Simple Summary
Around 5–10% of advanced melanoma patients progress early on anti-BRAF targeted therapy and 20–30% respond only with the stabilization of the disease. Presumably, these patients could benefit more from first-line immunotherapy. Resistance to BRAF/MEK inhibitors is generated by genetic and non-genetic factors inherent to a tumor or acquired during therapy. Some of them are well documented as a cause of treatment failure. They are potential predictive markers that could improve patients’ selection for both standard and also alternative therapy as some of them have therapeutic potential. Here, a summary of the most promising predictive and therapeutic targets is presented. This up-to-date knowledge may be useful for further study on implementing more accurate genetic/molecular tests in melanoma treatment.
Abstract
Melanoma is the most aggressive skin cancer, the number of which is increasing worldwide every year. It is completely curable in its early stage and fatal when spread to distant organs. In addition to new therapeutic strategies, biomarkers are an important element in the successful fight against this cancer. At present, biomarkers are mainly used in diagnostics. Some biological indicators also allow the estimation of the patient’s prognosis. Still, predictive markers are underrepresented in clinics. Currently, the only such indicator is the presence of the V600E mutation in the BRAF gene in cancer cells, which qualifies the patient for therapy with inhibitors of the MAPK pathway. The identification of response markers is particularly important given primary and acquired resistance to targeted therapies. Reliable predictive tests would enable the selection of patients who would have the best chance of benefiting from treatment. Here, up-to-date knowledge about the most promising genetic and non-genetic resistance-related factors is described. These are alterations in MAPK, PI3K/AKT, and RB signaling pathways, e.g., due to mutations in NRAS, RAC1, MAP2K1, MAP2K2, and NF1, but also other changes activating these pathways, such as the overexpression of HGF or EGFR. Most of them are also potential therapeutic targets and this issue is also addressed here.
1. Introduction
According to the National Institute of Health (NIH), a biomarker is a feature that can be objectively measured and estimated as an indicator of physiological and pathogenic biologic processes as well as an indicator of response to therapy [1]. In oncology, biomarkers are commonly used for the diagnosis of cancer, evaluation of disease stage and prognosis, as well as prediction and monitoring of treatment outcome [2]. Biomarkers differ in their nature. We can distinguish genetic markers (e.g., changes in the DNA sequence), biochemical markers (e.g., proteins and other substances in plasma), molecular markers (e.g., gene expression signatures), or cytological markers (e.g., lymphocyte infiltration). Concerning melanoma, diagnostic markers are mainly used to differentiate between benign and malignant lesions and to distinguish melanoma from other types of neoplasms, e.g., sarcoma. These are proteins that are less (e.g., S100 family) or more (e.g., MELAN-A/MART-1) specific for melanoma cells [3]. Some parameters of primary tumors are used to estimate the prognosis, e.g., Breslow thickness, ulceration, or mitotic index. The prognostic value is also well established for the level of lactate dehydrogenase (LDH) in the blood of patients with disseminated disease. A high level of this enzyme correlates with shorter overall survival [4]. The least numerous group are predictive markers.
Until 2011, there was no effective drug to prolong the survival of patients with advanced and unresectable cutaneous melanoma. Today, two main new therapeutic strategies are routinely used: molecularly targeted therapy (dabrafenib, vemurafenib, encorafenib, trametinib, cobimetinib, binimetinib) and immunotherapy (pembrolizumab, nivolumab, ipilimumab). Additionally, in the case of the presence of mutations in genes other than BRAF (B-Raf proto-oncogene, serine/threonine kinase), alternative targeted therapy may be considered, e.g., with imatinib, when a mutation in the c-KIT gene is present. It is also possible to treat injectable melanoma with the genetically modified oncolytic virus (talimogene laherparepvec) [5]. Adjuvant therapy with kinase inhibitors (dabrafenib and trametinib) and immunotherapy (pembrolizumab, nivolumab, ipilimumab) for high-risk melanoma are also registered [6]. Although many patients take advantage of these new therapies, some patients do not respond to the treatment, both targeted and immunological. The development of reliable markers of response would allow for better personalization of the treatment and consequently would lead to improved patient survival and lower costs of patient care.
2. Molecularly Targeted Therapy of Skin Melanoma
In 2011, the FDA approved the first targeted drug for the treatment of advanced melanoma, the BRAF protein inhibitor vemurafenib. BRAF is a kinase that belongs to the BRAF-MEK-ERK signaling pathway (MAPK pathways) and is mutated in approximately 60% of skin melanoma cases [7]. The mutant form is nearly five hundred times more active than the wild form of the protein, leading to continuous activation of the MAP kinase signaling pathway and unlimited tumor growth [8]. Detection of BRAFV600 mutation (with V600E being the most frequent aberration and V600K being the second one) in melanoma cells qualifies the patient for targeted therapy with BRAF, MEK inhibitors (BRAFi/MEKi).
Currently, there are three BRAF (vemurafenib, dabrafenib, encorafenib) and three MEK inhibitors (trametinib, kobimetinib, binimetinib), as well as their combinations approved for advanced melanoma treatment. The inhibitors selectively bind to the mutated kinases and inhibit their activity leading to decreased cell proliferation, cell cycle arrest, and induction of apoptosis [9]. Therapy with these drugs is well tolerated and improves the survival of most patients with advanced melanoma [10]. However, the primary and acquired resistance is a significant limitation of this therapeutic strategy and occurs in nearly 100% treated with monotherapy [11]. Primary resistance occurs when the patient, despite a diagnosed mutation in the BRAF oncogene, does not benefit from targeted therapy. It is estimated that 5% to 20% of patients do not respond satisfactorily to treatment despite the presence of mutations in the BRAF gene. The problem concerns approximately 5–15% of patients whose disease progression is observed at the first imaging examination after the initiation of monotherapy [12,13]. In the case of combined therapy (anti-BRAF and anti-MEK), primary resistance is observed in a few percent of patients (disease progression), while 20–30% of treated patients only benefit with disease stabilization [14,15]. The response to the treatment is a favorable prognostic factor affecting the time to progression and overall survival [16]. Unfortunately, if the course of the disease during molecularly targeted treatment is dynamic, the proportion of patients may not qualify for second-line immunotherapy due to poor general conditions. Palliative care remains the only option for such patients.
The possibility of estimating the probability of response to targeted therapy before treatment would allow for a better selection of therapy and thus increase the effectiveness of the treatment of advanced skin melanoma. Therefore, it is important to identify markers of primary or early resistance to this therapy. It seems especially useful as many genetic and molecular mediators of tumor resistance are also potential therapeutic targets.
3. Where Does Resistance to Targeted Therapy in Melanoma Cells Come from?
The main cause of primary resistance of melanoma to targeted therapy is the high heterogeneity and plasticity of this tumor, characterized by the presence of many cell clones within the tumor [17,18]. The formation of multiple clones is favored by the presence of numerous UV-induced mutations in melanoma cells [19], with an average number of about 50 mutations per million base pairs of DNA [20]. By comparison, the frequency of mutations in breast cancer is several per million bp. [19]. In addition to the genetic variability of melanoma, resistance to BRAF inhibitors can also be generated by the microenvironment and epigenetic changes [21]. Therefore, potential predictive markers can be divided into two types: genetic and non-genetic described below, presented in Table 1 and Figure 1.

Table 1.
The most important genetic and non-genetic factors that are involved in melanoma resistance to targeted therapy. The most promising, potential predictive markers are in bold.
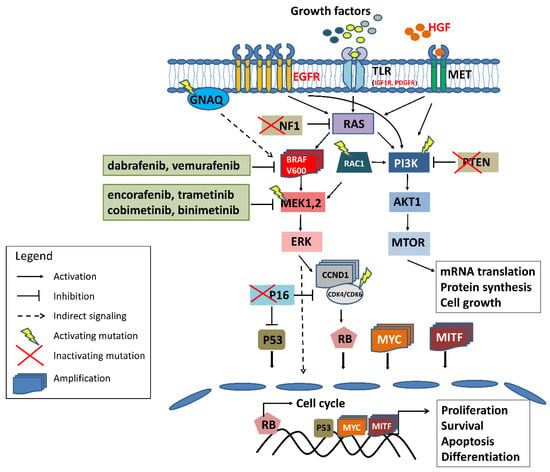
Figure 1.
The scheme of the most common and well-established genetic and molecular mechanisms of melanoma resistance to BRAF/MEK inhibitors. Red font denotes overexpression of the protein; in green frames are the inhibitors registered for advanced skin melanoma treatment. For more detailed information refer to the text.
4. Potential Genetic Predictive Markers in the Targeted Therapy of Melanoma
Resistance to targeted therapy results mainly from the reactivation of the MAPK pathway or activation of an alternative pathway regulating cell division, the PI3K/AKT pathway [52]. So far, several genetic changes which contribute to primary resistance have been identified. These are mutations in the following genes: RAC1, MAP2K1, MAP2K2, NF1, and PTEN. The potential predictive value may also have mutations in the RB1, PIK3CA, MYC, CDKN2A genes, and to a lesser extent CDK4, NRAS, and BRAF amplification [22,24,28].
4.1. RAC1
RAC1 is a signaling molecule that belongs to the RAS superfamily of small GTP-binding proteins. It plays an important role in key features of malignant tumors such as proliferation, metastasis, and resistance to therapies [53]. Mutations in RAC1 occur in skin melanoma with a frequency of 5–9% [54,55]. The most common mutation in this gene is a change in codon 29 (P29S). It is a “gain of function” mutation, which causes a constant activation of the MAPK signaling pathway, increasing the proliferation of melanoma cells [56], and leading to cell resistance to inhibitors of the kinase pathway MAP. It has been shown that cells with this mutation are several times more resistant to inhibitors of BRAF and MEK kinases compared to cells without this mutation. A similar effect was also observed in vivo in a mouse model [25,57]. RAC1P29 mutations are identified in tumors of patients with early resistance [22] and those treated with combined targeted therapy, whose time to progression (TTP) was shorter than the median TTP for this group of patients [24,29]. Analysis of TCGA data revealed significantly shorter relapse-free survival of patients with RAC1P29S mutation in comparison to those with wild-type RAC1 [57]. The above results and the fact that the RAC1P29 mutation is identified in almost one in ten patients suggest that this change should be a part of a potential predictive panel for skin-melanoma-targeted therapy [58]. Importantly, inhibitors for the RAC effector p21-activated kinase (PAK) might impede oncogenic signaling from mutated RAC1 [26], which makes this gene a potential therapeutic target.
4.2. MAP2K1 and MAP2K2
The reactivation of the MAPK pathway during therapy with BRAF and MEK kinase inhibitors is also the result of mutations in the MAP2K1 and MAP2K2 genes. The proteins encoded by these genes (MEK1 and MEK2) are the major kinases of the Ras-Raf-MEK-ERK pathway activated by the BRAF oncogene. Activating mutations in these genes may not only promote the development of melanoma but also reduce the effectiveness of BRAF/MEK inhibitors as a result of activation of the MAPK pathway despite the inhibition of BRAF kinase [59]. Changes in these genes are identified in samples of melanomas of patients with a worse response to targeted therapy (time to progression (TTP) < 4 months; [24] and in patients with primary and acquired resistance to BRAF inhibitors [22,29]. In vitro, functional studies have shown that selected mutations in the MAP2K2 and MAP2K1 genes can generate resistance to MAPK kinase inhibitors [22,59,60]. It seems that the effect of mutations in these genes may be different in the case of monotherapy and combination therapy (combined BRAF and MEK inhibitors); for example, MAP2K1C121S mutation reduces the effectiveness of the BRAF [59,60] and MEK [60] inhibitors used separately, but it seems to not affect the combined effect of both inhibitors [24]. Further research is needed to clarify this point. The results so far suggest that some mutations in the MAP2K1 and MAP2K2 genes may contribute to the primary resistance of melanoma cells to targeted therapy and should be further investigated both functionally and in clinics as a part of the genetic predictive panel. The most common mutations related to melanoma resistance are listed in Table 1.
4.3. NF1
Primary resistance resulting from the simultaneous reactivation of the MAPK pathway and activation of the PI3K/AKT pathway may be generated by a mutation in the NF1 (neurofibromin 1) gene [61]. Mutations in the NF1 gene leading to the inactivation of the encoded protein occur in melanoma with a frequency of 12–30% [62] and are the third most frequent mutation in melanoma (after BRAF and NRAS). The NF1 protein is a tumor suppressor that inhibits the activity of the NRAS oncogene. Lack of NF1 leads to increased activity of the NRAS protein, and thus the activation of both the MAPK pathway and the PI3K/AKT pathways. Therefore, the presence of a mutation in the NF1 gene together with a mutation in the BRAF gene may limit the effectiveness of BRAF/MEK inhibitors. It has been shown that the inhibition of NF1 expression reduces the sensitivity of cells to the BRAF inhibitor by up to 30-fold and to the MEK inhibitor several-fold [23]. Additionally, knockdown of this gene renders melanoma cells resistant to vemurafenib [63]. Mutations in the NF1 gene are also present in the tumor DNA of patients primarily resistant to BRAF inhibitors [23] and patients with early or acquired resistance to these drugs [22,28]. It seems that due to the relatively frequent occurrence of NF1 mutations in melanoma and the fact that the inactivation of this gene reduces the sensitivity of melanoma cells to both types of MAP kinase inhibitors (anti-BRAF and anti-MEK), genetic changes in this gene are a promising predictive marker in targeted therapy of skin melanoma. A limitation of such testing is the size of the gene (282,751 bp; mRNA—about 12,000 bp) and the lack of hot-spot mutations, which in the case of developing a predictive test will involve the sequencing of the entire gene and thus increased costs of the test.
4.4. PTEN
PTEN is a tumor suppressor protein that inhibits the activity of the PI3K/AKT pathway. Changes in the PTEN gene leading to protein inactivation occur in approximately 30–35% of melanoma cases [54]. When the pro-survival action of the MAPK pathway is inhibited by BRAF/MEK inhibitors, the PI3K/AKT pathway can take over this function, thereby reducing the effectiveness of the therapy. Genetic analyses have shown that PTEN mutations are more common in the tumors of patients who did not respond (primary resistance) or responded poorly (PFS < 6 months) compared to those who benefited from this therapy [22,24,64]. Additionally, a shorter progression-free time was observed in patients with a dysfunctional PTEN gene treated with a BRAF inhibitor compared to patients with a wild-type PTEN [64,65]. Some results of in vitro studies suggest that cells with the mutated PTEN gene are less sensitive to BRAF [66] or MEK [67] inhibitors. The potential mechanism involves the upregulation of PERK (EIF2AK3), inhibition of which restores the sensitivity of PTEN-impaired melanoma cells to BRAF inhibitor [68]. It has also been shown that cells lacking a functional PTEN gene are less sensitive to vemurafenib-induced apoptosis [69]. However, in this study, no difference in response to a BRAF inhibitor was observed between PTEN+ and PTEN− cells. Not all patients with impaired PTEN genes show a worse response to therapy with BRAF/MEK inhibitors [22,29]. Therefore, it seems that the lack of a functional PTEN protein may modulate the response to BRAF/MEK therapy, but it is not a sufficient genetic event to generate resistance to these drugs [61]. It is also possible that the effect of PTEN loss may be more relevant for monotherapy, while less important for combined treatment as was shown in the coBRIM study. PTEN loss was associated with shorter PFS only in patients treated with vemurafenib, but not cobimetinib and vemurafenib [70]. Nevertheless, the genetic analysis of this gene should certainly be included in a potential predictive test as it may suggest targeting PI3K/AKT pathway as an alternative to MAPK inhibition in case of resistance development.
4.5. CDKN2A
The impaired functioning of the CDKN2A gene may also have a potential impact on the sensitivity of melanoma cells to targeted therapy. It encodes two (frameshift) proteins: p16INK4a and ARF. Genetic aberrations in CDKN2A are identified in about 40–60% of melanomas [54,71], and almost all lines derived from tumors have a genetically impaired CDKN2A/B-ARF pathway [72]. Changes in this gene, especially deletions, most often lead to a simultaneous impairment of both the p53 pathway (via the ARF protein) and the RB pathway (via the p16INK4a protein). Hereditary mutation in CDKN2A strongly predisposes to skin melanoma and is diagnosed in about 25–50% of families with the aggregation of this cancer [73]. A mutation in the CDKN2A gene is identified in melanomas of patients whose time to progression was shorter than the median for patients treated with BRAF inhibitors [21,24,25]. Forschner [30] et al. described a case of a patient with melanoma harboring a homozygous deletion of CDKN2A, NRAS mutation, as well as amplification of CCNE1 and CDK6. The tumor was refractory to both combined immunotherapy and targeted therapy with BRAF/MEK inhibitors; however, it responded to combined MEK and CDK4/6 inhibition. Similar results were obtained in PDX models of BRAF-V600E-mutant melanoma refractory to BRAFi/MEKi therapy which harbored mutations in NRAS and CDKN2A [31]. CDKN2A mutations are also responsible for 11% of MAPK-reactivating mechanisms of resistance among disease-progressive melanomas described by Shi [74] et al., while a lower copy number of CDKN2A was associated with shorter progression-free survival of patients treated with dabrafenib [65]. The aforementioned data suggest that CDKN2A mutations may contribute to melanoma resistance; however, to my knowledge, there are no in vitro data confirming the influence of this gene dysfunction and individual mutations on the sensitivity of melanoma cells to BRAF/MEK inhibitors.
4.6. Other Resistance-Related Genes: PIK3CA, MYC, CDK4, MAP3K8
Mutations in PIK3CA [22,24,28,29] are identified in pre-treatment tumor samples of patients who respond poorly to targeted therapy, but their role in generating resistance has not yet been verified in melanoma functional studies. PIK3CAE545K was identified as a mutation that pre-existed in rare melanoma subpopulations before therapy and was proved to render resistance to MEK/CDK4 inhibitors [75]. Interestingly, PIK3CAH1047R mutation was also shown to generate resistance of BRAFV600E thyroid cancer to BRAF inhibition in a murine model [76].
The role of the CDK4 gene in generating resistance also requires investigation. In vitro studies indicate that mutations in the CDK4 do not per se induce resistance to BRAF/MEK inhibitors, but rather enhance resistance generated by increased expression of cyclin D1 (CCND1) [77]. Our genetic analysis, on the other hand, suggests that the CDK4R24 mutation may have an impact on the response to targeted therapy. Out of 37 analyzed cases, this mutation was present in three patients treated either with mono or combined therapy whose time to progression (TTP) was shorter than the median TTP for this group of patients [29].
There is also a rationale for the role of MYC in melanoma resistance to BRAF/MEK inhibitors. MYC has been proved to be a convergent downstream effector of resistance in melanoma caused by the reactivation of such pathways as ERK, PI3K, NOTCH1, and others. Its overexpression is related to both intrinsic and acquired resistance of melanoma cells to BRAF/MEK inhibitors [33]. In our study, we have detected VUS and amplification of this oncogene in samples of two patients with a very short time to progression (<3 months) [29]. This gene is definitely worth further study and inclusion in the predictive panel.
In the case of the MAP3K8 (COT) gene, functional studies have shown that its increased expression (as a result of amplification) causes the reactivation of the MAPK pathway and resistance of melanoma cells to MAPK inhibitors [32]. Increased expression of this protein is also present in some clinical specimens of resistant melanomas [32,78]. Interestingly, rearrangements and amplification of the MAP3K8 gene leading to increased levels of the truncated, active form of this protein are detected in approximately 15% of melanomas without driver mutations, such as BRAF, NRAS, or NF1 [79].
All the above-mentioned genes regulate the key signaling pathways in the development of melanoma, namely the MAP kinase pathway (NRAS, MEK1, MEK2, MAP3K8), the PI3K/AKT pathway (PTEN, RAC1, PIK3CA) and the RB pathway (CDKN2A). Mutations in the major genes of these pathways can lead to the reactivation of the MAPK pathway, activation of the alternative PI3K/AKT pathway, or increased proliferation of cells with impaired cell cycle control. These are the main mechanisms of melanoma cell resistance to targeted therapies but are not the only ones. In some cases, mutations in other genes, such as MITF, may play a key role. MITF is a transcription factor that regulates the expression of genes specific to melanocytes, which code for proteins involved in the production of melanin. The MITF pathway is dysregulated in approximately 15% of melanoma cases, and MITF amplification occurs in 10% of primary melanoma and 15% of metastatic diseases [80]. Mutations in this gene are also detected in patient samples both before therapy [22,29] and samples on-progression [22]. It has been shown that increased expression of the MITF oncogene reduces the sensitivity of melanoma cells to targeted therapy by several dozen times compared to cells with wild-type protein [22], although there are also contradictory results, which are described in the next chapter.
Another ‘melanoma’ gene potentially responsible for resistance to targeted therapies is the GNAQ. It belongs to the Gα1/Q pathway, deregulation of which is responsible for the development of eye melanoma [81]. It may also modulate the response to MAP kinase inhibitors. Turajlic [34] et al. described a case of a patient who experienced a very rapid progression during targeted therapy of skin melanoma presumably due to the simultaneous presence of mutations in GNAQ and PTEN genes.
4.7. The Role of BRAF and NRAS in the Acquisition of Melanoma Cells’ Resistance to MAPK Inhibitors
NRAS mutations and BRAF amplification are also often identified in genetic analysis of melanoma samples. In most cases, these changes are detected both in neoplasms that progressed during therapy [36] and melanoma samples collected before treatment from patients who responded to therapy [24]. Thus, it seems that both genetic alterations are more responsible for acquired resistance than for primary resistance. They may occur together with other genetic changes that generate resistance. It is estimated that in approximately 20% of cases, secondary resistance to BRAF/MEK inhibitors is multi-causal [11].
Alternative BRAF splicing is also associated with resistance to BRAFi/MEKi. Melanoma cells with BRAF deprived of exons 4–8 are selected during therapy, which leads to tumor resistance to vemurafenib [37]. The potential mechanism of BRAF splicing variant-related resistance is the enhanced association of BRAF ΔEx with its substrate MEK [82]. Splicing variants of the BRAF oncogene have been identified in the samples of patients with acquired resistance, while such forms were not present in the primary resistant samples [37,38]. This suggests that this mechanism of resistance is responsible for acquired insensitivity rather than for inherent resistance to the treatment.
4.8. Multi-Genetic Cause of Melanoma Resistance to Targeted Therapy
In most melanoma cases, there is more than one mutation [54]. The simultaneous presence of mutations in NRAS and MAP2K1 genes, NRAS mutation and the amplification of BRAF, as well as the presence of two mutations in the NRAS gene (Q61R, T58I) have been described concerning treatment resistance [22]. Additionally, the co-occurrence of GANQ and PTEN mutations was described in a case report of a patient with progressed disease [34]. In our study, we have identified more than one genetic alteration with a potential impact on the response to therapy in several patients, e.g., in a patient refractory to therapy, a mutation in the CDK4 gene and amplification of the MYC oncogene were present [29]. The significance of the presence of co-occurring mutations to therapy resistance requires further research, but the foregoing data suggest that combinations of mutations in MAP2K1, MAP2K2, RAC1, NF1, and mutated BRAF before therapy may be unfavorable for targeted therapy with BRAF/MEK inhibitors. Additionally, co-occurrence of other alterations namely PTEN deletions, MYC amplification, or CDK4 mutations may contribute to resistance to targeted therapy with BRAFi/MEKi, but the data are too scarce and inconclusive.
5. Potential Non-Genetic Predictive Markers in Targeted Therapy of Melanoma
Primary resistance is also influenced by factors regulating the gene expression and those related to the tumor microenvironment that modulates the response to therapy through molecules secreted by non-cancerous tumor cells and the state of hypoxia. In the context of the search for predictive biomarkers, the most useful are proteins that modulate the response to targeted therapy, and their expression changes depending on the degree of sensitivity/resistance of tumor cells to BRAF/MEK inhibitors. The most promising proteins are described below.
5.1. Hepatocyte Growth Factor (HGF)
One of the proteins secreted by tumor-associated stromal cells, the activity of which may promote the resistance of melanoma cells to inhibitors of the MAP kinase pathway, is the hepatocyte growth factor [40]. HGF is a ligand for the c-MET tyrosine kinase receptor. It is secreted by fibroblasts inhabiting the neoplastic tumor and, together with other proteins, determines the pro-neoplastic properties of these fibroblasts [83]. The HGF/c-MET pathway plays a key role in the proliferation, survival, metastasis, and resistance of melanoma cells to MAP kinase inhibitors [84]. It has been shown that HGF secreted by fibroblasts causes the primary insensitivity of melanoma cells to PLX4720 (BRAF inhibitor) as a result of the simultaneous activation of the MAPK and PI3K/AKT pathways. Increased expression of this protein in the extracellular matrix of neoplastic lesions correlates with resistance to therapies [40]. HGF rescues BRAF-mutated cell lines from growth inhibition caused by both mono and combination treatment with BRAF inhibitors and this effect is attenuated by the inhibition of MET signaling [85]. HGF also induces resistance to trametinib in uveal melanoma cells [86] and dasatinib (KIT inhibitor) in acral melanoma [87]. A correlation was also observed between the level of HGF in the blood and the progression-free time (PFS) and overall survival (OS) of patients treated with vemurafenib [88]. HGF is secreted not only by fibroblasts but also by melanoma cells stressed with such factors as drugs, and reduced glucose or oxygen concentration (hypoxia). Together with other proteins, it is responsible for the phenomenon of the so-called tolerance to stress factor-induced drugs (IDTCs, induced drug-tolerant cells), which is reversible after drug holidays [89].
It has been shown that the low oxygen concentration decreases the sensitivity of melanoma cells to vemurafenib several times, which is probably due to the increased activation of the PI3K/AKT and HGF/c-MET pathways [41]. The suggestion that the activation of the HGF/MET pathway under the influence of various factors (hypoxia, fibroblasts) may contribute to a lower sensitivity of melanoma cells to targeted therapies is also confirmed by the observations of an increased level of HGF (mRNA) expression in samples progressing during the therapy compared to samples before therapy [41]. More about the role of the HGF/MET pathway in acquiring resistance to targeted drugs in the treatment of lung and colon cancer, as well as melanoma, can be found in the review by Della Corte [42] et al. The aforementioned results argue for further evaluation of this protein as a predictive marker of melanoma response to targeted therapy.
5.2. Tyrosine Kinases Receptors
Activation of receptor tyrosine kinases (RTKs) and increased expression of growth factors may lead to the stimulation of the MAP kinase pathway and/or the PI3K/AKT pathway and is, therefore, one of the mechanisms of resistance to targeted therapy with BRAF/MEK inhibitors. In melanoma cells, the expression of receptor tyrosine kinases such as EGFR, PDGFRB, and ERBB3 increases upon exposition to BRAF/MEK inhibitors [44].
High expression of EGFR causes primary resistance to vemurafenib in colorectal cancers carrying BRAF oncogene mutations [90]. It cannot be ruled out that also in melanoma, strong expression of this receptor may contribute to early resistance to targeted drugs. In a study by Sun [44] et al., 6 out of 16 patients were characterized by the increased expression of EGFR in tumor samples obtained after targeted treatment. The authors suggest that EGFR expression causes adaptive tolerance to the drug, which is likely to disappear upon discontinuation of the drug. Increased expression of EGFR together with urokinase receptor (uPAR) was also observed in relapsed patients [91]. BRAF-mutant melanoma cells with increased expression of EGFR are less sensitive to BRAF inhibitors [92] and elevated expression of EGFR together with NGFR characterizes pre-resistant melanoma cells, which are selected during therapy and lead to treatment failure [93]. Additionally, the depletion of a negative regulator of EGFR protein degradation (ACK1) results in the accumulation of EGFR and melanoma cells which confers resistance to targeted therapy [94]. The involvement of proteins regulating EGFR in melanoma resistance indirectly proves the role of this receptor in response to targeted therapies [95,96]. It is plausible that EGFR mutations that occur in a certain percentage of melanomas [97,98] may contribute to primary resistance. Genetic studies on a large number of patients would allow the verification of this thesis. On the other hand, due to the significant role of epigenetics in the regulation of EGFR, evaluation of EGFR expression in tumor samples could be more informative than genetic testing. Previous studies on colon cancers suggest the usefulness of EGFR status in predicting response to BRAF inhibitors [99,100].
Insulin-like growth factor receptor 1 (IGF1R) is also involved in generating resistance of melanoma cells to inhibitors of the MAPK pathway. This protein is involved in the transformation of melanocytes [101] and the development of melanoma [102]. IGF1R is stabilized in melanoma cells by the activation of the BRAF oncogene, PTEN suppressor, or by contact with cancer-associated fibroblasts, which leads to its increased expression. Cells with IGF1R overexpression are more resistant to vemurafenib, and the downregulation of this protein leads to the sensitization of melanoma cells to the drug by inducing apoptosis [103]. Activation of the IGF1R–MEK5–Erk5 pathway was also proved to cause resistance to double treatment with BRAF/MEK inhibitors in melanoma cells [104]. Treating resistant melanoma cells with IGF1R inhibitors restores their BRAFi sensitivity [104,105,106], and a combination of BRAF/MEK inhibitors with IGF1R inhibitors significantly reduces melanoma cell growth in vitro and in vivo [107]. Increased expression of IGF1R protein was detected in melanoma samples of relapsed patients [47] suggesting its involvement in acquired resistance. It would be interesting to study the expression of this protein in pre-treatment melanoma samples in patients with primary resistance to BRAFi/MEKi. Possibly, insulin-like growth factor 1 (IGF1) also contributes to the resistance of melanoma cells to BRAF inhibitors. It is one of the proteins secreted by melanoma cells sensitive to the BRAF inhibitor during treatment. This unique therapy-induced secretome activates the PI3K/AKT pathway in resistant cells, leading to their growth and treatment failure [108].
PDGFB (PDGFRB) and PDGFA (PDGFRA) receptors may also play a certain role in generating resistance to MAPK inhibitors. These are tyrosine kinases that act as receptors for the PDGF family of growth factors that regulate such processes as embryonic development or wound healing [109]. They play an important role in the process of carcinogenesis, as well as the resistance of various cancers to therapies [110]. It has been shown that increased PDGFRB expression in melanoma cells makes them resistant to the BRAF inhibitor. High expression of this protein is also present in some treatment-resistant metastatic lesions [45,111]. Similar results were obtained for PDGFRA. Increased expression of this protein causes cell resistance to vemurafenib, while the inhibition of PDGFRA activity restores cell sensitivity to this inhibitor [46].
Functional studies of the effects of tyrosine kinase receptors on the sensitivity of melanoma cells to BRAF/MEK inhibitors indicate that these proteins may be involved in generating resistance to these drugs. However, further studies are needed to assess the potential relationship between the expression of the aforementioned proteins and the response to the targeted therapies concerning the usefulness of these proteins as predictive biomarkers.
5.3. Other Proteins Involved in the Generation of Resistance to Targeted Therapy in Melanoma
One of the proteins involved in the generation of resistance to BRAF/MEK inhibitors is the MITF oncogene. MITF is the major regulator of the plasticity of the melanoma cell phenotype. Its increased level induces cell proliferation, while the decrease in expression gives a signal to a more invasive phenotype regulated by the proteins WNT5A and NFĸB [49]. Lack of MITF expression generates not only acquired resistance but also significantly reduces the sensitivity to BRAF and MEK inhibitors of cells that have not been previously treated with these compounds. Cell resistance correlates with increased expression of AXL, EGFR, or PDGFRB tyrosine kinase receptors. It is suggested that a low MITF/AXL expression ratio may be a predictor of response to targeted therapy. Low MITF expression may also modulate the response to MAPK inhibitors in cooperation with NFĸB. Konieczkowski [50] et al. observed that low MITF expression and high NFĸB characterized melanoma cells resistant to MAPK inhibitors. Reduced expression of MITF during the development of resistance was also observed in the majority of melanoma cell populations derived from patients [112]. On the other hand, very high expression of MITF may also result in a decrease in the sensitivity of cells to MEK inhibitors [22,51]. The role of interplay between MITF, AXL, and other factors in the tumor microenvironment was comprehensively described in the review paper by Arozarena [113] et al. The influence of MITF on melanoma cell resistance is not fully understood [18]; therefore, for now, the use of this protein expression as a predictive marker is questionable. Assessing this protein together with other molecules, namely AXL and NFĸB, seems more rational. The issue is even more complicated by the fact that MITF regulates the genetic phenotype switching program that governs drug addiction [114]. The dependency of tumors on therapeutic drugs to which they have acquired resistance is a fascinating phenomenon [115]; however, it is beyond the scope of this review paper.
Other proteins that can modulate resistance to BRAF/MEK inhibitors are RIP1 [116] and FRA1 (FOSL1) [108]. RIP1 is a serine-threonine kinase involved in the regulation of inflammation and death. This protein has been shown to inhibit apoptosis of cells treated with MAPK pathway inhibitors by activating the NFĸB pathway, which, according to the authors, may be responsible for both primary and acquired resistance of cells to the above-mentioned inhibitors [116]. FRA1, on the other hand, regulates the secretion of compounds (TIS, therapy-induced secretome) in cells sensitive to BRAF/MEK inhibitors, which stimulate the growth of resistant cells [108]. This defensive stress response promotes the growth of cells resistant to the therapy and also helps circulating tumor cells to repopulate regressed tumors. Decreased FRA1 expression was observed in cell lines sensitive to targeted therapy (both melanoma and lung cancer) and patients’ tumors before treatment. Functional tests have confirmed the key role of this molecule in inducing resistance to MAPK inhibitors [108] as well as the addiction of melanoma cells to BRAFi/MEKi [114].
Recent studies suggest that, paradoxically, functional TP53 may contribute to melanoma resistance to targeted therapy. TP53 induces a therapy-resistant phenotype of slow-cycling cells in response to stress (e.g., therapy) and its inhibition promotes sensitivity to BRAF/MEK inhibitors [117]. The opposite role is suggested for other tumor suppressors RASSF1A. It is frequently silenced in melanoma cells and as a hub of many signaling pathways has the potential to modulate resistance to targeted therapy [118]. Other proteins with potential predictive capacity involve those regulating metabolism. Drug-tolerant melanoma cells acquire dependencies on fatty acid metabolism [119] and some proteins regulating this process are overexpressed in non-responding patients, e.g., PPAR [120]. Additionally, increased activity and expression of ABL1/2 kinases were proved to induce resistance to BRAF/MEK inhibitors and ABL kinase inhibitor, nilotinib caused the prolonged regression of resistant tumors [121]. STAT3–PAX3 signaling is also involved in generating the resistance of melanoma cells to BRAF inhibitors [122]. However, data on the aforementioned proteins and their role in resistance generation are scarce. Thus, they are not considered predictive markers, yet.
6. Conclusions
The existing clinical data show that patients with advanced and dynamic diseases should be treated with kinase inhibitors first, and immunotherapy should be considered only in the case of progression. Unfortunately, some patients after the failure of anti-BRAF therapy do not qualify for immunotherapy due to their poor general condition.
Therefore, apart from mutations in the BRAF gene, additional predictive factors are needed, that will allow the selection of patients for appropriate therapy. There are genetic and non-genetic mechanisms of primary and acquired resistance to BRAF and MEK kinase inhibitors (Figure 1). Some of them are common to primary and acquired resistance, such as mutations in the MEK1, PTEN, CDKN2A, or PIK3CA genes, while some seem to be specific to primary (RAC1, MEK1 mutations) or acquired (BRAF amplification, NRAS mutations).
From the clinical point of view, those that would allow better qualification of patients for targeted therapy and at the same time indicate potential new therapeutic targets in the treatment of advanced melanoma are important. The genes with the highest predictive potential are highlighted in Table 1. The table contains the most studied genetic and non-genetic factors.
Some of them have not been functionally tested and most of them lack large genetic and histological analyses of the clinical material. Therefore, in addition to research on cell models, integrated analysis of several or more potential predictive markers performed on a large group of patients is needed. Additionally, the global integration of genomic and clinical data would further help the identification of potential biomarkers [123]. This would allow for a reliable statistical analysis of the relationship between the presence of mutations in selected genes and the expression of selected proteins with the response to the treatment. Many potential genetic predictive markers are at the same time therapeutic targets as indicated in Table 1, so ideally, the predictive panel would also constitute a panel of alternative therapeutic targets. A predictive toolkit that, in my opinion, is worth further validation would consist of four tests:
- Targeted sequencing of seven genes, namely: BRAF, NRAS, NF1, RAC1, MAP2K1, MAP2K2, and MAP3K8 (in tumor or liquid biopsy sample)
- Copy number analysis of five genes, namely: PTEN, CDKN2A, MYC, MITF, and BRAF (in tumor or liquid biopsy sample)
- Protein level analysis of selected growth factors and receptors, namely: HGF, EGFR, PDGFRA, PDGFRB, and IGF1R (in a tumor sample).
- Optionally: determining HGF protein level in a blood sample and MITF protein expression in a tumor sample.
Based on the results of these tests, a board of specialists, i.e., a medical geneticist, molecular biologist, pathologist, and oncologist, would choose the best treatment option tailored to the individual genetic and protein profile of the patient. The genetic part of the test could also be used for further monitoring of melanoma evolution and adapting, if possible, the therapy to the current tumor genetic profile of a patient.
Funding
This research received no external funding.
Acknowledgments
I would like to thank Wiesława Widłak for reading the text and for her valuable remarks.
Conflicts of Interest
The author declares no conflict of interest.
References
- Atkinson, A.J.; Colburn, W.A.; DeGruttola, V.G.; DeMets, D.L.; Downing, G.J.; Hoth, D.F.; Oates, J.A.; Peck, C.C.; Schooley, R.T.; Spilker, B.A.; et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Therap. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Kamel, H.F.M.; Al-Amodi, H.S.A.B. Exploitation of Gene Expression and Cancer Biomarkers in Paving the Path to Era of Personalized Medicine. Genom. Proteom. Bioinf. 2017, 15, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Ohsie, S.J.; Sarantopoulos, G.P.; Cochran, A.J.; Binder, S.W. Immunohistochemical characteristics of melanoma. J. Cutan. Pathol. 2008, 35, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Pala, L.; Conforti, F.; Cocorocchio, E. Talimogene Laherparepvec (T-VEC): An Intralesional Cancer Immunotherapy for Advanced Melanoma. Cancers 2021, 13, 1383. [Google Scholar] [CrossRef] [PubMed]
- Olbryt, M. Molecular background of skin melanoma development and progression: Therapeutic implications. Postep. Derm. Alergol. 2019, 36, 129–138. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Gray-Schopfer, V.C.; Dias, S.D.; Marais, R. The role of B-RAF in melanoma. Cancer Metast. Rev. 2005, 24, 165–183. [Google Scholar] [CrossRef]
- Krämer, A.; Lu, J. Small Molecules in Oncology, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 77–91. [Google Scholar]
- Ugurel, S.; Rohmel, J.; Ascierto, P.A.; Flaherty, K.T.; Grob, J.J.; Hauschild, A.; Larkin, J.; Long, G.V.; Lorigan, P.; McArthur, G.A.; et al. Survival of patients with advanced metastatic melanoma: The impact of novel therapies-update 2017. Eur. J. Cancer 2017, 83, 247–257. [Google Scholar] [CrossRef]
- Zalesna, I.; Hartman, M.L.; Czyz, M. BRAF mutation in progression and therapy of melanoma, papillary thyroid carcinoma and colorectal adenocarcinoma. Postepy Hig. Med. Dosw. 2016, 70, 471–488. [Google Scholar] [CrossRef]
- Sosman, J.A.; Kim, K.B.; Schuchter, L.; Gonzalez, R.; Pavlick, A.C.; Weber, J.S.; McArthur, G.A.; Hutson, T.E.; Moschos, S.J.; Flaherty, K.T.; et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012, 366, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, A.; Grob, J.J.; Demidov, L.V.; Jouary, T.; Gutzmer, R.; Millward, M.; Rutkowski, P.; Blank, C.U.; Miller, W.H.; Kaempgen, E.; et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012, 380, 358–365. [Google Scholar] [CrossRef]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: A multicentre, double-blind, phase 3 randomised controlled trial. Lancet 2015, 386, 444–451. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined Vemurafenib and Cobimetinib in BRAF-Mutated Melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.D.; Larkin, J.; Ribas, A.; Flaherty, K.T.; McArthur, G.A.; Ascierto, P.A.; Dreno, B.; Yan, Y.; Wongchenko, M.; McKenna, E.; et al. Impact of depth of response on survival in patients treated with cobimetinib +/− vemurafenib: Pooled analysis of BRIM-2, BRIM-3, BRIM-7 and coBRIM. Br. J. Cancer 2019, 121, 522–528. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Hess, A.R.; Seftor, R.E. Molecular plasticity of human melanoma cells. Oncogene 2003, 22, 3070–3075. [Google Scholar] [CrossRef]
- Ahmed, F.; Haass, N.K. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity as a Mechanism of Melanoma Therapy Resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175. [Google Scholar] [CrossRef]
- Proietti, I.; Skroza, N.; Bernardini, N.; Tolino, E.; Balduzzi, V.; Marchesiello, A.; Michelini, S.; Volpe, S.; Mambrin, A.; Mangino, G.; et al. Mechanisms of Acquired BRAF Inhibitor Resistance in Melanoma: A Systematic Review. Cancers 2020, 12, 2801. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, S.R.; Theurillat, J.P.; Van Allen, E.; Wagle, N.; Hsiao, J.; Cowley, G.S.; Schadendorf, D.; Root, D.E.; Garraway, L.A. A Genome-Scale RNA Interference Screen Implicates NF1 Loss in Resistance to RAF Inhibition. Cancer Discov. 2013, 3, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef] [PubMed]
- Watson, I.R.; Li, L.R.; Cabeceiras, P.K.; Mahdavi, M.; Fang, Z.N.; Stemke-Hale, K.; Mills, G.B.; Chin, L. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014, 74, 4845–4852. [Google Scholar] [CrossRef] [PubMed]
- Araiza-Olivera, D.; Feng, Y.; Semenova, G.; Prudnikova, T.Y.; Rhodes, J.; Chernoff, J. Suppression of RAC1-driven malignant melanoma by group A PAK inhibitors. Oncogene 2018, 37, 944–952. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.B.; Wongchenko, M.J.; Robert, C.; Larkin, J.; Ascierto, P.A.; Dreno, B.; Maio, M.; Garbe, C.; Chapman, P.B.; Sosman, J.A.; et al. Genomic Features of Exceptional Response in Vemurafenib +/− Cobimetinib-treated Patients with BRAF(V600)-mutated Metastatic Melanoma. Clin. Cancer Res. 2019, 25, 3239–3246. [Google Scholar] [CrossRef]
- Wheler, J.; Yelensky, R.; Falchook, G.; Kim, K.B.; Hwu, P.; Tsimberidou, A.M.; Stephens, P.J.; Hong, D.; Cronin, M.T.; Kurzrock, R. Next generation sequencing of exceptional responders with BRAF-mutant melanoma: Implications for sensitivity and resistance. BMC Cancer 2015, 15, 61. [Google Scholar] [CrossRef][Green Version]
- Olbryt, M.; Piglowski, W.; Rajczykowski, M.; Pfeifer, A.; Student, S.; Fiszer-Kierzkowska, A. Genetic Profiling of Advanced Melanoma: Candidate Mutations for Predicting Sensitivity and Resistance to Targeted Therapy. Target. Oncol. 2020, 15, 101–113. [Google Scholar] [CrossRef]
- Forschner, A.; Sinnberg, T.; Mroz, G.; Schroeder, C.; Reinert, C.P.; Gatidis, S.; Bitzer, M.; Eigentler, T.; Garbe, C.; Niessner, H.; et al. Case Report: Combined CDK4/6 and MEK Inhibition in Refractory CDKN2A and NRAS Mutant Melanoma. Front. Oncol. 2021, 11, 643156. [Google Scholar] [CrossRef]
- Nassar, K.W.; Hintzsche, J.D.; Bagby, S.M.; Espinoza, V.; Langouet-Astrie, C.; Amato, C.M.; Chimed, T.S.; Fujita, M.; Robinson, W.; Tan, A.C.; et al. Targeting CDK4/6 Represents a Therapeutic Vulnerability in Acquired BRAF/MEK Inhibitor-Resistant Melanoma. Mol. Cancer Ther. 2021, 20, 2049–2060. [Google Scholar] [CrossRef]
- Johannessen, C.M.; Boehm, J.S.; Kim, S.Y.; Thomas, S.R.; Wardwell, L.; Johnson, L.A.; Emery, C.M.; Stransky, N.; Cogdill, A.P.; Barretina, J.; et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010, 468, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Singleton, K.R.; Crawford, L.; Tsui, E.; Manchester, H.E.; Maertens, O.; Liu, X.J.; Liberti, M.V.; Magpusao, A.N.; Stein, E.M.; Tingley, J.P.; et al. Melanoma Therapeutic Strategies that Select against Resistance by Exploiting MYC-Driven Evolutionary Convergence. Cell Rep. 2017, 21, 2796–2812. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Furney, S.J.; Stamp, G.; Rana, S.; Ricken, G.; Oduko, Y.; Saturno, G.; Springer, C.; Hayes, A.; Gore, M.; et al. Whole-genome sequencing reveals complex mechanisms of intrinsic resistance to BRAF inhibition. Ann. Oncol. 2014, 25, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Lovly, C.M.; Flavin, M.; Panageas, K.S.; Ayers, G.D.; Zhao, Z.G.; Iams, W.T.; Colgan, M.; DeNoble, S.; Terry, C.R.; et al. Impact of NRAS Mutations for Patients with Advanced Melanoma Treated with Immune Therapies. Cancer Immunol. Res. 2015, 3, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Menzies, A.M.; Zimmer, L.; Eroglu, Z.; Ye, F.; Zhao, S.; Rizos, H.; Sucker, A.; Scolyer, R.A.; Gutzmer, R.; et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 2015, 51, 2792–2799. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.J.; Ng, C.; Moriceau, G.; Shi, H.B.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Pupo, G.M.; Boyd, S.C.; Fung, C.; Carlino, M.S.; Menzies, A.M.; Pedersen, B.; Johansson, P.; Hayward, N.K.; Kefford, R.F.; Scolyer, R.A.; et al. Clinical significance of intronic variants in BRAF inhibitor resistant melanomas with altered BRAF transcript splicing. Biomark Res. 2017, 5, 17. [Google Scholar] [CrossRef]
- Ajiro, M.; Zheng, Z.M. Vemurafenib-resistant BRAF selects alternative branch points different from its wild-type BRAF in intron 8 for RNA splicing. Cell Biosci. 2015, 5, 70. [Google Scholar] [CrossRef]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.Y.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef]
- Qin, Y.; Roszik, J.; Chattopadhyay, C.; Hashimoto, Y.; Liu, C.W.; Cooper, Z.A.; Wargo, J.A.; Hwu, P.; Ekmekcioglu, S.; Grimm, E.A. Hypoxia-Driven Mechanism of Vemurafenib Resistance in Melanoma. Mol. Cancer Ther. 2016, 15, 2442–2454. [Google Scholar] [CrossRef]
- Della Corte, C.M.; Fasano, M.; Papaccio, F.; Ciardiello, F.; Morgillo, F. Role of HGF-MET Signaling in Primary and Acquired Resistance to Targeted Therapies in Cancer. Biomedicines 2014, 2, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Pucciarelli, D.; Lengger, N.; Takacova, M.; Csaderova, L.; Bartosova, M.; Breiteneder, H.; Pastorekova, S.; Hafner, C. Hypoxia increases the heterogeneity of melanoma cell populations and affects the response to vemurafenib. Mol. Med. Rep. 2016, 13, 3281–3288. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, L.; Huang, S.; Heynen, G.J.; Prahallad, A.; Robert, C.; Haanen, J.; Blank, C.; Wesseling, J.; Willems, S.M.; et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014, 508, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Shi, H.B.; Wang, Q.; Kong, X.J.; Koya, R.C.; Lee, H.; Chen, Z.G.; Lee, M.K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance toB-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Sabbatino, F.; Wang, Y.Y.; Wang, X.H.; Flaherty, K.T.; Pepin, D.; Scognamiglio, G.; Yu, L.; Cooper, Z.A.; Pepe, S.; Kirkwood, J.M.; et al. PDGFR alpha up-regulation mediated by Sonic Hedgehog Pathway activation leads to BRAF inhibitor resistance in melanoma cells with BRAF mutation. Oncotarget 2014, 5, 1926–1941. [Google Scholar] [CrossRef]
- Villanueva, J.; Vultur, A.; Lee, J.T.; Somasundaram, R.; Fukunaga-Kalabis, M.; Cipolla, A.K.; Wubbenhorst, B.; Xu, X.W.; Gimotty, P.A.; Kee, D.; et al. Acquired Resistance to BRAF Inhibitors Mediated by a RAF Kinase Switch in Melanoma Can Be Overcome by Cotargeting MEK and IGF-1R/PI3K. Cancer Cell 2010, 18, 683–695. [Google Scholar] [CrossRef]
- Wang, J.; Sinnberg, T.; Niessner, H.; Dolker, R.; Sauer, B.; Kempf, W.E.; Meier, F.; Leslie, N.; Schittek, B. PTEN regulates IGF-1R-mediated therapy resistance in melanoma. Pigm. Cell Melanoma Res. 2015, 28, 572–589. [Google Scholar] [CrossRef]
- Muller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.Y.; Kong, X.J.; Possik, P.A.; Cornelissen-Steijger, P.D.M.; Foppen, M.H.G.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712. [Google Scholar] [CrossRef]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A Melanoma Cell State Distinction Influences Sensitivity to MAPK Pathway Inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef]
- Najem, A.; Krayem, M.; Sales, F.; Hussein, N.; Badran, B.; Robert, C.; Awada, A.; Journe, F.; Ghanem, G.E. P53 and MITF/Bcl-2 identified as key pathways in the acquired resistance of NRAS-mutant melanoma to MEK inhibition. Eur. J. Cancer 2017, 83, 154–165. [Google Scholar] [CrossRef]
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many ways to resistance: How melanoma cells evade targeted therapies. BBA Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Aske, J.C.; Dey, N. RAC1 Takes the Lead in Solid Tumors. Cells 2019, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.R.; Place, C.; et al. A Landscape of Driver Mutations in Melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Krauthammer, M.; Kong, Y.; Ha, B.H.; Evans, P.; Bacchiocchi, A.; McCusker, J.P.; Cheng, E.; Davis, M.J.; Goh, G.; Choi, M.; et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012, 44, 1006. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.Y.; Miller, D.M.; Tsao, H. Somatic Driver Mutations in Melanoma. Cancer Am. Cancer Soc. 2017, 123, 2104–2117. [Google Scholar] [CrossRef]
- Lionarons, D.A.; Hancock, D.C.; Rana, S.; East, P.; Moore, C.; Murillo, M.M.; Carvalho, J.; Spencer-Dene, B.; Herbert, E.; Stamp, G.; et al. RAC1(P29S) Induces a Mesenchymal Phenotypic Switch via Serum Response Factor to Promote Melanoma Development and Therapy Resistance. Cancer Cell 2019, 36, 68. [Google Scholar] [CrossRef]
- Colon-Bolea, P.; Garcia-Gomez, R.; Casar, B. RAC1 Activation as a Potential Therapeutic Option in Metastatic Cutaneous Melanoma. Biomolecules 2021, 11, 1554. [Google Scholar] [CrossRef]
- Yang, H.; Kircher, D.A.; Kim, K.H.; Grossmann, A.H.; VanBrocklin, M.W.; Holmen, S.L.; Robinson, J.P. Activated MEK cooperates with Cdkn2a and Pten loss to promote the development and maintenance of melanoma. Oncogene 2017, 36, 3842–3851. [Google Scholar] [CrossRef]
- Wagle, N.; Emery, C.; Berger, M.F.; Davis, M.J.; Sawyer, A.; Pochanard, P.; Kehoe, S.M.; Johannessen, C.M.; MacConaill, L.E.; Hahn, W.C.; et al. Dissecting Therapeutic Resistance to RAF Inhibition in Melanoma by Tumor Genomic Profiling. J. Clin. Oncol. 2011, 29, 3085–3096. [Google Scholar] [CrossRef]
- Manzano, J.L.; Layos, L.; Buges, C.; Gil, M.D.; Vila, L.; Martinez-Balibrea, E.; Martinez-Cardus, A. Resistant mechanisms to BRAF inhibitors in melanoma. Ann. Transl. Med. 2016, 4, 1–9. [Google Scholar] [CrossRef]
- Philpott, C.; Tovell, H.; Frayling, I.M.; Cooper, D.N.; Upadhyaya, M. The NF1 somatic mutational landscape in sporadic human cancers. Hum. Genom. 2017, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, M.; Feddersen, C.R.; Varzavand, A.; Zhu, E.Y.; Foley, T.; Zhao, L.; Holt, K.H.; Milhem, M.; Piper, R.; Stipp, C.S.; et al. Functional Genomic Screening Independently Identifies CUL3 as a Mediator of Vemurafenib Resistance via Src-Rac1 Signaling Axis. Front. Oncol. 2020, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Catalanotti, F.; Cheng, D.T.; Shoushtari, A.N.; Johnson, D.B.; Panageas, K.S.; Momtaz, P.; Higham, C.; Won, H.H.; Harding, J.J.; Merghoub, T.; et al. PTEN Loss-of-Function Alterations Are Associated With Intrinsic Resistance to BRAF Inhibitors in Metastatic Melanoma. JCO Precis. Oncol. 2017, 1, 54. [Google Scholar] [CrossRef] [PubMed]
- Nathanson, K.L.; Martin, A.M.; Wubbenhorst, B.; Greshock, J.; Letrero, R.; D’Andrea, K.; O’Day, S.; Infante, J.R.; Falchook, G.S.; Arkenau, H.T.; et al. Tumor Genetic Analyses of Patients with Metastatic Melanoma Treated with the BRAF Inhibitor Dabrafenib (GSK2118436). Clin. Cancer Res. 2013, 19, 4868–4878. [Google Scholar] [CrossRef]
- Zuo, Q.; Liu, J.; Huang, L.P.; Qin, Y.F.; Hawley, T.; Seo, C.; Merlino, G.; Yu, Y.L. AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma. Oncogene 2018, 37, 3275–3289. [Google Scholar] [CrossRef]
- Byron, S.A.; Loch, D.C.; Wellens, C.L.; Wortmann, A.; Wu, J.Y.; Wang, J.; Nomoto, K.; Pollock, P.M. Sensitivity to the MEK inhibitor E6201 in melanoma cells is associated with mutant BRAF and wildtype PTEN status. Mol. Cancer 2012, 11, 75. [Google Scholar] [CrossRef]
- Qin, Y.F.; Zuo, Q.; Huang, L.; Huang, L.P.; Merlino, G.; Yu, Y.L. PERK mediates resistance to BRAF inhibition in melanoma with impaired PTEN. Npj Precis. Oncol. 2021, 5, 68. [Google Scholar] [CrossRef]
- Paraiso, K.H.; Rebecca, V.; Fedorenko, I.V.; Xiang, Y.; Wood, E.; Anderson, A.R.; Sondak, V.K.; Abei, E.V.; Koomen, J.M.; Messina, J.L.; et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011, 71, 2750–2760. [Google Scholar] [CrossRef]
- Wongchenko, M.J.; Ribas, A.; Ascierto, P.A.; Dreno, B.; di Giacomo, A.M.; Garbe, C.; Chang, I.; Hsu, E.; Rooney, I.; Lu, W.; et al. Effects of Molecular Heterogeneity on Survival of Patients with BRAF(V600)-Mutated Melanoma Treated with Vemurafenib with or without Cobimetinib in the coBRIM Study. JCO Precis. Oncol. 2018, 2, 1–18. [Google Scholar] [CrossRef]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Brocker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Walker, G.J.; Flores, J.F.; Glendening, J.M.; Lin, A.H.T.; Markl, I.D.C.; Fountain, J.W. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Gene Chromosome Cancer 1998, 22, 157–163. [Google Scholar] [CrossRef]
- Nelson, A.A.; Tsao, H. Melanoma and genetics. Clin. Dermatol. 2009, 27, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Hugo, W.; Kong, X.J.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.Q.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Chen, P.L.; Song, P.; McQuade, J.L.; Liang, R.J.; Liu, M.G.; Roh, W.; Duose, D.Y.; Carapeto, F.C.L.; Li, J.; et al. A Preexisting Rare PIK3CA(E545K) Subpopulation Confers Clinical Resistance to MEK plus CDK4/6 Inhibition in NRAS Melanoma and Is Dependent on S6K1 Signaling. Cancer Discov. 2018, 8, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Roelli, M.A.; Ruffieux-Daidie, D.; Stooss, A.; ElMokh, O.; Phillips, W.A.; Dettmer, M.S.; Charles, R.P. PIK3CA(H1047R)-induced paradoxical ERK activation results in resistance to BRAF(V600E) specific inhibitors in BRAFV600E PIK3CA(H1047R) double mutant thyroid tumors. Oncotarget 2017, 8, 103207–103222. [Google Scholar] [CrossRef] [PubMed]
- Smalley, K.S.M.; Lioni, M.; Palma, M.D.; Xiao, M.; Desai, B.; Egyhazi, S.; Hansson, J.; Wu, H.; King, A.J.; Van Belle, P.; et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol. Cancer Ther. 2008, 7, 2876–2883. [Google Scholar] [CrossRef]
- Monsma, D.J.; Cherba, D.M.; Eugster, E.E.; Dylewski, D.L.; Davidson, P.T.; Peterson, C.A.; Borgman, A.S.; Winn, M.E.; Dykema, K.J.; Webb, C.P.; et al. Melanoma patient derived xenografts acquire distinct Vemurafenib resistance mechanisms. Am. J. Cancer Res. 2015, 5, 1507. [Google Scholar]
- Lehmann, B.D.; Shaver, T.M.; Johnson, D.B.; Li, Z.; Gonzalez-Ericsson, P.I.; Anchez, V.S.; Shyr, Y.; Sanders, M.E.; Pietenpol, J.A. Identification of Targetable Recurrent MAP3K8 Rearrangements in Melanomas Lacking Known Driver Mutations. Mol. Cancer Res. 2019, 17, 1842–1853. [Google Scholar] [CrossRef]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.Y.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef]
- Smit, K.N.; Jager, M.J.; de Klein, A.; Kili, E. Uveal melanoma: Towards a molecular understanding. Prog. Retin. Eye Res. 2019, 75, 100800. [Google Scholar] [CrossRef]
- Vido, M.J.; Le, K.; Hartsough, E.J.; Aplin, A.E. BRAF Splice Variant Resistance to RAF Inhibitor Requires Enhanced MEK Association. Cell Rep. 2018, 25, 1501. [Google Scholar] [CrossRef] [PubMed]
- Noriega-Guerra, H.; Freitas, V.M. Extracellular Matrix Influencing HGF/c-MET Signaling Pathway: Impact on Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3300. [Google Scholar] [CrossRef] [PubMed]
- Czyz, M. HGF/c-MET Signaling in Melanocytes and Melanoma. Int. J. Mol. Sci. 2018, 19, 3844. [Google Scholar] [CrossRef] [PubMed]
- Caenepeel, S.; Cooke, K.; Wadsworth, S.; Huang, G.; Robert, L.; Moreno, B.H.; Parisi, G.; Cajulis, E.; Kendall, R.; Beltran, P.; et al. MAPK pathway inhibition induces MET and GAB1 levels, priming BRAF mutant melanoma for rescue by hepatocyte growth factor. Oncotarget 2017, 8, 17795–17809. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Terai, M.; Kageyama, K.; Ozaki, S.; McCue, P.A.; Sato, T.; Aplin, A.E. Paracrine Effect of NRG1 and HGF Drives Resistance to MEK Inhibitors in Metastatic Uveal Melanoma. Cancer Res. 2015, 75, 2737–2748. [Google Scholar] [CrossRef]
- Oba, J.; Kim, S.H.; Wang, W.L.; Macedo, M.P.; Carapeto, F.; McKean, M.A.; Van Arnam, J.; Eterovic, A.K.; Sen, S.; Kale, C.R.; et al. Targeting the HGF/MET Axis Counters Primary Resistance to KIT Inhibition in KIT-Mutant Melanoma. JCO Precis. Oncol. 2018, 2, 55. [Google Scholar] [CrossRef]
- Wilson, T.R.; Fridlyand, J.; Ribas, A.; Li, J.; Koeppen, H.; Merchant, M.; Neve, R.; Settleman, J.; Yan, Y.B.; Penuel, E.; et al. Widespread potential for growth factor-driven resistance to anti-cancer kinase inhibitors. Nature 2012, 487, 505–509. [Google Scholar] [CrossRef]
- Menon, D.R.; Das, S.; Krepler, C.; Vultur, A.; Rinner, B.; Schauer, S.; Kashofer, K.; Wagner, K.; Zhang, G.; Rad, E.B.; et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene 2015, 34, 4448–4459. [Google Scholar] [CrossRef]
- Prahallad, A.; Sun, C.; Huang, S.D.; Di Nicolantonio, F.; Salazar, R.; Zecchin, D.; Beijersbergen, R.L.; Bardelli, A.; Bernards, R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012, 483, 100–103. [Google Scholar] [CrossRef]
- Laurenzana, A.; Margheri, F.; Biagioni, A.; Chilla, A.; Pimpinelli, N.; Ruzzolini, J.; Peppicelli, S.; Andreucci, E.; Calorini, L.; Serrati, S.; et al. EGFR/uPAR interaction as druggable target to overcome vemurafenib acquired resistance in melanoma cells. Ebiomedicine 2019, 39, 194–206. [Google Scholar] [CrossRef]
- Molnar, E.; Garay, T.; Donia, M.; Baranyi, M.; Rittler, D.; Berger, W.; Timar, J.; Grusch, M.; Hegedus, B. Long-Term Vemurafenib Exposure Induced Alterations of Cell Phenotypes in Melanoma: Increased Cell Migration and Its Association with EGFR Expression. Int. J. Mol. Sci. 2019, 20, 4484. [Google Scholar] [CrossRef] [PubMed]
- Torre, E.A.; Arai, E.; Bayatpour, S.; Jiang, C.L.; Beck, L.E.; Emert, B.L.; Shaffer, S.M.; Mellis, I.A.; Fane, M.E.; Alicea, G.M.; et al. Genetic screening for single-cell variability modulators driving therapy resistance. Nat. Genet. 2021, 53. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.Y.; Njauw, C.N.; Guhan, S.; Kumar, R.; Reddy, B.; Rajadurai, A.; Flaherty, K.; Tsao, H. Loss of ACK1 Upregulates EGFR and Mediates Resistance to BRAF Inhibition. J. Investig. Dermatol. 2021, 141, 1317. [Google Scholar] [CrossRef] [PubMed]
- Billing, O.; Holmgren, Y.; Nosek, D.; Hedman, H.; Hemmingsson, O. LRIG1 is a conserved EGFR regulator involved in melanoma development, survival and treatment resistance. Oncogene 2021, 40, 3707–3718. [Google Scholar] [CrossRef]
- Bhattarai, P.Y.; Kim, G.; Poudel, M.; Lim, S.C.; Choi, H.S. METTL3 induces PLX4032 resistance in melanoma by promoting m(6)A-dependent EGFR translation. Cancer Lett. 2021, 522, 44–56. [Google Scholar] [CrossRef]
- Siroy, A.E.; Boland, G.M.; Milton, D.R.; Roszik, J.; Frankian, S.; Malke, J.; Haydu, L.; Prieto, V.G.; Tetzlaff, M.; Ivan, D.; et al. Beyond BRAF(V600): Clinical Mutation Panel Testing by Next-Generation Sequencing in Advanced Melanoma. J. Investig. Dermatol. 2015, 135, 508–515. [Google Scholar] [CrossRef]
- Miraflor, A.P.; de Abreu, F.B.; Peterson, J.D.; Ernstoff, M.S.; Amos, C.I.; Wells, W.A.; Tsongalis, G.J.; Yan, S. Somatic Mutation Analysis in Melanoma Using Targeted Next-Generation Sequencing. J. Mol. Diagn. 2017, 16, 756. [Google Scholar] [CrossRef]
- Lawal, B.; Wang, Y.C.; Wu, A.T.H.; Huang, H.S. Pro-Oncogenic c-Met/EGFR, Biomarker Signatures of the Tumor Microenvironment are Clinical and Therapy Response Prognosticators in Colorectal Cancer, and Therapeutic Targets of 3-Phenyl-2H-benzo[e](1,3)-Oxazine-2,4(3H)-Dione Derivatives. Front. Pharmacol. 2021, 12, 691234. [Google Scholar] [CrossRef]
- Capdevila, J.; Arques, O.; Hernandez Mora, J.R.; Matito, J.; Caratu, G.; Mancuso, F.M.; Landolfi, S.; Barriuso, J.; Jimenez-Fonseca, P.; Lopez Lopez, C.; et al. Epigenetic EGFR Gene Repression Confers Sensitivity to Therapeutic BRAFV600E Blockade in Colon Neuroendocrine Carcinomas. Clin. Cancer Res. 2020, 26, 902–909. [Google Scholar] [CrossRef]
- Teh, J.L.F.; Shah, R.; Shin, S.S.; Wen, Y.; Mehnert, J.M.; Goydos, J.; Chen, S.Z. Metabotropic glutamate receptor 1 mediates melanocyte transformation via transactivation of insulin-like growth factor 1 receptor. Pigm. Cell Melanoma Res. 2014, 27, 621–629. [Google Scholar] [CrossRef]
- Karasic, T.B.; Hei, T.K.; Ivanov, V.N. Disruption of IGF-1R signaling increases TRAIL-induced apoptosis: A new potential therapy for the treatment of melanoma. Exp. Cell Res. 2010, 316, 1994–2007. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Ding, N.; Li, Y.J.; Cheng, H.; Wang, D.; Yang, Q.; Deng, Y.H.; Yang, Y.R.; Li, Y.M.; Ruan, X.Y.; et al. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget 2015, 6, 20636–20649. [Google Scholar] [CrossRef] [PubMed]
- Benito-Jardon, L.; Diaz-Martinez, M.; Arellano-Sanchez, N.; Vaquero-Morales, P.; Esparis-Ogando, A.; Teixido, J. Resistance to MAPK Inhibitors in Melanoma Involves Activation of the IGF1R-MEK5-Erk5 Pathway. Cancer Res. 2019, 79, 2244–2256. [Google Scholar] [CrossRef] [PubMed]
- Bugide, S.; Parajuli, K.R.; Chava, S.; Pattanayak, R.; Manna, D.L.D.; Shrestha, D.; Yang, E.S.; Cai, G.; Johnson, D.B.; Gupta, R. Loss of HAT1 expression confers BRAFV600E inhibitor resistance to melanoma cells by activating MAPK signaling via IGF1R. Oncogenesis 2020, 9, 44. [Google Scholar] [CrossRef]
- Strub, T.; Ghiraldini, F.G.; Carcamo, S.; Li, M.; Wroblewska, A.; Singh, R.; Goldberg, M.S.; Hasson, D.; Wang, Z.C.; Gallagher, S.J.; et al. SIRT6 haploinsufficiency induces BRAF(V600E) melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat. Commun. 2018, 9, 3440. [Google Scholar] [CrossRef]
- Patel, H.; Mishra, R.; Yacoub, N.; Alanazi, S.; Kilroy, M.K.; Garrett, J.T. IGF1R/IR Mediates Resistance to BRAF and MEK Inhibitors in BRAF-Mutant Melanoma. Cancers 2021, 13, 5863. [Google Scholar] [CrossRef]
- Obenauf, A.C.; Zou, Y.; Ji, A.L.; Vanharanta, S.; Shu, W.; Shi, H.; Kong, X.; Bosenberg, M.C.; Wiesner, T.; Rosen, N.; et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 2015, 520, 368–372. [Google Scholar] [CrossRef]
- Roskoski, R. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol. Res. 2018, 129, 65–83. [Google Scholar] [CrossRef]
- Wang, Y.; Appiah-Kubi, K.; Wu, M.; Yao, X.Y.; Qian, H.; Wu, Y.; Chen, Y.C. The platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are major players in oncogenesis, drug resistance, and attractive oncologic targets in cancer. Growth Factors 2016, 34, 64–71. [Google Scholar] [CrossRef]
- Vella, L.J.; Behren, A.; Coleman, B.; Greening, D.W.; Hill, A.F.; Cebon, J. Intercellular Resistance to BRAF Inhibition Can Be Mediated by Extracellular Vesicle-Associated PDGFR beta. Neoplasia 2017, 19, 932–940. [Google Scholar] [CrossRef]
- Hartman, M.L.; Sztiller-Sikorska, M.; Gajos-Michniewicz, A.; Czyz, M. Dissecting Mechanisms of Melanoma Resistance to BRAF and MEK Inhibitors Revealed Genetic and Non-Genetic Patient- and Drug-Specific Alterations and Remarkable Phenotypic Plasticity. Cells 2020, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.J.; Kuilman, T.; Shahrabi, A.; Oshuizen, J.B.; Kemper, K.; Song, J.Y.; Niessen, H.W.M.; Rozeman, E.A.; Foppen, M.H.G.; Lank, C.U.B. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 2017, 550, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.; Moriceau, G.; Sun, L.; Lomeli, S.; Piva, M.; Damoiseaux, R.; Holmen, S.L.; Sharpless, N.E.; Hugo, W.; Lo, R.S. Exploiting Drug Addiction Mechanisms to Select against MAPKi-Resistant Melanoma. Cancer Discov. 2018, 8, 74–93. [Google Scholar] [CrossRef] [PubMed]
- Lei, F.X.; Jin, L.; Liu, X.Y.; Lai, F.; Yan, X.G.; Farrelly, M.; Guo, S.T.; Zhao, X.H.; Zhang, X.D. RIP1 protects melanoma cells from apoptosis induced by BRAF/MEK inhibitors. Cell Death Dis. 2018, 9, 679. [Google Scholar] [CrossRef]
- Webster, M.R.; Fane, M.E.; Alicea, G.M.; Basu, S.; Kossenkov, A.V.; Marino, G.E.; Douglass, S.M.; Kaur, A.; Ecker, B.L.; Gnanapradeepan, K.; et al. Paradoxical Role for Wild-Type p53 in Driving Therapy Resistance in Melanoma. Mol. Cell 2020, 77, 633–644.e5. [Google Scholar] [CrossRef]
- McKenna, S.; Garcia-Gutierrez, L. Resistance to Targeted Therapy and RASSF1A Loss in Melanoma: What Are We Missing? Int. J. Mol. Sci. 2021, 22, 5115. [Google Scholar] [CrossRef]
- Alkaraki, A.; McArthur, G.A.; Sheppard, K.E.; Smith, L.K. Metabolic Plasticity in Melanoma Progression and Response to Oncogene Targeted Therapies. Cancers 2021, 13, 5810. [Google Scholar] [CrossRef]
- Shen, S.S.; Faouzi, S.; Souquere, S.; Roy, S.; Routier, E.; Libenciuc, C.; Andre, F.; Pierron, G.; Scoazec, J.Y.; Robert, C. Melanoma Persister Cells Are Tolerant to BRAF/MEK Inhibitors via ACOX1-Mediated Fatty Acid Oxidation. Cell Rep. 2020, 33, 108421. [Google Scholar] [CrossRef]
- Tripathi, R.; Liu, Z.L.; Jain, A.; Lyon, A.; Meeks, C.; Richards, D.; Liu, J.P.; He, D.H.; Wang, C.; Nespi, M.; et al. Combating acquired resistance to MAPK inhibitors in melanoma by targeting Abl1/2-mediated reactivation of MEK/ERK/MYC signaling. Nat. Commun. 2020, 11, 5463. [Google Scholar] [CrossRef]
- Liu, F.; Cao, J.X.; Wu, J.X.; Sullivan, K.; Shen, J.; Ryu, B.; Xu, Z.X.; Wei, W.Y.; Cui, R.T. Stat3-Targeted Therapies Overcome the Acquired Resistance to Vemurafenib in Melanomas. J. Investig. Dermatol. 2013, 133, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Das Sahu, A.; Lee, J.S.; Wang, Z.Y.; Zhang, G.; Iglesias-Bartolome, R.; Tian, T.; We, Z.; Miao, B.; Nair, N.U.; Ponomarova, O.; et al. Genome-wide prediction of synthetic rescue mediators of resistance to targeted and immunotherapy. Mol. Syst. Biol. 2019, 15, e8323. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).