Hepatocellular Carcinoma. Hepatology 2012, 56, 1342–1351. [Google Scholar] [CrossRef]
- Bolm, L.; Petruch, N.; Sivakumar, S.; Annels, N.E.; Frampton, A.E. Gene of the Month: T-Cell Immunoreceptor with Immunoglobulin and ITIM Domains (TIGIT). J. Clin. Pathol. 2022, 75, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, J.-M.; Zarour, H.M. TIGIT in Cancer Immunotherapy. J. Immunother. Cancer 2020, 8, e000957. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Ao, X.; Yang, Y.; Chen, Z.; Xu, X. Soluble Immune Checkpoints in Cancer: Production, Function and Biological Significance. J. Immunother. Cancer 2018, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Nelson, H.H.; Kelsey, K.T. Epigenetic Epidemiology as a Tool to Understand the Role of Immunity in Chronic Disease. Epigenomics 2016, 8, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Amin, N.; McGrath, A.; Chen, Y.-P.P. Evaluation of Deep Learning in Non-Coding RNA Classification. Nat. Mach. Intell. 2019, 1, 246–256. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long Non-Coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The Hallmarks of Cancer. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.A. Treating Disease at the RNA Level with Oligonucleotides. N. Engl. J. Med. 2019, 380, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Ahn, Y.; Kook, H.; Kim, Y.-K. The Roles of Non-Coding RNAs in Vascular Calcification and Opportunities as Therapeutic Targets. Pharmacol. Ther. 2021, 218, 107675. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Rinn, J.L. Discovery and Annotation of Long Noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The Emergence of LncRNAs in Cancer Biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-Coding RNAs in Mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Pillay, S.; Takahashi, H.; Carninci, P.; Kanhere, A. Antisense RNAs during Early Vertebrate Development Are Divided in Groups with Distinct Features. Genome Res. 2021, 31, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, Y.; Xie, Y.; Ma, X. The Role of Long Non-Coding RNAs in Immunotherapy Resistance. Front. Oncol. 2019, 9, 1292. [Google Scholar] [CrossRef]
- Tonner, P.; Srinivasasainagendra, V.; Zhang, S.; Zhi, D. Detecting Transcription of Ribosomal Protein Pseudogenes in Diverse Human Tissues from RNA-Seq Data. BMC Genom. 2012, 13, 412. [Google Scholar] [CrossRef]
- Calin, G.A.; Liu, C.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved Regions Encoding NcRNAs Are Altered in Human Leukemias and Carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A Class of Long Noncoding RNAs Synthesized at Enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nie, H.; He, X.; Liao, Z.; Zhou, Y.; Zhou, J.; Ou, C. The Emerging Role of Super Enhancer-Derived Noncoding RNAs in Human Cancer. Theranostics 2020, 10, 11049–11062. [Google Scholar] [CrossRef] [PubMed]
- Chellini, L.; Frezza, V.; Paronetto, M.P. Dissecting the Transcriptional Regulatory Networks of Promoter-Associated Noncoding RNAs in Development and Cancer. J. Exp. Clin. Cancer Res. 2020, 39, 51. [Google Scholar] [CrossRef] [PubMed]
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (LncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Noncoding RNA 2021, 7, 36. [Google Scholar] [CrossRef]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genom. 2017, 2017, 6218353. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and Characterizing Circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Efficient Backsplicing Produces Translatable Circular MRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef]
- Diallo, L.H.; Tatin, F.; David, F.; Godet, A.-C.; Zamora, A.; Prats, A.-C.; Garmy-Susini, B.; Lacazette, E. How Are CircRNAs Translated by Non-Canonical Initiation Mechanisms? Biochimie 2019, 164, 45–52. [Google Scholar] [CrossRef]
- Osca-Verdegal, R.; Beltrán-García, J.; Górriz, J.L.; Martínez Jabaloyas, J.M.; Pallardó, F.V.; García-Giménez, J.L. Use of Circular RNAs in Diagnosis, Prognosis and Therapeutics of Renal Cell Carcinoma. Front. Cell Dev. Biol. 2022, 10, 879814. [Google Scholar] [CrossRef]
- Fang, Z.; Jiang, C.; Li, S. The Potential Regulatory Roles of Circular RNAs in Tumor Immunology and Immunotherapy. Front. Immunol. 2021, 11, 617583. [Google Scholar] [CrossRef]
- Cheng, J.-T.; Wang, L.; Wang, H.; Tang, F.-R.; Cai, W.-Q.; Sethi, G.; Xin, H.-W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Tsagakis, I.; Douka, K.; Birds, I.; Aspden, J.L. Long Non-Coding RNAs in Development and Disease: Conservation to Mechanisms. J. Pathol. 2020, 250, 480–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Yamazaki, T. Long Noncoding RNAs and Their Applications: Focus on Architectural RNA (ArcRNA), a Class of LncRNA. In Applied RNA Bioscience; Springer: Singapore, 2018; pp. 161–187. [Google Scholar]
- Pathania, A.S.; Challagundla, K.B. Exosomal Long Non-Coding RNAs: Emerging Players in the Tumor Microenvironment. Mol. Ther. Nucleic Acids 2020, 23, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
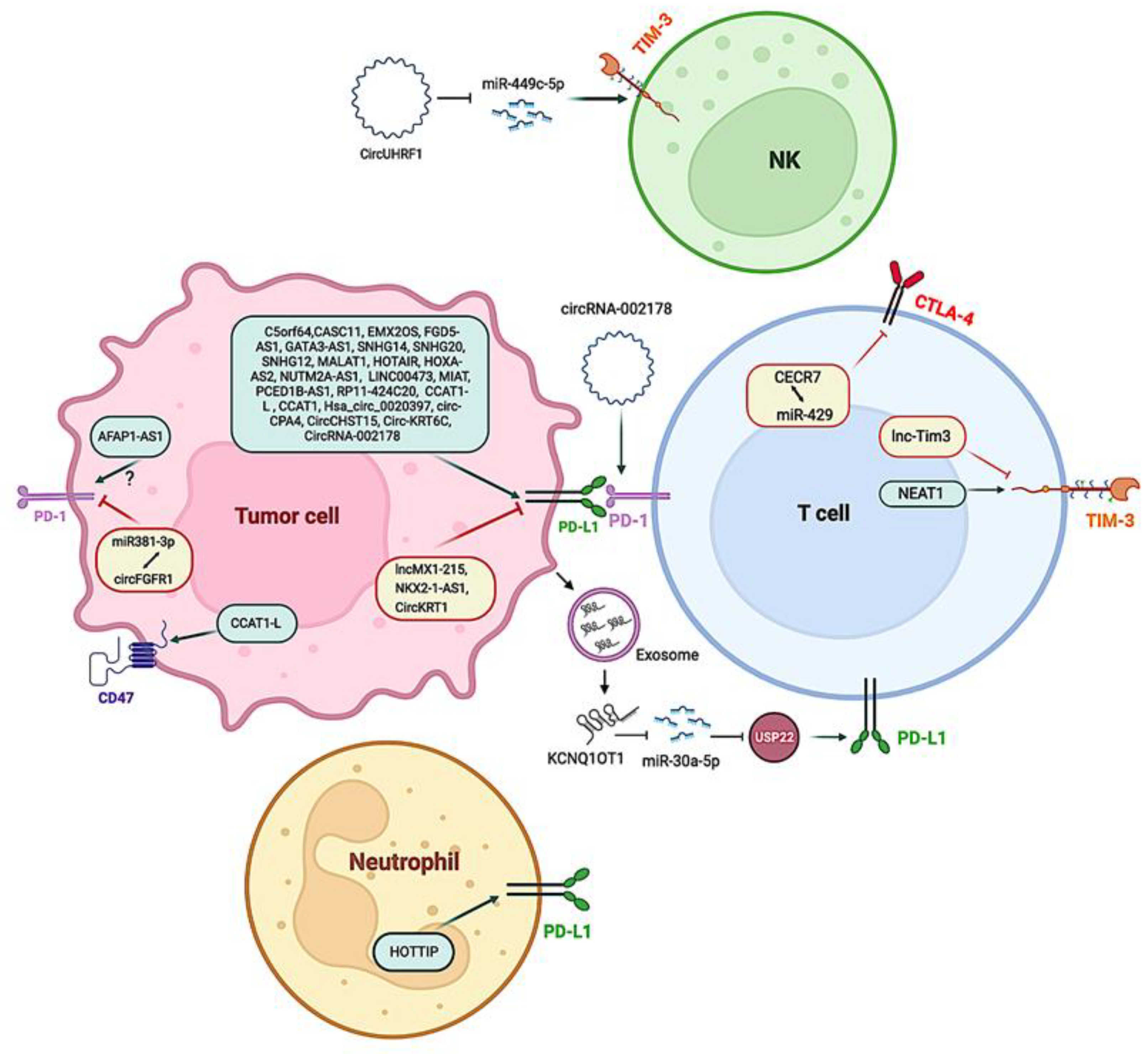
- Tang, Y.; He, Y.; Shi, L.; Yang, L.; Wang, J.; Lian, Y.; Fan, C.; Zhang, P.; Guo, C.; Zhang, S.; et al. Co-Expression of AFAP1-AS1 and PD-1 Predicts Poor Prognosis in Nasopharyngeal Carcinoma. Oncotarget 2017, 8, 39001–39011. [Google Scholar] [CrossRef]
- Strazza, M.; Azoulay-Alfaguter, I.; Peled, M.; Adam, K.; Mor, A. Transmembrane Adaptor Protein PAG Is a Mediator of PD-1 Inhibitory Signaling in Human T Cells. Commun. Biol. 2021, 4, 672. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, X.; Wang, Y.; Wang, Y.; Yan, T.; Wan, J.; Wang, K.; Du, J. Long Non-Coding RNA C5orf64 Is a Potential Indicator for Tumor Microenvironment and Mutation Pattern Remodeling in Lung Adenocarcinoma. Genomics 2021, 113, 291–304. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, Transcriptional and Post-Translational Regulation of the Programmed Death Protein Ligand 1 in Cancer: Biology and Clinical Correlations. Oncogene 2018, 37, 4639–4661. [Google Scholar] [CrossRef]
- Shklovskaya, E.; Rizos, H. Spatial and Temporal Changes in PD-L1 Expression in Cancer: The Role of Genetic Drivers, Tumor Microenvironment and Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 7139. [Google Scholar] [CrossRef]
- Oshi, M.; Maiti, A.; Wu, R.; Tokumaru, Y.; Murthy, V.; Yan, L.; Ishikawa, T.; Endo, I.; Hait, N.; Takabe, K. MiR-150 Expression in Breast Cancer Attracts and Activates Immune Cells, and Is Associated with Better Patient Outcome. J. Clin. Oncol. 2022, 40, 570. [Google Scholar] [CrossRef]
- An, S.-Q.; He, K.; Liu, F.; Ding, Q.-S.; Wei, Y.-L.; Xia, Z.-L.; Duan, X.-P.; Huang, R.; Li, B.-W.; Wang, H.-H.; et al. Low MicroRNA150 Expression Is Associated with Activated Carcinogenic Pathways and a Poor Prognosis in Patients with Breast Cancer. Oncol. Rep. 2021, 45, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Akbay, E.A.; Koyama, S.; Carretero, J.; Altabef, A.; Tchaicha, J.H.; Christensen, C.L.; Mikse, O.R.; Cherniack, A.D.; Beauchamp, E.M.; Pugh, T.J.; et al. Activation of the PD-1 Pathway Contributes to Immune Escape in EGFR-Driven Lung Tumors. Cancer Discov. 2013, 3, 1355–1363. [Google Scholar] [CrossRef]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 Expression in the Tumor Microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, C.; Chang, Y.; Zhang, Z.; Hu, Y.; Zhang, F.; Lu, Y.; Zheng, L.; Zhang, W.; Li, X.; et al. Long Non-Coding RNA CASC11 Interacts with HnRNP-K and Activates the WNT/β-Catenin Pathway to Promote Growth and Metastasis in Colorectal Cancer. Cancer Lett. 2016, 376, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Liu, Y.; Li, X.; Chen, S.; Xie, R.; Chen, D.; Gao, H.; Wang, G.; Cai, B.; Yang, X. Long Noncoding RNA CASC11 Promotes Hepatocarcinogenesis and HCC Progression through EIF4A3-Mediated E2F1 Activation. Clin. Transl. Med. 2020, 10, e220. [Google Scholar] [CrossRef]
- Mafi, S.; Mansoori, B.; Taeb, S.; Sadeghi, H.; Abbasi, R.; Cho, W.C.; Rostamzadeh, D. MTOR-Mediated Regulation of Immune Responses in Cancer and Tumor Microenvironment. Front. Immunol. 2021, 12, 774103. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, D.; Jian, Z.; Chen, G.G.; Lai, P.B.S. Cancer Specific Long Noncoding RNAs Show Differential Expression Patterns and Competing Endogenous RNA Potential in Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0141042. [Google Scholar] [CrossRef]
- Yao, K.; Wang, Q.; Jia, J.; Zhao, H. A Competing Endogenous RNA Network Identifies Novel MRNA, MiRNA and LncRNA Markers for the Prognosis of Diabetic Pancreatic Cancer. Tumor Biol. 2017, 39, 1010428317707882. [Google Scholar] [CrossRef]
- Li, S.; Chen, S.; Wang, B.; Zhang, L.; Su, Y.; Zhang, X. A Robust 6-LncRNA Prognostic Signature for Predicting the Prognosis of Patients With Colorectal Cancer Metastasis. Front. Med. 2020, 7, 56. [Google Scholar] [CrossRef]
- Lu, J.; Fei, F.; Wu, C.; Mei, J.; Xu, J.; Lu, P. ZEB1: Catalyst of Immune Escape during Tumor Metastasis. Biomed. Pharmacother. 2022, 153, 113490. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Janji, B.; Abdou, A.; Hasmim, M.; Terry, S.; Tan, T.Z.; Mami-Chouaib, F.; Thiery, J.P.; Chouaib, S. The Immune Checkpoint Ligand PD-L1 Is Upregulated in EMT-Activated Human Breast Cancer Cells by a Mechanism Involving ZEB-1 and MiR-200. Oncoimmunology 2017, 6, e1263412. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Xu, S.; Ruan, H.; Wang, T.; Song, W.; Qian, L.; Chen, K. MiR-195/-16 Family Enhances Radiotherapy via T Cell Activation in the Tumor Microenvironment by Blocking the PD-L1 Immune Checkpoint. CPB 2018, 48, 801–814. [Google Scholar] [CrossRef]
- Wei, S.; Wang, K.; Huang, X.; Zhao, Z.; Zhao, Z. LncRNA MALAT1 Contributes to Non-Small Cell Lung Cancer Progression via Modulating MiR-200a-3p/Programmed Death-Ligand 1 Axis. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419859699. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.; Yuan, C.; Han, T.; Cui, J.; Jiao, F.; Wang, L. A Novel Feedback Loop between High MALAT-1 and Low MiR-200c-3p Promotes Cell Migration and Invasion in Pancreatic Ductal Adenocarcinoma and Is Predictive of Poor Prognosis. BMC Cancer 2018, 18, 1032. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Zhang, J.; Liu, Y.; Qi, Q. LncRNA SNHG14/MiR-5590-3p/ZEB1 Positive Feedback Loop Promoted Diffuse Large B Cell Lymphoma Progression and Immune Evasion through Regulating PD-1/PD-L1 Checkpoint. Cell Death Dis. 2019, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, C.; Li, W.; Sun, X.; Liu, Q.; Wang, D. Nano-Coated Si-SNHG14 Regulated PD-L1 Expression and Decreased Epithelial-Mesenchymal Transition in Nasopharyngeal Carcinoma Cells. J. Biomed. Nanotechnol. 2021, 17, 1993–2002. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the Tumor Microenvironment in PD-L1/PD-1-Mediated Tumor Immune Escape. Mol Cancer 2019, 18, 10. [Google Scholar] [CrossRef]
- Liu, Q.; Zhu, A.; Gao, W.; Gui, F.; Zou, Y.; Zhou, X.; Hong, Z. MiR-5590-3p Inhibits the Proliferation and Metastasis of Renal Cancer Cells by Targeting ROCK2 to Inhibit Proliferation, Migration and Invasion. Oncol. Lett. 2022, 24, 377. [Google Scholar] [CrossRef]
- Wu, N.; Han, Y.; Liu, H.; Jiang, M.; Chu, Y.; Cao, J.; Lin, J.; Liu, Y.; Xu, B.; Xie, X. MiR-5590-3p Inhibited Tumor Growth in Gastric Cancer by Targeting DDX5/AKT/m-TOR Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 1491–1497. [Google Scholar] [CrossRef]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS Induces Proliferation, Invasion and Sphere Formation of Ovarian Cancer Cells via Regulating the MiR-654-3p/AKT3/PD-L1 Axis. Cancer Manag. Res. 2020, 12, 2141–2154. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-X.; Tan, Y.-Z.; He, G.-C.; Zhang, Q.-L.; Liu, P. EMX2OS Plays a Prognosis-Associated Enhancer RNA Role in Gastric Cancer. Medicine 2021, 100, e27535. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. MiR-142-5p Regulates Tumor Cell PD-L1 Expression and Enhances Anti-Tumor Immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Niu, R.; Shao, X.; Shao, X. FGD5-AS1 Promotes Cisplatin Resistance of Human Lung Adenocarcinoma Cell via the MiR-142-5p/PD-L1 Axis. Int. J. Mol. Med. 2021, 47, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, W.-C.; Shen, W.-H.; Xu, K.; Hu, Y.-Y.; Han, G.-H.; Liu, Y.-B. Thalidomide Suppresses Angiogenesis and Immune Evasion via LncRNA FGD5-AS1/MiR-454-3p/ZEB1 Axis-Mediated VEGFA Expression and PD-1/PD-L1 Checkpoint in NSCLC. Chem. Biol. Interact. 2021, 349, 109652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, N.; Song, P.; Fu, Y.; Ren, Y.; Li, Z.; Wang, J. LncRNA GATA3-AS1 Facilitates Tumour Progression and Immune Escape in Triple-Negative Breast Cancer through Destabilization of GATA3 but Stabilization of PD-L1. Cell Prolif. 2020, 53, e12855. [Google Scholar] [CrossRef]
- Shao, W.; Ding, Q.; Yugang, G.; Wang, Z.; Xu, Q.; Guo, Y.; Xing, J. A Pan-Cancer Landscape of HOX-Related LncRNAs and Their Association with Prognosis and Tumor Microenvironment. Front. Mol. Biosci. 2021, 8, 1077. [Google Scholar] [CrossRef]
- Shang, A.; Wang, W.; Gu, C.; Chen, C.; Zeng, B.; Yang, Y.; Ji, P.; Sun, J.; Wu, J.; Lu, W.; et al. Long Non-Coding RNA HOTTIP Enhances IL-6 Expression to Potentiate Immune Escape of Ovarian Cancer Cells by Upregulating the Expression of PD-L1 in Neutrophils. J. Exp. Clin. Cancer Res. 2019, 38, 411. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Hailemichael, Y.; Johnson, D.H.; Abdel-Wahab, N.; Foo, W.C.; Bentebibel, S.-E.; Daher, M.; Haymaker, C.; Wani, K.; Saberian, C.; Ogata, D.; et al. Interleukin-6 Blockade Abrogates Immunotherapy Toxicity and Promotes Tumor Immunity. Cancer Cell 2022, 40, 509–523. [Google Scholar] [CrossRef]
- Qian, M.; Ling, W.; Ruan, Z. Long Non-Coding RNA SNHG12 Promotes Immune Escape of Ovarian Cancer Cells through Their Crosstalk with M2 Macrophages. Aging 2020, 12, 17122. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Shen, Q.; Ma, W. Long Noncoding RNA Hotair Promotes the Progression and Immune Escape in Laryngeal Squamous Cell Carcinoma through MicroRNA-30a/GRP78/PD-L1 Axis. J. Immunol. Res. 2022, 2022, 5141426. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; You, H.; Yu, S. Long Non-Coding RNA HOXA-AS2 Promotes the Expression Levels of Hypoxia-Inducible Factor-1α and Programmed Death-Ligand 1, and Regulates Nasopharyngeal Carcinoma Progression via MiR-519. Oncol. Lett. 2020, 20, 245. [Google Scholar] [CrossRef]
- Wang, J.; Yu, Z.; Wang, J.; Shen, Y.; Qiu, J.; Zhuang, Z. LncRNA NUTM2A-AS1 Positively Modulates TET1 and HIF-1A to Enhance Gastric Cancer Tumorigenesis and Drug Resistance by Sponging MiR-376a. Cancer Med. 2020, 9, 9499–9510. [Google Scholar] [CrossRef]
- Xian, D.; Niu, L.; Zeng, J.; Wang, L. LncRNA KCNQ1OT1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating PD-L1 Ubiquitination via MiR-30a-5p/USP22. Front. Cell Dev. Biol. 2021, 9, 653808. [Google Scholar] [CrossRef]
- Chen, Q.-H.; Li, B.; Liu, D.-G.; Zhang, B.; Yang, X.; Tu, Y.-L. LncRNA KCNQ1OT1 Sponges MiR-15a to Promote Immune Evasion and Malignant Progression of Prostate Cancer via up-Regulating PD-L1. Cancer Cell Int. 2020, 20, 394. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Ma, X.; Yuan, Z.; Hu, M. KCNQ1OT1 Contributes to Sorafenib Resistance and Programmed Death-ligand-1-mediated Immune Escape via Sponging MiR-506 in Hepatocellular Carcinoma Cells. Int. J. Mol. Med. 2020, 46, 1794–1804. [Google Scholar] [CrossRef]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.; Li, Y.; Li, W.; Zhang, H.; Song, J. Repression of LncRNA NEAT1 Enhances the Antitumor Activity of CD8+ T Cells against Hepatocellular Carcinoma via Regulating MiR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef]
- Naveed, A.; Cooper, J.A.; Li, R.; Hubbard, A.; Chen, J.; Liu, T.; Wilton, S.D.; Fletcher, S.; Fox, A.H. NEAT1 PolyA-Modulating Antisense Oligonucleotides Reveal Opposing Functions for Both Long Non-Coding RNA Isoforms in Neuroblastoma. Cell. Mol. Life Sci. 2021, 78, 2213–2230. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, B.; Qiu, J.; Ke, X.; Shen, S.; Wang, X.; Tang, N. LncRNA MIAT Targets MiR-411-5p/STAT3/PD-L1 Axis Mediating Hepatocellular Carcinoma Immune Response. Int. J. Exp. Pathol. 2022, 103, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Shan, D.; Shang, Y.; Hu, T. MicroRNA-411 Inhibits Cervical Cancer Progression by Directly Targeting STAT3. Oncol. Res. 2019, 27, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, H.; Millien, G.; McNally, L.; Gower, A.C.; Tagne, J.-B.; Cao, Y.; Ramirez, M.I. NKX2-1-AS1 Negatively Regulates CD274/PD-L1, Cell-Cell Interaction Genes, and Limits Human Lung Carcinoma Cell Migration. Sci. Rep. 2018, 8, 14418. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Zimta, A.-A.; Harangus, A.; Iurca, I.; Irimie, A.; Coza, O.; Berindan-Neagoe, I. The Function of Non-Coding RNAs in Lung Cancer Tumorigenesis. Cancers 2019, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, X.; Zhang, L.; Chen, Y.; Dong, R.; Zhang, J.; Zhao, J.; Guo, X.; Yang, G.; Li, Y.; et al. MiR-194-5p down-Regulates Tumor Cell PD-L1 Expression and Promotes Anti-Tumor Immunity in Pancreatic Cancer. Int. Immunopharmacol. 2021, 97, 107822. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Yu, J.; Chen, L.; Tao, T.; Yi, S.; Hanley, S.J.; Yue, J.; Watari, H.; Sakuragi, N. Control of PD-L1 Expression by MiR-140/142/340/383 and Oncogenic Activation of the OCT4–MiR-18a Pathway in Cervical Cancer. Oncogene 2018, 37, 5257–5268. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual Targeting of PD-L1 and PD-L2 by PCED1B-AS1 via Sponging Hsa-MiR-194-5p Induces Immunosuppression in Hepatocellular Carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef]
- Ma, H.; Chang, H.; Yang, W.; Lu, Y.; Hu, J.; Jin, S. A Novel IFNα-Induced Long Noncoding RNA Negatively Regulates Immunosuppression by Interrupting H3K27 Acetylation in Head and Neck Squamous Cell Carcinoma. Mol. Cancer 2020, 19, 4. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Song, H. A Disparate Role of RP11-424C20.2/UHRF1 Axis through Control of Tumor Immune Escape in Liver Hepatocellular Carcinoma and Thymoma. Aging 2019, 11, 6422–6439. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, F.; Su, C.; Xie, P.; Xu, L. Upregulation of Long Noncoding RNA SNHG20 Promotes Cell Growth and Metastasis in Esophageal Squamous Cell Carcinoma via Modulating ATM-JAK-PD-L1 Pathway. J. Cell. Biochem. 2019, 120, 11642–11650. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, X.; Xue, X.; Li, L.; Hu, Y. A Long Noncoding RNA Sensitizes Genotoxic Treatment by Attenuating ATM Activation and Homologous Recombination Repair in Cancers. PLoS Biol. 2020, 18, e3000666. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yin, Y.; Ju, H.; Xu, X.; Liu, W.; Fu, Q.; Hu, J.; Zhang, X.; Sun, B. Long Non-Coding RNA Lnc-Tim3 Exacerbates CD8 T Cell Exhaustion via Binding to Tim-3 and Inducing Nuclear Translocation of Bat3 in HCC. Cell Death Dis. 2018, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Xu, K.; Jin, X.; Li, J.; Shi, Y.; Zhang, M.; Jin, X.; Li, Y.; Xu, J.; Li, X. LncSpA: LncRNA Spatial Atlas of Expression across Normal and Cancer Tissues. Cancer Res. 2020, 80, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An Integrated Database of NcRNA Interactions. Nucleic Acids Res. 2020, 48, D160–D165. [Google Scholar] [CrossRef]
- Wu, M.; Shen, J. From Super-Enhancer Non-Coding RNA to Immune Checkpoint: Frameworks to Functions. Front. Oncol. 2019, 9, 1307. [Google Scholar] [CrossRef]
- Xiang, J.-F.; Yin, Q.-F.; Chen, T.; Zhang, Y.; Zhang, X.-O.; Wu, Z.; Zhang, S.; Wang, H.-B.; Ge, J.; Lu, X.; et al. Human Colorectal Cancer-Specific CCAT1-L LncRNA Regulates Long-Range Chromatin Interactions at the MYC Locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, Y.; Xie, J. Co-Activation of Super-Enhancer-Driven CCAT1 by TP63 and SOX2 Promotes Squamous Cancer Progression. Nat. Commun. 2018, 9, 3619. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Hogg, S.J.; Vervoort, S.J.; Deswal, S.; Ott, C.J.; Li, J.; Cluse, L.A.; Beavis, P.A.; Darcy, P.K.; Martin, B.P.; Spencer, A.; et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017, 18, 2162–2174. [Google Scholar] [CrossRef]
- Zhang, X.-L.; Xu, L.-L.; Wang, F. Hsa_circ_0020397 Regulates Colorectal Cancer Cell Viability, Apoptosis and Invasion by Promoting the Expression of the MiR-138 Targets TERT and PD-L1. Cell Biol. Int. 2017, 41, 1056–1064. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Pei, X.; Li, K.-S.; Jin, L.-N.; Wang, F.; Wu, J.; Zhang, X.-M. Circular RNA CircFGFR1 Promotes Progression and Anti-PD-1 Resistance by Sponging MiR-381-3p in Non-Small Cell Lung Cancer Cells. Mol. Cancer 2019, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Xue, M.; Jiang, J.; Zhang, Y.; Gao, X. Circular RNA Circ-CPA4/Let-7 MiRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-Small Cell Lung Cancer (NSCLC). J. Exp. Clin. Cancer Res. 2020, 39, 149. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jia, Y.; Wang, B.; Yang, S.; Du, K.; Luo, Y.; Li, Y.; Zhu, B. Circular RNA CHST15 Sponges MiR-155-5p and MiR-194-5p to Promote the Immune Escape of Lung Cancer Cells Mediated by PD-L1. Front. Oncol. 2021, 11, 595609. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Hou, Z.; Liu, W.; Yu, Z.; Liang, Z.; Chen, S. Circ-Keratin 6c Promotes Malignant Progression and Immune Evasion of Colorectal Cancer through MicroRNA-485-3p/Programmed Cell Death Receptor Ligand 1 Axis. J. Pharmacol. Exp. Ther. 2021, 377, 358–367. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Gao, C.; Huang, X.-Y.; Lu, J.-C.; Guo, X.-J.; Shi, G.-M.; Cai, J.-B.; Ke, A.-W. Cancer Cell-Derived Exosomal CircUHRF1 Induces Natural Killer Cell Exhaustion and May Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.; Wang, Y.; Ren, F.; Sun, D.; Yan, Y.; Kong, X.; Bu, J.; Liu, M.; Xu, S. CircRNA-002178 Act as a CeRNA to Promote PDL1/PD1 Expression in Lung Adenocarcinoma. Cell Death Dis. 2020, 11, 32. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, W.; Wang, Y.; Qin, M.; Ji, Y. CircKRT1 Drives Tumor Progression and Immune Evasion in Oral Squamous Cell Carcinoma by Sponging MiR-495-3p to Regulate PDL1 Expression. Cell Biol. Int. 2021, 45, 1423–1435. [Google Scholar] [CrossRef]
- Schalper, K.A.; Carvajal-Hausdorf, D.; McLaughlin, J.; Altan, M.; Velcheti, V.; Gaule, P.; Sanmamed, M.F.; Chen, L.; Herbst, R.S.; Rimm, D.L. Differential Expression and Significance of PD-L1, IDO-1, and B7-H4 in Human Lung Cancer. Clin. Cancer Res. 2017, 23, 370–378. [Google Scholar] [CrossRef]
- Garbo, S.; Maione, R.; Tripodi, M.; Battistelli, C. Next RNA Therapeutics: The Mine of Non-Coding. Int. J. Mol. Sci. 2022, 23, 7471. [Google Scholar] [CrossRef]
- Hu, Q.; Ye, Y.; Chan, L.-C.; Li, Y.; Liang, K.; Lin, A.; Egranov, S.D.; Zhang, Y.; Xia, W.; Gong, J.; et al. Oncogenic LncRNA Downregulates Cancer Cell Antigen Presentation and Intrinsic Tumor Suppression. Nat. Immunol. 2019, 20, 835–851. [Google Scholar] [CrossRef]

| Immune Checkpoint | Cellular Expression | Ligand | Cellular Expression | Effects on Tumor Microenvironment | Ref. |
|---|---|---|---|---|---|
| CTLA-4 (CD152) | Tregs, Teffs, B cells, NK cells, NKT cells, and DCs | CD80 (B7-1), CD86 (B7-2) | APCs | Inhibits T cell activation by binding to its ligand. Inhibits IL-2 production and influences naive CD4+ T cell differentiation. | [29] |
| PD-1 | Tregs, Teffs, B cells, NK cells, mast cells, and some subsets | PD-L1 (B7-H1), PD-L2 (B7-DC) | Tumor cells, non-lymphoid, and non-hematopoietic cells | Inhibits effector T cell activation and promotes Treg cell generation | [30] |
| LAG-3 | Tregs, Teffs, B cells, NK cells, and DCs | MHC II | APCs | Has a synergistic impact with PD-1 to inhibit immune responses by suppressing T cell activation and cytokine production, thereby ensuring immune homeostasis | [31,32] |
| TIM-3 (HAVCR2) | Tregs, Teffs, NK cells, and some subsets of myeloid cells | Galectin-9, CEACAM1, Soluble HMGB1, PtdSer | Some myeloid subsets; Tregs, Teffs, NK cells, and some subsets of myeloid cells; Released by tumor cells or activated DCs: Apoptotic cells | Favors tumor escape to immune cells. Inhibits T cell responses. CD8+-T cells lose the ability to secrete cytokines IFNγ, IL-2, and TNFα | [33,34] |
| TIGIT | Tregs, Teffs, and NK cells | CD112 (PVRL2; nectin-2), CD155 (PVR) | DCs, APCs, and tumor cells | Suppresses the activation of TILs | [35,36] |
| BTLA (CD272) | T cells, B cells, macrophages, and NK cells | HVEM, TNFRSF14 | - | Suppresses pathway for T cell, B cell, or NK. | [37,38] |
| IDO-1 | EC, fibroblasts, macrophages, DCs, and PBMCs | GITR, ICOS, CD200 | DCs | Increases intratumoral infiltration and impairs cytotoxic T cell function. In DC, decreases antigen uptake and downregulates CD40/CD80 | [39] |
| Immune Checkpoint | Cellular Expression | Ligand | Cellular Expression | Effects in Tumor Microenvironment | Ref. |
|---|---|---|---|---|---|
| sCTLA-4 | Monocytes, immature DCs, and Treg cells | CD80, CD86 | APCs | Inhibits T cell responses | [56] |
| sPD-1 | PBMCs | PD-L1/2 | Tumor cells | Blocks PD-L/PD-1 interactions, Activates CD8+ T cells | [57] |
| sPD-L1 | Mature DCs | PD-1 | T cells | Combines with PD-1, inhibits T cell responses, and reduces T cell proliferation | [58,59] |
| sPD-L2 | Activated leukocytes | PD-1 | - | Unknown function | [60] |
| sCD80 (sB7–1) | unstimulated B cells and monocytes, and activated T and B cells | CTLA-4, CD28 | T cells | Inhibits PD-1/PD-L1 pathway, T cell proliferation, and IL-2 production | [61] |
| sCD86 (sB7–2) | Constitutively expressed on APCs, monocytes, DC, and certain cancer cells | CTLA-4, CD28 | T cells | Inhibits T cell responses | [61] |
| sB7-H3 | Monocytes, DCs, and activated T cells | B7-H3R | T cells | Promotes IL-8 and VEGF expression, increasing invasion and metastases of pancreatic carcinoma cells | [62,63] |
| sCD137 (s4-1BB) | Activated PBMCs | CD137L (4-1BBL) | T cells | Inhibits CD137/CD137L pathway | [64] |
| Name | Ensembl ID | Chromosome | Strand | Class | Mechanism of Action Related to Immune Evasion in Cancer |
|---|---|---|---|---|---|
| AFAP1-AS1 | ENSG00000272620 | 4 | + | Intergenic | AFAP-AS1/PAG/PD-1 |
| C5orf64 | ENSG00000178722 | 5 | + | Intergenic | C5orf64/miRNA-150/EGFR/PD-L1 |
| CASC11 | ENSG00000249375 | 8 | − | Intergenic | CASC11/EIF4A3/E2F1/NF-κB/PD-L1 |
| CASC11/EIF4A3/E2F1/PI3K/AKT/mTOR/PD-L1 | |||||
| CECR7 | ENSG00000237438 | 22 | + | Intergenic | CECR7/miR-429/CTLA4 |
| EMX2OS | ENSG00000229847 | 10 | − | Antisense | EMXOS/miR-654-3p/AKT3/PD-L1 |
| FGD5-AS1 | ENSG00000225733 | 3 | − | Antisense | FGD5-AS1/miR-454-3p/ZEB1/PD-L1 |
| FGD5-AS1/miR-142/PD-L1 | |||||
| GATA3-AS1 | ENSG00000197308 | 10 | − | Antisense | GATA3-AS1/miR-676-5p/COPS5/PD-L1 stability |
| HOTAIR | ENSG00000228630 | 12 | − | Intergenic | HOTAIR/miR-30a-5p/GRP78/PD-L1 stability |
| HOTTIP | ENSG00000243766 | 7 | + | Antisense | HOTTIP/c-jun/IL-6 |
| HOXA-AS2 | ENSG00000253552 | 7 | + | Antisense | HOXA-AS2/miR-519/PD-L1 |
| HOXA-AS2/miR-519/HIF-1a/PD-L1 | |||||
| KCNQ1OT1 | ENSG00000269821 | 11 | − | Antisense | KCNQ1OT1/miR-30a-5p/USP22/PD-L1 stability |
| KCNQ1OT1/miR-15a/PD-L1 | |||||
| KCNQ1OT1/miR-506/PD-L1 | |||||
| LINC00473 | ENSG00000223414 | 6 | − | Intronic | LINC00473/miR-195-5p/PD-L1 |
| Lnc-Tim3 | ENST00000443947.1 | 7 | + | Intergenic | lnc-Tim3/TIM3 blocking |
| LncMX1–215 | lncMX1–215/GCN5/H3K27ac in PD-L1 and Galectin-9 | ||||
| MALAT1 | ENSG00000251562 | 11 | + | Intergenic | MALAT1/miR-200c/ZEB1/PD-L1 |
| MALAT1/mir-200a/PD-L1 stability | |||||
| MALAT1/miR-195/PD-L1 stability | |||||
| MIAT | ENSG00000225783 | 22 | + | Intergenic | MIAT/miR-411-5p/STAT3/PD-L1 |
| NEAT1 | ENSG00000245532 | 11 | + | Intergenic | NEAT1/miR-155/TIM3 |
| NKX2-1-AS1 | ENSG00000253563 | 14 | + | Pseudogene | NKX2-1-AS1/NKX2-1/PD-L1 |
| NUTM2A-AS1 | ENSG00000223482 | 10 | − | Intergenic | NUTM2A-AS1/miR-376a/PD-L1 |
| PCED1B-AS1 | ENSG00000247774 | 12 | − | Antisense | PCED1B-AS1/miR-149-5p/PD-L1 and PD-L2 |
| SNHG12 | ENSG00000197989 | 1 | − | Antisense | SNHG20/IL-6R/IL6 |
| SNHG14 | ENSG00000224078 | 15 | + | Antisense | SNHG14/miR-5590-3p/ZEB1/PD-L1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saadi, W.; Fatmi, A.; Pallardó, F.V.; García-Giménez, J.L.; Mena-Molla, S. Long Non-Coding RNAs as Epigenetic Regulators of Immune Checkpoints in Cancer Immunity. Cancers 2023, 15, 184. https://doi.org/10.3390/cancers15010184
Saadi W, Fatmi A, Pallardó FV, García-Giménez JL, Mena-Molla S. Long Non-Coding RNAs as Epigenetic Regulators of Immune Checkpoints in Cancer Immunity. Cancers. 2023; 15(1):184. https://doi.org/10.3390/cancers15010184
Chicago/Turabian StyleSaadi, Wiam, Ahlam Fatmi, Federico V. Pallardó, José Luis García-Giménez, and Salvador Mena-Molla. 2023. "Long Non-Coding RNAs as Epigenetic Regulators of Immune Checkpoints in Cancer Immunity" Cancers 15, no. 1: 184. https://doi.org/10.3390/cancers15010184
APA StyleSaadi, W., Fatmi, A., Pallardó, F. V., García-Giménez, J. L., & Mena-Molla, S. (2023). Long Non-Coding RNAs as Epigenetic Regulators of Immune Checkpoints in Cancer Immunity. Cancers, 15(1), 184. https://doi.org/10.3390/cancers15010184





