Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells
Abstract
Simple Summary
Abstract
1. Introduction
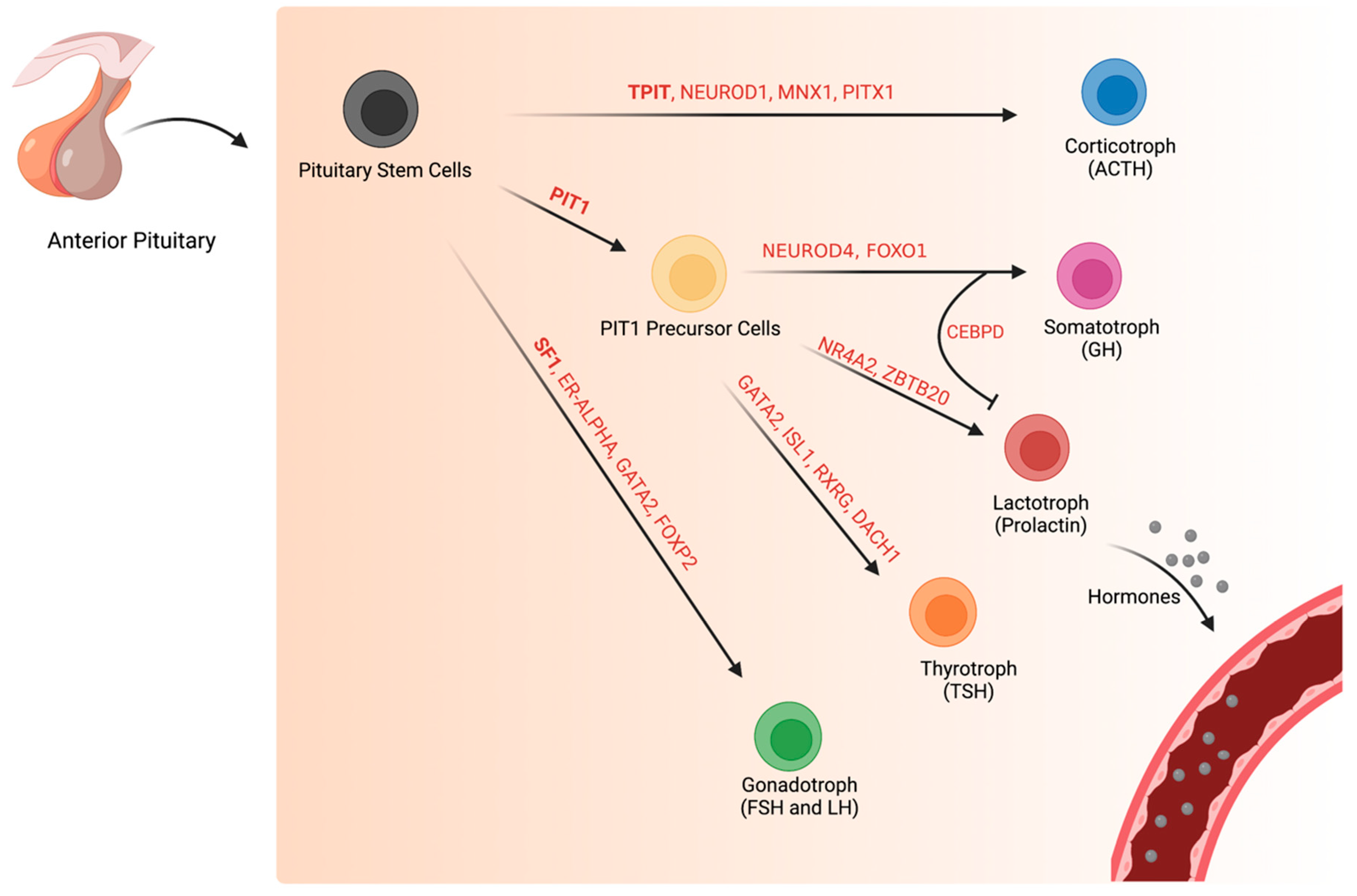
2. Normal Pituitary Gland during and after Development
2.1. Normal Corticotroph (TPIT-Lineage) Transcription Factors during and after Development
2.2. Development of Normal Pituitary Cells of the PIT1-Lineage
2.3. Normal Somatotroph Transcriptional Pathways during and after Development
2.4. Normal Lactotroph Transcriptional Pathways during and after Development
2.5. Normal Thyrotroph Transcriptional Pathways during and after Development
2.6. Normal Gonadotroph (SF1-Lineage) Transcriptional Factors during and after Development
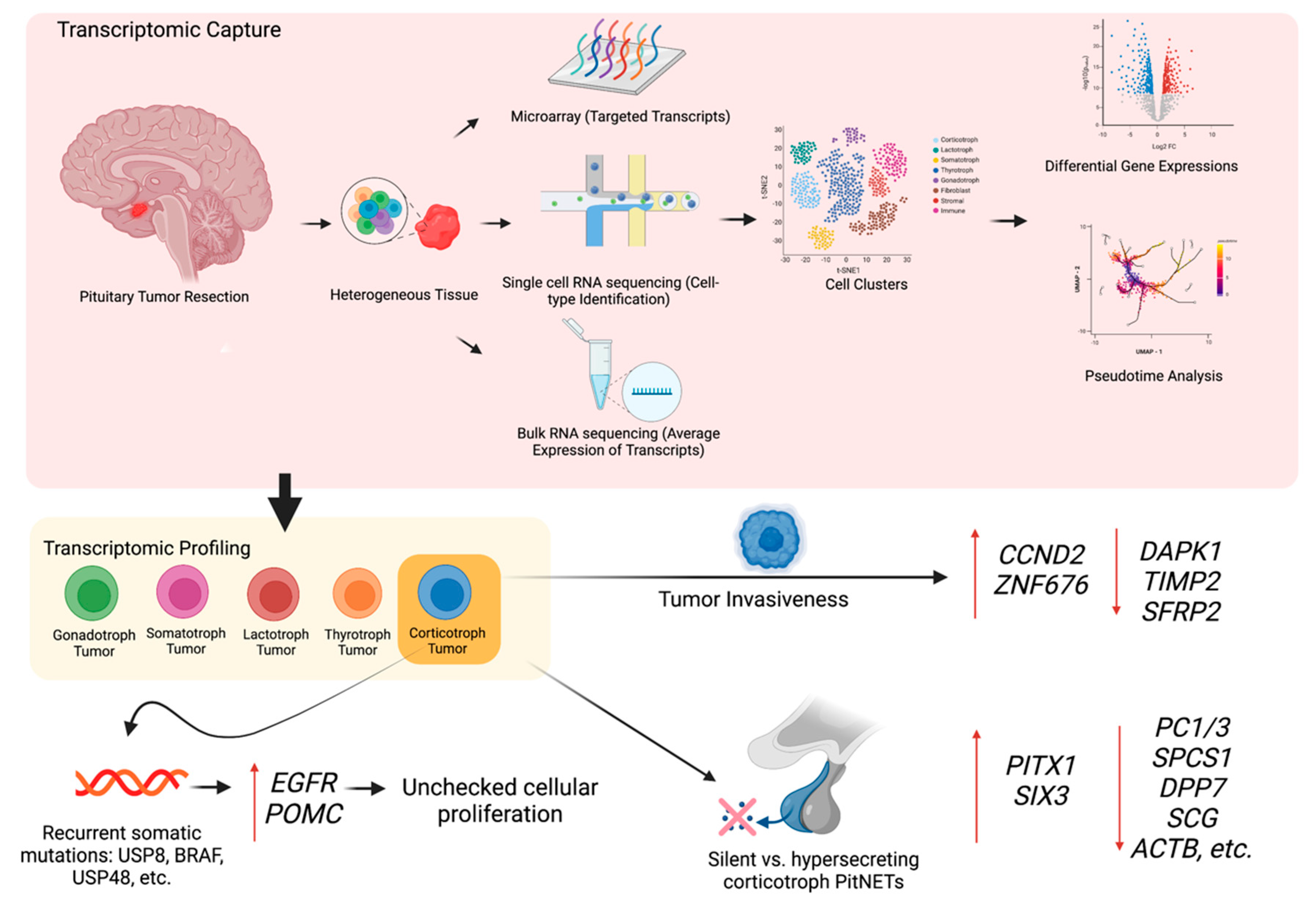
3. The Transcriptomic Landscape of PitNETs
3.1. Corticotroph PitNETs
3.2. Somatotroph PitNETs
3.3. Lactotroph PitNETs
3.4. Thyrotrophic PitNETs
3.5. Gonadotroph PitNETs and Null Cell PitNETs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yoo, E.-S.; Yu, J.; Sohn, J.-W. Neuroendocrine control of appetite and metabolism. Exp. Mol. Med. 2021, 53, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Hoermann, R.; Midgley, J.E.M.; Larisch, R.; Dietrich, J.W. Homeostatic Control of the Thyroid–Pituitary Axis: Perspectives for Diagnosis and Treatment. Front. Endocrinol. 2015, 6, 177. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, S.; Asa, S.L. Mechanisms of disease: The pathogenesis of pituitary tumors. Nat. Clin. Pract. Endocrinol. Metab. 2006, 2, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Molitch, M.E. Diagnosis and Treatment of Pituitary Adenomas: A Review. JAMA 2017, 317, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Drummond, J.; Roncaroli, F.; Grossman, A.B.; Korbonits, M. Clinical and Pathological Aspects of Silent Pituitary Adenomas. J. Clin. Endocrinol. Metab. 2019, 104, 2473–2489. [Google Scholar] [CrossRef]
- Spada, A.; Mantovani, G.; Lania, A.G.; Treppiedi, D.; Mangili, F.; Catalano, R.; Carosi, G.; Sala, E.; Peverelli, E. Pituitary Tumors: Genetic and Molecular Factors Underlying Pathogenesis and Clinical Behavior. NEN 2022, 112, 15–33. [Google Scholar] [CrossRef]
- Melmed, S. Pathogenesis of pituitary tumors. Nat. Rev. Endocrinol. 2011, 7, 257–266. [Google Scholar] [CrossRef]
- Zheng, A.C.; Wang, E.J.; Aghi, M.K. Recent advancements in the molecular biology of pituitary adenomas. Expert Rev. Endocrinol. Metab. 2022, 17, 293–304. [Google Scholar] [CrossRef]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef]
- Jia, Q.; Chu, H.; Jin, Z.; Long, H.; Zhu, B. High-throughput single-cell sequencing in cancer research. Sig. Transduct. Target Ther. 2022, 7, 1–20. [Google Scholar] [CrossRef]
- Zhu, X.; Gleiberman, A.S.; Rosenfeld, M.G. Molecular Physiology of Pituitary Development: Signaling and Transcriptional Networks. Physiol. Rev. 2007, 87, 933–963. [Google Scholar] [CrossRef] [PubMed]
- Lamolet, B.; Pulichino, A.-M.; Lamonerie, T.; Gauthier, Y.; Brue, T.; Enjalbert, A.; Drouin, J. A Pituitary Cell-Restricted T Box Factor, Tpit, Activates POMC Transcription in Cooperation with Pitx Homeoproteins. Cell 2001, 104, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Pulichino, A.-M.; Vallette-Kasic, S.; Tsai, J.P.-Y.; Couture, C.; Gauthier, Y.; Drouin, J. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 2003, 17, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, Y.; Ma, X.; Yong, J.; Yan, L.; Yang, M.; Ren, J.; Tang, F.; Wen, L.; Qiao, J. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat. Commun. 2020, 11, 5275. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Goto, M.; Hojo, M.; Kita, A.; Kitagawa, M.; Ohtsuka, T.; Kageyama, R.; Miyamoto, S. The proneural bHLH genes Mash1, Math3 and NeuroD are required for pituitary development. J. Mol. Endocrinol. 2018, 61, 127–138. [Google Scholar] [CrossRef]
- Lamolet, B.; Poulin, G.; Chu, K.; Guillemot, F.; Tsai, M.-J.; Drouin, J. Tpit-Independent Function of NeuroD1(BETA2) in Pituitary Corticotroph Differentiation. Mol. Endocrinol. 2004, 18, 995–1003. [Google Scholar] [CrossRef]
- Zhang, Z.; Zamojski, M.; Smith, G.R.; Willis, T.L.; Yianni, V.; Mendelev, N.; Pincas, H.; Seenarine, N.; Amper, M.A.S.; Vasoya, M.; et al. Single nucleus transcriptome and chromatin accessibility of postmortem human pituitaries reveal diverse stem cell regulatory mechanisms. Cell Rep. 2022, 38, 110467. [Google Scholar] [CrossRef]
- Harrison, K.A.; Druey, K.M.; Deguchi, Y.; Tuscano, J.M.; Kehrl, J.H. A novel human homeobox gene distantly related to proboscipedia is expressed in lymphoid and pancreatic tissues. J. Biol. Chem. 1994, 269, 19968–19975. [Google Scholar] [CrossRef]
- Arber, S.; Han, B.; Mendelsohn, M.; Smith, M.; Jessell, T.M.; Sockanathan, S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 1999, 23, 659–674. [Google Scholar] [CrossRef]
- Toda, M.; Tamura, R.; Toda, M. Recent Progress in Stem Cell Research of the Pituitary Gland and Pituitary Adenoma. Endocrines 2020, 1, 49–57. [Google Scholar] [CrossRef]
- hu, X.; Zhang, J.; Tollkuhn, J.; Ohsawa, R.; Bresnick, E.H.; Guillemot, F.; Kageyama, R.; Rosenfeld, M.G. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006, 20, 2739–2753. [Google Scholar]
- Kapali, J.; Kabat, B.E.; Schmidt, K.L.; Stallings, C.E.; Tippy, M.; Jung, D.O.; Edwards, B.S.; Nantie, L.B.; Raeztman, L.T.; Navratil, A.M.; et al. Foxo1 Is Required for Normal Somatotrope Differentiation. Endocrinology 2016, 157, 4351–4363. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zhou, J.; Mizutani, J.; Fukuoka, H.; Ren, S.-G.; Gutierrez-Hartmann, A.; Koeffler, H.P.; Melmed, S. CEBPD Suppresses Prolactin Expression and Prolactinoma Cell Proliferation. Mol. Endocrinol. 2011, 25, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Ruf-Zamojski, F.; Zhang, Z.; Zamojski, M.; Smith, G.R.; Mendelev, N.; Liu, H.; Nudelman, G.; Moriwaki, M.; Pincas, H.; Castanon, R.G.; et al. Single nucleus multi-omics regulatory landscape of the murine pituitary. Nat. Commun. 2021, 12, 2677. [Google Scholar] [CrossRef]
- Cheung, L.Y.M.; George, A.S.; McGee, S.R.; Daly, A.Z.; Brinkmeier, M.L.; Ellsworth, B.S.; Camper, S.A. Single-Cell RNA Sequencing Reveals Novel Markers of Male Pituitary Stem Cells and Hormone-Producing Cell Types. Endocrinology 2018, 159, 3910–3924. [Google Scholar] [CrossRef] [PubMed]
- CastaÑo, J.P.; Ramírez, J.L.; Garrido-Gracia, J.C.; Gracia-Navarro, F. Somatotrope Heterogeneity and Its Involvement in Growth Hormone (GH) Regulation. In Sex-Steroid Interactions with Growth Hormone; Veldhuis, J.D., Giustina, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 183–191. [Google Scholar] [CrossRef]
- Bradford, A.P.; Brodsky, K.S.; Diamond, S.E.; Kuhn, L.C.; Liu, Y.; Gutierrez-Hartmann, A. The Pit-1 Homeodomain and β-Domain Interact with Ets-1 and Modulate Synergistic Activation of the Rat Prolactin Promoter *. J. Biol. Chem. 2000, 275, 3100–3106. [Google Scholar] [CrossRef] [PubMed]
- Peel, M.T.; Ho, Y.; Liebhaber, S.A. Transcriptome Analyses of Female Somatotropes and Lactotropes Reveal Novel Regulators of Cell Identity in the Pituitary. Endocrinology 2018, 159, 3965–3980. [Google Scholar] [CrossRef]
- Cao, D.; Ma, X.; Cai, J.; Luan, J.; Liu, A.-J.; Yang, R.; Cao, Y.; Zhu, X.; Zhang, H.; Chen, Y.-X.; et al. ZBTB20 is required for anterior pituitary development and lactotrope specification. Nat. Commun. 2016, 7, 11121. [Google Scholar] [CrossRef]
- Han, Q.; Yan, X.; Ye, Y.; Han, L.; Ma, X.; Wang, T.; Cao, D.; Zhang, W.J. ZBTB20 Regulates Prolactin Expression and Lactotrope Function in Adult Mice. Endocrinology 2022, 163, bqac181. [Google Scholar] [CrossRef]
- Fletcher, P.A.; Smiljanic, K.; Maso Prévide, R.; Iben, J.R.; Li, T.; Rokic, M.B.; Sherman, A.; Coon, S.L.; Stojilkovic, S.S. Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells. Front. Endocrinol. 2019, 10, 623. [Google Scholar] [CrossRef]
- Durand, D.; Pampillo, M.; Caruso, C.; Lasaga, M. Role of metabotropic glutamate receptors in the control of neuroendocrine function. Neuropharmacology 2008, 55, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Hu, P.; Peel, M.T.; Chen, S.; Camara, P.G.; Epstein, D.J.; Wu, H.; Liebhaber, S.A. Single-cell transcriptomic analysis of adult mouse pituitary reveals sexual dimorphism and physiologic demand-induced cellular plasticity. Protein Cell 2020, 11, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Dasen, J.S.; O’Connell, S.M.; Flynn, S.E.; Treier, M.; Gleiberman, A.S.; Szeto, D.P.; Hooshmand, F.; Aggarwal, A.K.; Rosenfeld, M.G. Reciprocal Interactions of Pit1 and GATA2 Mediate Signaling Gradient–Induced Determination of Pituitary Cell Types. Cell 1999, 97, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.Z.; Dudley, L.A.; Peel, M.T.; Liebhaber, S.A.; Parker, S.C.J.; Camper, S.A. Multi-omic profiling of pituitary thyrotropic cells and progenitors. BMC Biol. 2021, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.A.; Saunders, T.L.; Wood, W.M.; Owens, K.; Parlow, A.F.; Camper, S.A.; Ridgway, E.C.; Gordon, D.F. Pituitary-Specific Gata2 Knockout: Effects on Gonadotrope and Thyrotrope Function. Mol. Endocrinol. 2006, 20, 1366–1377. [Google Scholar] [CrossRef]
- Castinetti, F.; Brinkmeier, M.L.; Mortensen, A.H.; Vella, K.R.; Gergics, P.; Brue, T.; Hollenberg, A.N.; Gan, L.; Camper, S.A. ISL1 Is Necessary for Maximal Thyrotrope Response to Hypothyroidism. Mol. Endocrinol. 2015, 29, 1510–1521. [Google Scholar] [CrossRef][Green Version]
- Zhao, L.; Bakke, M.; Krimkevich, Y.; Cushman, L.J.; Parlow, A.F.; Camper, S.A.; Parker, K.L. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development 2001, 128, 147–154. [Google Scholar] [CrossRef]
- Pacini, V.; Petit, F.; Querat, B.; Laverriere, J.-N.; Cohen-Tannoudji, J.; L’hôte, D. Identification of a pituitary ERα-activated enhancer triggering the expression of Nr5a1, the earliest gonadotrope lineage-specific transcription factor. Epigenetics Chromatin 2019, 12, 48. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, X. The molecular pathogenesis of pituitary adenomas: An update. Endocrinol. Metab. 2013, 28, 245–254. [Google Scholar] [CrossRef]
- Mete, O.; Lopes, M.B. Overview of the 2017 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2017, 28, 228–243. [Google Scholar] [CrossRef]
- Asa, S.L.; Mete, O.; Perry, A.; Osamura, R.Y. Overview of the 2022 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2022, 33, 6–26. [Google Scholar] [CrossRef]
- Theodoropoulou, M.; Reincke, M.; Fassnacht, M.; Komada, M. Decoding the genetic basis of Cushing’s disease: USP8 in the spotlight. Eur. J. Endocrinol. 2015, 173, M73–M83. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.-Y.; Song, Z.-J.; Chen, J.-H.; Wang, Y.-F.; Li, S.-Q.; Zhou, L.-F.; Mao, Y.; Li, Y.-M.; Hu, R.-G.; Zhang, Z.-Y.; et al. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res. 2015, 25, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Reincke, M.; Sbiera, S.; Hayakawa, A.; Theodoropoulou, M.; Osswald, A.; Beuschlein, F.; Meitinger, T.; Mizuno-Yamasaki, E.; Kawaguchi, K.; Saeki, Y.; et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat. Genet. 2015, 47, 31–38. [Google Scholar] [CrossRef]
- Song, Z.-J.; Reitman, Z.J.; Ma, Z.-Y.; Chen, J.-H.; Zhang, Q.-L.; Shou, X.-F.; Huang, C.-X.; Wang, Y.-F.; Li, S.-Q.; Mao, Y.; et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res. 2016, 26, 1255–1259. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, E.; Iura, T.; Mukai, A.; Yoshimori, T.; Kitamura, N.; Komada, M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 2005, 16, 5163–5174. [Google Scholar] [CrossRef] [PubMed]
- Niendorf, S.; Oksche, A.; Kisser, A.; Löhler, J.; Prinz, M.; Schorle, H.; Feller, S.; Lewitzky, M.; Horak, I.; Knobeloch, K.-P. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell Biol. 2007, 27, 5029–5039. [Google Scholar] [CrossRef]
- Fukuoka, H.; Cooper, O.; Ben-Shlomo, A.; Mamelak, A.; Ren, S.-G.; Bruyette, D.; Melmed, S. EGFR as a therapeutic target for human, canine, and mouse ACTH-secreting pituitary adenomas. J. Clin. Invest. 2011, 121, 4712–4721. [Google Scholar] [CrossRef]
- Chen, J.; Jian, X.; Deng, S.; Ma, Z.; Shou, X.; Shen, Y.; Zhang, Q.; Song, Z.; Li, Z.; Peng, H.; et al. Identification of recurrent USP48 and BRAF mutations in Cushing’s disease. Nat. Commun. 2018, 9, 3171. [Google Scholar] [CrossRef]
- Sbiera, S.; Perez-Rivas, L.G.; Taranets, L.; Weigand, I.; Flitsch, J.; Graf, E.; Monoranu, C.-M.; Saeger, W.; Hagel, C.; Honegger, J.; et al. Driver mutations in USP8 wild-type Cushing’s disease. Neuro-Oncol. 2019, 21, 1273–1283. [Google Scholar] [CrossRef]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.-L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic Classification of Pituitary Neuroendocrine Tumors. Cancer Cell 2020, 37, 123–134.e5. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Li, C.; Jiang, Z.; Zhang, S.; Li, Q.; Liu, X.; Zhou, Y.; Li, R.; Wei, L.; Li, L.; et al. Single-cell transcriptome and genome analyses of pituitary neuroendocrine tumors. Neuro-Oncol. 2021, 23, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Klein, A.M. Lineage tracing meets single-cell omics: Opportunities and challenges. Nat. Rev. Genet. 2020, 21, 410–427. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, L.J.T.; Lerario, A.M.; de Castro, M.; Martins, C.S.; Bronstein, M.D.; Machado, M.C.; Trarbach, E.B.; Villares Fragoso, M.C.B. Transcriptome Analysis Showed a Differential Signature between Invasive and Non-invasive Corticotrophinomas. Front. Endocrinol. 2017, 8, 55. [Google Scholar] [CrossRef]
- Bourboulia, D.; Stetler-Stevenson, W.G. Matrix MetalloProteinases (MMPs) andTissue Inhibitors of MetalloProteinases (TIMPs): Positive and negative regulators intumor cell adhesion. Semin. Cancer Biol. 2010, 20, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Inbal, B.; Shani, G.; Cohen, O.; Kissil, J.L.; Kimchi, A. Death-Associated Protein Kinase-Related Protein 1, a Novel Serine/Threonine Kinase Involved in Apoptosis. Mol. Cell Biol. 2000, 20, 1044–1054. [Google Scholar] [CrossRef]
- Martins, C.S.; Santana-Lemos, B.A.; Saggioro, F.P.; Neder, L.; Machado, H.R.; Moreira, A.C.; Calado, R.T.; de Castro, M. Telomere length and telomerase expression in pituitary tumors. J. Endocrinol. Invest. 2015, 38, 1243–1246. [Google Scholar] [CrossRef]
- Alzoubi, H.; Minasi, S.; Gianno, F.; Antonelli, M.; Belardinilli, F.; Giangaspero, F.; Jaffrain-Rea, M.-L.; Buttarelli, F.R. Alternative Lengthening of Telomeres (ALT) and Telomerase Reverse Transcriptase Promoter Methylation in Recurrent Adult and Primary Pediatric Pituitary Neuroendocrine Tumors. Endocr. Pathol. 2022, 33, 494–505. [Google Scholar] [CrossRef]
- Ren, J.; Jian, F.; Jiang, H.; Sun, Y.; Pan, S.; Gu, C.; Chen, X.; Wang, W.; Ning, G.; Bian, L.; et al. Decreased expression of SFRP2 promotes development of the pituitary corticotroph adenoma by upregulating Wnt signaling. Int. J. Oncol. 2018, 52, 1934–1946. [Google Scholar] [CrossRef]
- Wu, Q.; Yin, X.; Zhao, W.; Xu, W.; Chen, L. Downregulation of SFRP2 facilitates cancer stemness and radioresistance of glioma cells via activating Wnt/β-catenin signaling. PLoS ONE 2021, 16, e0260864. [Google Scholar] [CrossRef]
- Zhang, D.; Hugo, W.; Bergsneider, M.; Wang, M.B.; Kim, W.; Vinters, H.V.; Heaney, A.P. Single Cell RNA Sequencing in Silent Corticotroph Tumors Confirms Impaired POMC Processing and Provides New Insights into Their Invasive Behavior. Eur. J. Endocrinol. 2022, 187, 49–64. [Google Scholar] [CrossRef]
- Eieland, A.K.; Normann, K.R.; Sundaram, A.Y.M.; Nyman, T.A.; Øystese, K.A.B.; Lekva, T.; Berg, J.P.; Bollerslev, J.; Olarescu, N.C. Distinct Pattern of Endoplasmic Reticulum Protein Processing and Extracellular Matrix Proteins in Functioning and Silent Corticotroph Pituitary Adenomas. Cancers 2020, 12, 2980. [Google Scholar] [CrossRef]
- Asuzu, D.T.; Alvarez, R.; Fletcher, P.A.; Mandal, D.; Johnson, K.; Wu, W.; Elkahloun, A.; Clavijo, P.; Allen, C.; Maric, D.; et al. Pituitary adenomas evade apoptosis via noxa deregulation in Cushing’s disease. Cell Reports 2022, 40, 111223. [Google Scholar] [CrossRef]
- Landis, C.A.; Masters, S.B.; Spada, A.; Pace, A.M.; Bourne, H.R.; Vallar, L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 1989, 340, 692–696. [Google Scholar] [CrossRef]
- SPADA, A.; AROSIO, M.; BOCHICCHIO, D.; BAZZONI, N.; VALLAR, L.; BASSETTI, M.; FAGLIA, G. Clinical, Biochemical, and Morphological Correlates in Patients Bearing Growth Hormone-Secreting Pituitary Tumors with or without Constitutively Active Adenylyl Cyclase. J. Clin. Endocrinol. Metab. 1990, 71, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Drange, M.R.; Melmed, S. Molecular Pathogenesis of Acromegaly. Pituitary 1999, 2, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, L.S.; Liu, J.; Sakamoto, A.; Xie, T.; Chen, M. Minireview: GNAS: Normal and Abnormal Functions. Endocrinology 2004, 145, 5459–5464. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Takahashi, Y. Genetic and Epigenetic Pathogenesis of Acromegaly. Cancers 2022, 14, 3861. [Google Scholar] [CrossRef]
- Efstathiadou, Z.A.; Bargiota, A.; Chrisoulidou, A.; Kanakis, G.; Papanastasiou, L.; Theodoropoulou, A.; Tigas, S.K.; Vassiliadi, D.A.; Alevizaki, M.; Tsagarakis, S. Impact of gsp mutations in somatotroph pituitary adenomas on growth hormone response to somatostatin analogs: A meta-analysis. Pituitary 2015, 18, 861–867. [Google Scholar] [CrossRef]
- Barlier, A.; Gunz, G.; Zamora, A.J.; Morange-Ramos, I.; Figarella-Branger, D.; Dufour, H.; Enjalbert, A.; Jaquet, P. Pronostic and therapeutic consequences of Gs alpha mutations in somatotroph adenomas. J. Clin. Endocrinol. Metab. 1998, 83, 1604–1610. [Google Scholar]
- Adams, E.F.; Lei, T.; Buchfelder, M.; Petersen, B.; Fahlbusch, R. Biochemical characteristics of human pituitary somatotropinomas with and without gsp mutations: In vitro cell culture studies. J. Clin. Endocrinol. Metab. 1995, 80, 2077–2081. [Google Scholar] [PubMed]
- Salomon, M.P.; Wang, X.; Marzese, D.M.; Hsu, S.C.; Nelson, N.; Zhang, X.; Matsuba, C.; Takasumi, Y.; Ballesteros-Merino, C.; Fox, B.A.; et al. The Epigenomic Landscape of Pituitary Adenomas Reveals Specific Alterations and Differentiates Among Acromegaly, Cushing’s Disease and Endocrine-Inactive Subtypes. Clin. Cancer Res. 2018, 24, 4126–4136. [Google Scholar] [CrossRef]
- Gadelha, M.R.; Bronstein, M.D.; Brue, T.; Coculescu, M.; Fleseriu, M.; Guitelman, M.; Pronin, V.; Raverot, G.; Shimon, I.; Lievre, K.K.; et al. Pasireotide versus continued treatment with octreotide or lanreotide in patients with inadequately controlled acromegaly (PAOLA): A randomised, phase 3 trial. Lancet Diabetes Endocrinol. 2014, 2, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Rocheville, M.; Lange, D.C.; Kumar, U.; Patel, S.C.; Patel, R.C.; Patel, Y.C. Receptors for dopamine and somatostatin: Formation of hetero-oligomers with enhanced functional activity. Science 2000, 288, 154–157. [Google Scholar] [CrossRef]
- Ben-Shlomo, A.; Liu, N.-A.; Melmed, S. Somatostatin and dopamine receptor regulation of pituitary somatotroph adenomas. Pituitary 2017, 20, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Colao, A.; Filippella, M.; Pivonello, R.; Somma, C.D.; Faggiano, A.; Lombardi, G. Combined therapy of somatostatin analogues and dopamine agonists in the treatment of pituitary tumours. Eur. J. Endocrinol. 2007, 156, S57–S63. [Google Scholar] [CrossRef]
- Cantone, M.C.; Dicitore, A.; Vitale, G. Somatostatin-Dopamine Chimeric Molecules in Neuroendocrine Neoplasms. J. Clin. Med. 2021, 10, 501. [Google Scholar] [CrossRef]
- Fontanals-Cirera, B.; Hasson, D.; Vardabasso, C.; Di Micco, R.; Agrawal, P.; Chowdhury, A.; Gantz, M.; de Pablos-Aragoneses, A.; Morgenstern, A.; Wu, P.; et al. Harnessing BET Inhibitor Sensitivity Reveals AMIGO2 as a Melanoma Survival Gene. Mol. Cell 2017, 68, 731–744.e9. [Google Scholar] [CrossRef]
- Li, C.; Xie, W.; Rosenblum, J.S.; Zhou, J.; Guo, J.; Miao, Y.; Shen, Y.; Wang, H.; Gong, L.; Li, M.; et al. Somatic SF3B1 hotspot mutation in prolactinomas. Nat. Commun. 2020, 11, 2506. [Google Scholar] [CrossRef]
- apaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 Mutation in Myelodysplasia with Ring Sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Harbour, J.W.; Roberson, E.D.O.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gong, Q.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark. Res. 2020, 8, 38. [Google Scholar] [CrossRef]
- Guo, J.; Li, C.; Fang, Q.; Liu, Y.; Wang, D.; Chen, Y.; Xie, W.; Zhang, Y. The SF3B1R625H mutation promotes prolactinoma tumor progression through aberrant splicing of DLG1. J. Exp. Clin. Cancer Res. 2022, 41, 26. [Google Scholar] [CrossRef]
- Saiardi, A.; Bozzi, Y.; Baik, J.H.; Borrelli, E. Antiproliferative role of dopamine: Loss of D2 receptors causes hormonal dysfunction and pituitary hyperplasia. Neuron 1997, 19, 115–126. [Google Scholar] [CrossRef]
- Asa, S.L.; Kelly, M.A.; Grandy, D.K.; Low, M.J. Pituitary lactotroph adenomas develop after prolonged lactotroph hyperplasia in dopamine D2 receptor-deficient mice. Endocrinology 1999, 140, 5348–5355. [Google Scholar] [CrossRef] [PubMed]
- Bossé, R.; Fumagalli, F.; Jaber, M.; Giros, B.; Gainetdinov, R.R.; Wetsel, W.C.; Missale, C.; Caron, M.G. Anterior pituitary hypoplasia and dwarfism in mice lacking the dopamine transporter. Neuron 1997, 19, 127–138. [Google Scholar] [CrossRef]
- Friedman, E.; Adams, E.F.; Höög, A.; Gejman, P.V.; Carson, E.; Larsson, C.; De Marco, L.; Werner, S.; Fahlbusch, R.; Nordenskjöld, M. Normal structural dopamine type 2 receptor gene in prolactin-secreting and other pituitary tumors. J. Clin. Endocrinol. Metab. 1994, 78, 568–574. [Google Scholar]
- Biagetti, B.; Simò, R. Molecular Pathways in Prolactinomas: Translational and Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 11247. [Google Scholar] [CrossRef]
- Caccavelli, L.; Feron, F.; Morange, I.; Rouer, E.; Benarous, R.; Dewailly, D.; Jaquet, P.; Kordon, C.; Enjalbert, A. Decreased expression of the two D2 dopamine receptor isoforms in bromocriptine-resistant prolactinomas. Neuroendocrinology 1994, 60, 314–322. [Google Scholar] [CrossRef]
- Wu, Z.B.; Zheng, W.M.; Su, Z.P.; Chen, Y.; Wu, J.S.; Wang, C.D.; Lin, C.; Zeng, Y.J.; Zhuge, Q.C. Expression of D2RmRNA isoforms and ERmRNA isoforms in prolactinomas: Correlation with the response to bromocriptine and with tumor biological behavior. J. Neurooncol. 2010, 99, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Caccavelli, L.; Morange-Ramos, I.; Kordon, C.; Jaquet, P.; Enjalbert, A. Alteration of Gα Subunits mRNA Levels in Bromocriptine Resistant Prolactinomas. J. Neuroendocrinol. 1996, 8, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, I.; Rasolonjanahary, R.; Gunz, G.; Bertrand, P.; Delivet, S.; Jedynak, C.P.; Kordon, C.; Peillon, F.; Jaquet, P.; Enjalbert, A. Resistance to Bromocriptine in Prolactinomas. J. Clin. Endocrinol. Metab. 1989, 69, 10. [Google Scholar] [CrossRef] [PubMed]
- Passos, V.Q.; Fortes, M.A.H.Z.; Giannella-Neto, D.; Bronstein, M.D. Genes Differentially Expressed in Prolactinomas Responsive and Resistant to Dopamine Agonists. NEN 2009, 89, 163–170. [Google Scholar] [CrossRef]
- Iorentini, C.; Guerra, N.; Facchetti, M.; Finardi, A.; Tiberio, L.; Schiaffonati, L.; Spano, P.; Missale, C. Nerve Growth Factor Regulates Dopamine D2 Receptor Expression in Prolactinoma Cell Lines via p75NGFR-Mediated Activation of Nuclear Factor-κB. Mol. Endocrinol. 2002, 16, 353–366. [Google Scholar]
- Missale, C.; Boroni, F.; Losa, M.; Giovanelli, M.; Zanellato, A.; Toso, R.D.; Balsari, A.; Spano, P. Nerve growth factor suppresses the transforming phenotype of human prolactinomas. Proc. Natl. Acad. Sci. USA 1993, 90, 7961. [Google Scholar] [CrossRef]
- Wierinckx, A.; Auger, C.; Devauchelle, P.; Reynaud, A.; Chevallier, P.; Jan, M.; Perrin, G.; Fèvre-Montange, M.; Rey, C.; Figarella-Branger, D.; et al. A diagnostic marker set for invasion, proliferation, and aggressiveness of prolactin pituitary tumors. Endocr. Relat. Cancer 2007, 14, 887–900. [Google Scholar] [CrossRef]
- Raverot, G.; Wierinckx, A.; Dantony, E.; Auger, C.; Chapas, G.; Villeneuve, L.; Brue, T.; Figarella-Branger, D.; Roy, P.; Jouanneau, E.; et al. Prognostic factors in prolactin pituitary tumors: Clinical, histological, and molecular data from a series of 94 patients with a long postoperative follow-up. J. Clin. Endocrinol. Metab. 2010, 95, 1708–1716. [Google Scholar] [CrossRef]
- Zhang, X.; Horwitz, G.A.; Prezant, T.R.; Valentini, A.; Nakashima, M.; Bronstein, M.D.; Melmed, S. Structure, Expression, and Function of Human Pituitary Tumor-Transforming Gene (PTTG). Mol. Endocrinol. 1999, 13, 156–166. [Google Scholar] [CrossRef]
- Fong, M.Y.; Farghaly, H.; Kakar, S.S. Tumorigenic potential of pituitary tumor transforming gene (PTTG) in vivoinvestigated using a transgenic mouse model, and effects of cross breeding with p53 (+/−) transgenic mice. BMC Cancer 2012, 12, 532. [Google Scholar] [CrossRef]
- Heaney, A.P.; Horwitz, G.A.; Wang, Z.; Singson, R.; Melmed, S. Early involvement of estrogen-induced pituitary tumor transforming gene and fibroblast growth factor expression in prolactinoma pathogenesis. Nat. Med. 1999, 5, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Mead, T.J. ADAMTS6: Emerging roles in cardiovascular, musculoskeletal and cancer biology. Front Mol. Biosci. 2022, 9, 1023511. [Google Scholar] [CrossRef]
- Sapkota, S.; Horiguchi, K.; Tosaka, M.; Yamada, S.; Yamada, M. Whole-Exome Sequencing Study of Thyrotropin-Secreting Pituitary Adenomas. J. Clin. Endocrinol. Metab. 2017, 102, 566–575. [Google Scholar] [CrossRef]
- Dong, Q.; Brucker-Davis, F.; Weintraub, B.D.; Smallridge, R.C.; Carr, F.E.; Battey, J.; Spiegel, A.M.; Shenker, A. Screening of candidate oncogenes in human thyrotroph tumors: Absence of activating mutations of the G alpha q, G alpha 11, G alpha s, or thyrotropin-releasing hormone receptor genes. J. Clin. Endocrinol. Metab. 1996, 81, 1134–1140. [Google Scholar] [PubMed]
- Ando, S.; Sarlis, N.J.; Oldfield, E.H.; Yen, P.M. Somatic Mutation of TRβ Can Cause a Defect in Negative Regulation of TSH in a TSH-Secreting Pituitary Tumor. J. Clin. Endocrinol. Metab. 2001, 86, 5572–5576. [Google Scholar] [CrossRef][Green Version]
- Tagami, T.; Usui, T.; Shimatsu, A.; Beniko, M.; Yamamoto, H.; Moriyama, K.; Naruse, M. Aberrant Expression of Thyroid Hormone Receptor β Isoform May Cause Inappropriate Secretion of TSH in a TSH-Secreting Pituitary Adenoma. J. Clin. Endocrinol. Metab. 2011, 96, E948–E952. [Google Scholar] [CrossRef]
- Balogun, J.A.; Monsalves, E.; Juraschka, K.; Parvez, K.; Kucharczyk, W.; Mete, O.; Gentili, F.; Zadeh, G. Null cell adenomas of the pituitary gland: An institutional review of their clinical imaging and behavioral characteristics. Endocr. Pathol. 2015, 26, 63–70. [Google Scholar] [CrossRef]
- Haddad, A.F.; Young, J.S.; Oh, T.; Pereira, M.P.; Joshi, R.S.; Pereira, K.M.; Osorio, R.C.; Donohue, K.C.; Peeran, Z.; Sudhir, S.; et al. Clinical characteristics and outcomes of null-cell versus silent gonadotroph adenomas in a series of 1166 pituitary adenomas from a single institution. Neurosurg. Focus 2020, 48, E13. [Google Scholar] [CrossRef]
- Galland, F.; Lacroix, L.; Saulnier, P.; Dessen, P.; Meduri, G.; Bernier, M.; Gaillard, S.; Guibourdenche, J.; Fournier, T.; Evain-Brion, D.; et al. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocr.-Relat. Cancer 2010, 17, 361–371. [Google Scholar] [CrossRef]
- Letellier, E.; Schmitz, M.; Ginolhac, A.; Rodriguez, F.; Ullmann, P.; Qureshi-Baig, K.; Frasquilho, S.; Antunes, L.; Haan, S. Loss of Myosin Vb in colorectal cancer is a strong prognostic factor for disease recurrence. Br. J. Cancer 2017, 117, 1689–1701. [Google Scholar] [CrossRef] [PubMed]
- an, L.; Han, H.; Zuo, H.; Chen, Z.; Du, Y.; Zhao, W.; Gu, J.; Zhang, Z. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int. J. Cancer 2010, 126, 53–64. [Google Scholar]
- Dittmer, J. Biological effects and regulation of IGFBP5 in breast cancer. Front. Endocrinol. 2022, 13, 983793. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.Y.; Osorio, R.C.; Jung, J.; Carrete, L.; Choudhary, N.; Lad, M.; Saha, A.; Aghi, M.K. Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells. Cancers 2023, 15, 110. https://doi.org/10.3390/cancers15010110
Oh JY, Osorio RC, Jung J, Carrete L, Choudhary N, Lad M, Saha A, Aghi MK. Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells. Cancers. 2023; 15(1):110. https://doi.org/10.3390/cancers15010110
Chicago/Turabian StyleOh, Jun Y., Robert C. Osorio, Jangham Jung, Luis Carrete, Nikita Choudhary, Meeki Lad, Atul Saha, and Manish K. Aghi. 2023. "Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells" Cancers 15, no. 1: 110. https://doi.org/10.3390/cancers15010110
APA StyleOh, J. Y., Osorio, R. C., Jung, J., Carrete, L., Choudhary, N., Lad, M., Saha, A., & Aghi, M. K. (2023). Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells. Cancers, 15(1), 110. https://doi.org/10.3390/cancers15010110




