Germline Variants in Angiogenesis-Related Genes Contribute to Clinical Outcome in Head and Neck Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. SNP Identification
2.3. Statistical Analysis
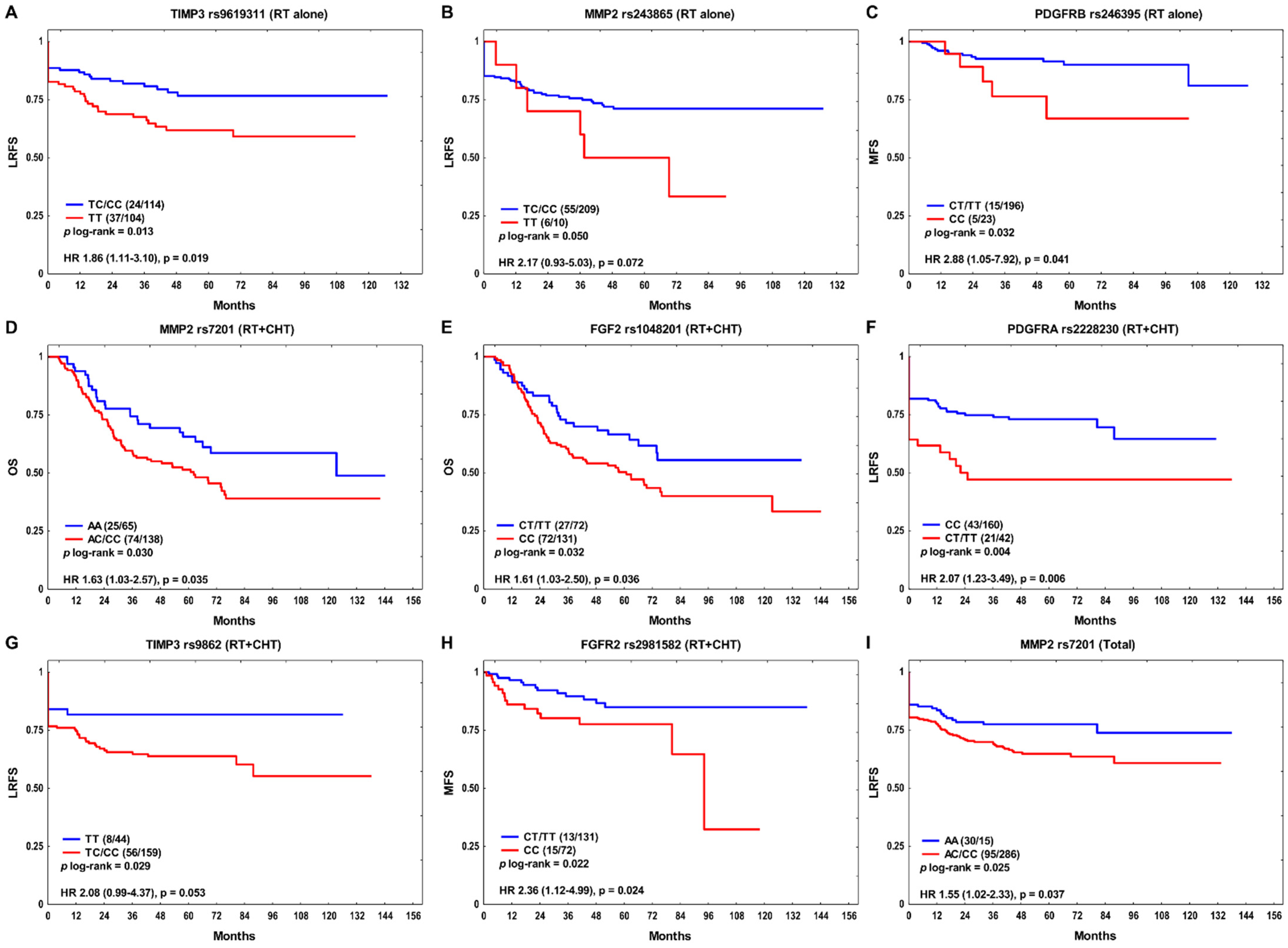
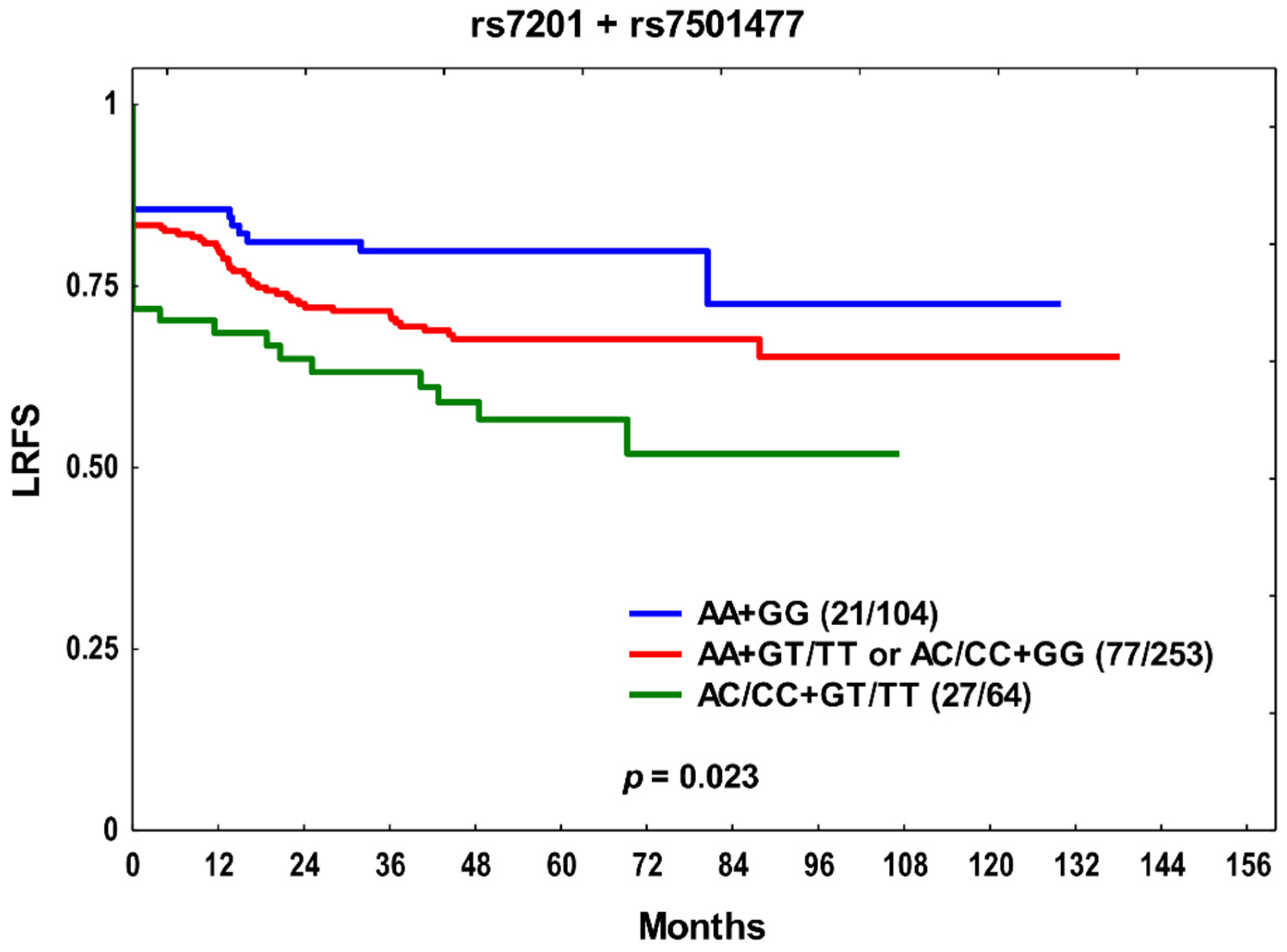
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Cohen, E.E. State-of-the-art management of locally advanced head and neck cancer. Br. J. Cancer 2005, 92, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Kelleher, D.K.; Höckel, M. Oxygen status of malignant tumors: Pathogenesis of hypoxia and significance for tumor therapy. Semin. Oncol. 2001, 28, 29–35. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Folkman, J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 1996, 86, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef]
- Murakami, M.; Simons, M. Fibroblast growth factor regulation of neovascularization. Curr. Opin. Hematol. 2008, 15, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Östman, A. PDGF receptors in tumor stroma: Biological effects and associations with prognosis and response to treatment. Adv. Drug Deliv. Rev. 2017, 121, 117–123. [Google Scholar] [CrossRef]
- Jing, Q.; Wang, Y.; Liu, H.; Deng, X.; Jiang, L.; Liu, R.; Song, H.; Li, J. FGFs: Crucial factors that regulate tumour initiation and progression. Cell Prolif. 2016, 49, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [Green Version]
- Heldin, C.H.; Lennartsson, J.; Westermark, B. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J. Intern. Med. 2018, 283, 16–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akl, M.R.; Nagpal, P.; Ayoub, N.M.; Tai, B.; Prabhu, S.A.; Capac, C.M.; Gliksman, M.; Goy, A.; Suh, K.S. Molecular and clinical significance of fibroblast growth factor 2 (FGF2 /bFGF) in malignancies of solid and hematological cancers for personalized therapies. Oncotarget 2016, 7, 44735–44762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deryugina, E.I.; Quigley, J.P. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: Contrasting, overlapping and compensatory functions. Biochim. Et Biophys. Acta 2010, 1803, 103–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rundhaug, J.E. Matrix metalloproteinases and angiogenesis. J. Cell. Mol. Med. 2005, 9, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.H.; Ebrahem, Q.; Moore, N.; Murphy, G.; Claesson-Welsh, L.; Bond, M.; Baker, A.; Anand-Apte, B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): Inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 2003, 9, 407–415. [Google Scholar] [CrossRef]
- Bourboulia, D.; Jensen-Taubman, S.; Stetler-Stevenson, W.G. TIMP-2: An Endogenous Angiogenesis Inhibitor with Distinct Antitumoral Properties. Treat. Strateg. Hematol. 2012, 2, 31–35. [Google Scholar]
- Rosenthal, E.L.; Matrisian, L.M. Matrix metalloproteases in head and neck cancer. Head Neck 2006, 28, 639–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, A.K.; Pandya, S.; Ghosh, K.; Nadkarni, A. Matrix metalloproteinase and its drug targets therapy in solid and hematological malignancies: An overview. Mutat. Res. 2013, 753, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Buysschaert, I.; Schmidt, T.; Roncal, C.; Carmeliet, P.; Lambrechts, D. Genetics, epigenetics and pharmaco-(epi)genomics in angiogenesis. J. Cell. Mol. Med. 2008, 12, 2533–2551. [Google Scholar] [CrossRef] [PubMed]
- Butkiewicz, D.; Gdowicz-Kłosok, A.; Krześniak, M.; Rutkowski, T.; Krzywon, A.; Cortez, A.J.; Domińczyk, I.; Składowski, K. Association of Genetic Variants in ANGPT/TEK and VEGF/VEGFR with Progression and Survival in Head and Neck Squamous Cell Carcinoma Treated with Radiotherapy or Radiochemotherapy. Cancers 2020, 12, 1506. [Google Scholar] [CrossRef] [PubMed]
- Butkiewicz, D.; Krześniak, M.; Drosik, A.; Giglok, M.; Gdowicz-Kłosok, A.; Kosarewicz, A.; Rusin, M.; Masłyk, B.; Gawkowska-Suwińska, M.; Suwiński, R. The VEGFR2, COX-2 and MMP-2 polymorphisms are associated with clinical outcome of patients with inoperable non-small cell lung cancer. Int. J. Cancer 2015, 137, 2332–2342. [Google Scholar] [CrossRef] [PubMed]
- Ensembl Database 103. Available online: http://www.ensembl.org/ (accessed on 25 February 2021).
- Li, D.; Zhang, H.; Ma, L.; Han, Y.; Xu, M.; Wang, Z.; Jiang, H.; Zhang, W.; Wang, L.; Pan, Y. Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Sci. Rep. 2016, 6, 31054. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yang, L.; Xu, T.; Si, L.; Cui, C.; Sheng, X.; Chi, Z.; Mao, L.; Lian, B.; Tang, B.; et al. A Functional Synonymous Variant in PDGFRA Is Associated with Better Survival in Acral Melanoma. J. Cancer 2020, 11, 2945–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrigós, C.; Espinosa, M.; Salinas, A.; Osman, I.; Medina, R.; Taron, M.; Molina-Pinelo, S.; Duran, I. Single nucleotide polymorphisms as prognostic and predictive biomarkers in renal cell carcinoma. Oncotarget 2017, 8, 106551–106564. [Google Scholar] [CrossRef] [Green Version]
- Estevez-Garcia, P.; Castaño, A.; Martin, A.C.; Lopez-Rios, F.; Iglesias, J.; Muñoz-Galván, S.; Lopez-Calderero, I.; Molina-Pinelo, S.; Pastor, M.D.; Carnero, A.; et al. PDGFRα/β and VEGFR2 polymorphisms in colorectal cancer: Incidence and implications in clinical outcome. BMC Cancer 2012, 12, 514. [Google Scholar] [CrossRef] [Green Version]
- Price, S.J.; Greaves, D.R.; Watkins, H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: Role of Sp1 in allele-specific transcriptional regulation. J. Biol. Chem. 2001, 276, 7549–7558. [Google Scholar] [CrossRef] [Green Version]
- Tsai, E.M.; Wang, Y.S.; Lin, C.S.; Lin, W.Y.; His, E.; Wu, M.T.; Juo, S.H. A microRNA-520 mirSNP at the MMP2 gene influences susceptibility to endometriosis in Chinese women. J. Hum. Genet. 2013, 58, 202–209. [Google Scholar] [CrossRef]
- Peterson, N.B.; Beeghly-Fadiel, A.; Gao, Y.T.; Long, J.; Cai, Q.; Shu, X.O.; Zheng, W. Polymorphisms in tissue inhibitors of metalloproteinases-2 and -3 and breast cancer susceptibility and survival. Int. J. Cancer 2009, 125, 844–850. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Huang, P.Y.; Tang, L.Q.; Chen, Q.Y.; Zhang, Y.; Zhang, L.; Guo, L.; Luo, D.H.; Mo, H.Y.; Xiang, Y.Q.; et al. Functional polymorphisms of matrix metalloproteinase-9 and survival in patients with locoregionally advanced nasopharyngeal carcinoma treated with chemoradiotherapy. Med. Oncol. 2013, 30, 685. [Google Scholar] [CrossRef]
- Jin, G.; Miao, R.; Hu, Z.; Xu, L.; Huang, X.; Chen, Y.; Tian, T.; Wei, Q.; Boffetta, P.; Shen, H. Putative functional polymorphisms of MMP9 predict survival of NSCLC in a Chinese population. Int. J. Cancer 2009, 124, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, G.; Huang, S.; Luo, A.; Jing, X.; Li, G.; Zhou, Y.; Zhao, X. Association between TIMP-2 gene polymorphism and breast cancer in Han Chinese women. BMC Cancer 2019, 19, 446. [Google Scholar] [CrossRef] [PubMed]
- Su, C.W.; Huang, Y.W.; Chen, M.K.; Su, S.C.; Yang, S.F.; Lin, C.W. Polymorphisms and Plasma Levels of Tissue Inhibitor of Metalloproteinase-3: Impact on Genetic Susceptibility and Clinical Outcome of Oral Cancer. Medicine 2015, 94, e2092. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.T.; Hsieh, M.J.; Chiou, H.L.; Lee, H.L.; Hsin, M.C.; Liou, Y.S.; Yang, C.C.; Yang, S.F.; Kuo, W.H. TIMP-3-1296 T>C and TIMP-4 -55 T>C gene polymorphisms play a role in the susceptibility of hepatocellular carcinoma among women. Tumor Biol. 2014, 35, 8999–9007. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Liu, L.C.; Hsiao, C.L.; Su, C.H.; Wang, H.C.; Ji, H.X.; Tsai, C.W.; Maa, M.C.; Bau, D.T. The contributions of the tissue inhibitor of metalloproteinase-1 genotypes to triple negative breast cancer risk. Biomedicine 2016, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Luizon, M.R.; Palei, A.C.; Sandrim, V.C.; Amaral, L.M.; Machado, J.S.; Lacchini, R.; Cavalli, R.C.; Duarte, G.; Tanus-Santos, J.E. Tissue inhibitor of matrix metalloproteinase-1 polymorphism, plasma TIMP-1 levels, and antihypertensive therapy responsiveness in hypertensive disorders of pregnancy. Pharm. J. 2014, 14, 535–541. [Google Scholar] [CrossRef]
- Campbell, T.M.; Castro, M.A.A.; de Santiago, I.; Fletcher, M.N.C.; Halim, S.; Prathalingam, R.; Ponder, B.A.J.; Meyer, K.B. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 2016, 37, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Schulz, S.; Köhler, K.; Schagdarsurengin, U.; Greiser, P.; Birkenmeier, G.; Müller-Werdan, U.; Werdan, K.; Gläser, C. The human FGF2 level is influenced by genetic predisposition. Int. J. Cardiol. 2005, 101, 265–271. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, S.K.; Park, H.J.; Chung, D.H.; Park, H.K.; Lee, J.S.; Kwon, K.H.; Chung, J.H. PDGFRA promoter polymorphisms are associated with the risk of papillary thyroid cancer. Mol. Med. Rep. 2012, 5, 1267–1270. [Google Scholar] [CrossRef] [Green Version]
- Duan, B.; Hu, J.; Liu, H.; Wang, Y.; Li, H.; Liu, S.; Xie, J.; Owzar, K.; Abbruzzese, J.; Hurwitz, H.; et al. Genetic variants in the platelet-derived growth factor subunit B gene associated with pancreatic cancer risk. Int. J. Cancer 2018, 142, 1322–1331. [Google Scholar] [CrossRef] [Green Version]
- Stensrud, M.J.; Hernán, M.A. Why Test for Proportional Hazards? JAMA 2020, 323, 1401–1402. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.F.; Papasian, C.J.; Deng, H.W. Polymorphisms in predicted miRNA binding sites and osteoporosis. J. Bone Miner. Res. 2011, 26, 72–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, G.; Xiao, G.; Luo, C.; Tang, Z.; Teng, Z.; Peng, X. Correlation Between SNPs at the 3’UTR of the FGF2 Gene and Their Interaction with Environmental Factors in Han Chinese Diabetic Peripheral Neuropathy Patients. J. Mol. Neurosci. 2021, 71, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Dietz, A.; Rudat, V.; Conradt, C.; Weidauer, H.; Ho, A.; Moehler, T. Prognostic relevance of serum levels of the angiogenic peptide bFGF in advanced carcinoma of the head and neck treated by primary radiochemotherapy. Head Neck 2000, 22, 666–673. [Google Scholar] [CrossRef]
- Rades, D.; Seibold, N.D.; Gebhard, M.P.; Noack, F.; Bruchhage, K.L.; Schild, S.E. Fibroblast growth factor 2 is of prognostic value for patients with locally advanced squamous cell carcinoma of the head and neck. Strahlenther. Onkol. 2014, 190, 68–74. [Google Scholar] [CrossRef]
- Mariz, B.A.L.A.; Soares, C.D.; de Carvalho, M.G.F.; Jorge-Júnior, J. FGF-2 and FGFR-1 might be independent prognostic factors in oral tongue squamous cell carcinoma. Histopathology 2019, 74, 311–320. [Google Scholar] [CrossRef]
- Bremnes, R.M.; Camps, C.; Sirera, R. Angiogenesis in non-small cell lung cancer: The prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer 2006, 51, 143–158. [Google Scholar] [CrossRef]
- Hu, M.; Hu, Y.; He, J.; Li, B. Prognostic Value of Basic Fibroblast Growth Factor (bFGF) in Lung Cancer: A Systematic Review with Meta-Analysis. PLoS ONE 2016, 11, e0147374. [Google Scholar] [CrossRef]
- He, L.; Meng, Y.; Zhang, Z.; Liu, Y.; Wang, X. Downregulation of basic fibroblast growth factor increases cisplatin sensitivity in A549 non-small cell lung cancer cells. J. Cancer Res. Ther. 2018, 14, 1519–1524. [Google Scholar] [CrossRef]
- McDermott, S.C.; Rodriguez-Ramirez, C.; McDermott, S.P.; Wicha, M.S.; Nör, J.E. FGFR signaling regulates resistance of head and neck cancer stem cells to cisplatin. Oncotarget 2018, 9, 25148–25165. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wu, C.; Lu, G.; Hu, Z.; Chen, Q.; Du, X. FGF/FGFR signaling pathway involved resistance in various cancer types. J. Cancer 2020, 11, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Hunt, R.; Sauna, Z.E.; Ambudkar, S.V.; Gottesman, M.M.; Kimchi-Sarfaty, C. Silent (synonymous) SNPs: Should we care about them? Methods Mol. Biol. 2009, 578, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yin, J.; Zha, L.; Wang, Z. Clinicopathological significance of platelet-derived growth factor B, platelet-derived growth factor receptor-β, and E-cadherin expression in gastric carcinoma. Contemp. Oncol. 2013, 17, 150–155. [Google Scholar] [CrossRef]
- Steller, E.J.; Raats, D.A.; Koster, J.; Rutten, B.; Govaert, K.M.; Emmink, B.L.; Snoeren, N.; van Hooff, S.R.; Holstege, F.C.; Maas, C.; et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia 2013, 15, 204–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avril, S.; Dincer, Y.; Malinowsky, K.; Wolff, C.; Gündisch, S.; Hapfelmeier, A.; Boxberg, M.; Bronger, H.; Becker, K.F.; Schmalfeldt, B. Increased PDGFR-beta and VEGFR-2 protein levels are associated with resistance to platinum-based chemotherapy and adverse outcome of ovarian cancer patients. Oncotarget 2017, 8, 97851–97861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strell, C.; Stenmark Tullberg, A.; Jetne Edelmann, R.; Akslen, L.A.; Malmström, P.; Fernö, M.; Holmberg, E.; Östman, A.; Karlsson, P. Prognostic and predictive impact of stroma cells defined by PDGFRb expression in early breast cancer: Results from the randomized SweBCG91RT trial. Breast Cancer Res. Treat. 2021, 187, 45–55. [Google Scholar] [CrossRef]
- Lin, L.H.; Lin, J.S.; Yang, C.C.; Cheng, H.W.; Chang, K.W.; Liu, C.J. Overexpression of Platelet-Derived Growth Factor and Its Receptor Are Correlated with Oral Tumorigenesis and Poor Prognosis in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 2360. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cao, R.; Zhang, Y.; Jia, T.; Cao, Y.; Wahlberg, E. Differential roles of PDGFR-alpha and PDGFR-beta in angiogenesis and vessel stability. FASEB J. 2009, 23, 153–163. [Google Scholar] [CrossRef]
- Ribeiro, A.L.; Okamoto, O.K. Combined effects of pericytes in the tumor microenvironment. Stem Cells Int. 2015, 2015, 868475. [Google Scholar] [CrossRef] [Green Version]
- Sulzbacher, I.; Birner, P.; Träxler, M.; Marberger, M.; Haitel, A. Expression of platelet-derived growth factor-alpha alpha receptor is associated with tumor progression in clear cell renal cell carcinoma. Am. J. Clin. Pathol. 2003, 120, 107–112. [Google Scholar] [CrossRef]
- Carvalho, I.; Milanezi, F.; Martins, A.; Reis, R.M.; Schmitt, F. Overexpression of platelet-derived growth factor receptor alpha in breast cancer is associated with tumour progression. Breast Cancer Res. 2005, 7, R788–R795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, H.S.; Gokavarapu, S.; Tian, Z.; Li, J.; Xu, Q.; Zhang, C.P.; Cao, W. PDGFRA mRNA overexpression is associated with regional metastasis and reduced survival in oral squamous cell carcinoma. J. Oral Pathol. Med. 2018, 47, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.K.; Singh, M.; Bharti, A.C.; Asotra, K.; Sundaram, S.; Mehrotra, R. Genetic polymorphisms of matrix metalloproteinases and their inhibitors in potentially malignant and malignant lesions of the head and neck. J. Biomed. Sci. 2010, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, B.; Cao, L.; Ma, X.; Wang, W.; Wang, D.; Yu, L. Meta-analysis of association between matrix metalloproteinases 2, 7 and 9 promoter polymorphisms and cancer risk. Mutagenesis 2010, 25, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Langers, A.M.; Sier, C.F.; Hawinkels, L.J.; Kubben, F.J.; van Duijn, W.; van der Reijden, J.J.; Lamers, C.B.; Hommes, D.W.; Verspaget, H.W. MMP-2 geno-phenotype is prognostic for colorectal cancer survival, whereas MMP-9 is not. Br. J. Cancer 2008, 98, 1820–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grieu, F.; Li, W.Q.; Iacopetta, B. Genetic polymorphisms in the MMP-2 and MMP-9 genes and breast cancer phenotype. Breast Cancer Res. Treat. 2004, 88, 197–204. [Google Scholar] [CrossRef]
- Xie, B.; Zhang, Z.; Wang, H.; Chen, Z.; Wang, Y.; Liang, H.; Yang, G.; Yang, X.; Zhang, H. Genetic polymorphisms in MMP 2, 3, 7, and 9 genes and the susceptibility and clinical outcome of cervical cancer in a Chinese Han population. Tumour Biol. 2016, 37, 4883–4888. [Google Scholar] [CrossRef]
- Srivastava, P.; Kapoor, R.; Mittal, R.D. Association of single nucleotide polymorphisms in promoter of matrix metalloproteinase-2, 8 genes with bladder cancer risk in Northern India. Urol. Oncol. 2013, 31, 247–254. [Google Scholar] [CrossRef]
- Tsai, C.W.; Hsu, H.M.; Wang, Y.C.; Chang, W.S.; Shih, L.C.; Sun, K.T.; Hung, Y.W.; Yang, Y.C.; Gong, C.L.; Bau, D.T. Contribution of MMP2 Promoter Genotypes to Oral Cancer Susceptibility, Recurrence and Metastasis in Taiwan. Anticancer Res. 2018, 38, 6821–6826. [Google Scholar] [CrossRef]
- Charoenrat, P.; Khantapura, P. The role of genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes in head and neck cancer. Oral Oncol. 2006, 42, 257–267. [Google Scholar] [CrossRef]
- Zhang, C.; Li, C.; Zhu, M.; Zhang, Q.; Xie, Z.; Niu, G.; Song, X.; Jin, L.; Li, G.; Zheng, H. Meta-analysis of MMP2, MMP3, and MMP9 promoter polymorphisms and head and neck cancer risk. PLoS ONE 2013, 8, e62023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Reilly, M.S.; Wiederschain, D.; Stetler-Stevenson, W.G.; Folkman, J.; Moses, M.A. Regulation of angiostatin production by matrix metalloproteinase-2 in a model of concomitant resistance. J. Biol. Chem. 1999, 274, 29568–29571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Pérez, J.M.; Martínez-Rodríguez, N.; Vargas-Alarcón, G.; Vallejo, M.; Monroy-Muñoz, I.E.; Posadas-Romero, C.; Kimura-Hayama, E.; Juárez-Cedillo, T.; Fragoso, J.M.; Pérez-Hernández, N. TIMP2 gene polymorphisms are associated with hypertension in patients with myocardial infarction. J. Genet. 2014, 93, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guo, L.; Wang, S.; Yu, Q.; Lu, J. Association of Smoking and XPG, CYP1A1, OGG1, ERCC5, ERCC1, MMP2, and MMP9 Gene Polymorphisms with the early detection and occurrence of Laryngeal Squamous Carcinoma. J. Cancer 2018, 9, 968–977. [Google Scholar] [CrossRef]
- Liu, Y.; Cai, H.; Liu, J.; Fan, H.; Wang, Z.; Wang, Q.; Shao, M.; Sun, X.; Diao, J.; Liu, Y.; et al. A miR-151 binding site polymorphism in the 3′-untranslated region of the cyclin E1 gene associated with nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2013, 432, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Juttermann, R.; Soloway, P.D. TIMP-2 is required for efficient activation of proMMP-2 in vivo. J. Biol. Chem. 2000, 275, 26411–26415. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Cui, J. Anti-angiogenesis: Opening a new window for immunotherapy. Life Sci. 2020, 258, 118163. [Google Scholar] [CrossRef]
- Solimando, A.G.; Summa, S.; Vacca, A.; Ribatti, D. Cancer-Associated Angiogenesis: The Endothelial Cell as a Checkpoint for Immunological Patrolling. Cancers 2020, 12, 3380. [Google Scholar] [CrossRef]
| SNP | Genotype | RT Alone | RT + CT | Total | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | ||
| OS | |||||||
| rs1048201 | CC | 0.76 (0.49–1.17) | 0.208 | 1.66 (1.03–2.68) | 0.039 | 1.10 (0.81–1.48) | 0.544 |
| LRFS | |||||||
| rs2228230 | CT/TT | 0.83 (0.44–1.57) | 0.572 | 2.49 (1.42–4.36) | 0.001 | 1.41 (0.93–2.14) | 0.106 |
| rs243865 | TT | 2.92 (1.20–7.11) | 0.019 | 0.74 (0.23–2.42) | 0.620 | 1.38 (0.69–2.73) | 0.360 |
| rs7201 | AC/CC | 1.50 (0.82–2.75) | 0.191 | 1.54 (0.84–2.81) | 0.159 | 1.59 (1.04–2.42) | 0.032 |
| rs7501477 | GT/TT | 1.41 (0.79–2.52) | 0.250 | 1.57 (0.91–2.72) | 0.107 | 1.49 (1.01–2.21) | 0.045 |
| rs9862 | TC/CC | 0.91 (0.51–1.63) | 0.749 | 2.12 (0.98–4.57) | 0.055 | 1.18 (0.76–1.82) | 0.459 |
| MFS | |||||||
| rs246395 | CC | 3.06 (1.05–8.95) | 0.041 | 0.36 (0.08–1.70) | 0.198 | 1.29 (0.56–2.99) | 0.548 |
| rs1048201 | CC | 3.08 (0.92–10.25) | 0.067 | 1.31 (0.57–3.01) | 0.519 | 1.70 (0.89–3.24) | 0.111 |
| RT Alone | RT + CT | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | HR (95% CI) | p | Variables | HR (95% CI) | p | Variables | HR (95% CI) | p | |
| OS | Stage N > 0 SPC Metastasis Local recurrence | 2.24 (1.49–3.36) 2.29 (1.37–3.84) 1.89 (1.08–3.32) 3.95 (2.62–5.97) | 0.0001 0.0016 0.026 <1 × 10−6 | rs1048201 CC Alcohol: ever HPSCC Local recurrence Regional recurrence SPC | 1.61 (1.01–2.55) 2.11 (1.18–3.74) 2.01 (1.25–3.24) 5.51 (3.33–9.11) 1.73 (1.06–2.84) 2.07 (1.09–3.93) | 0.044 0.011 0.004 <1 × 10−6 0.029 0.026 | Alcohol: ever Stage N > 0 Local recurrence Regional recurrence Metastasis SPC | 1.51 (1.06–2.16) 1.81 (1.31–2.49) 4.84 (3.52–6.67) 1.49 (1.01–2.18) 1.72 (1.17–2.54) 2.32 (1.56–3.46) | 0.024 0.0003 <1 × 10−6 0.044 0.006 4 × 10−5 |
| LRFS | rs243865 TT Stage T3–4 Stage N > 0 Non-OPSCC | 2.92 (1.23–6.94) 2.97 (1.72–5.14) 2.19 (1.23–3.91) 2.29 (1.20–4.39) | 0.015 0.0001 0.008 0.012 | rs2228230 CT/TT Non-OPSCC | 2.26 (1.33–3.84) 1.74 (1.05–2.87) | 0.003 0.032 | rs7201 AC/CC Stage T3–4 Stage N > 0 Non-OPSCC | 1.56 (1.02–2.37) 1.69 (1.14–2.50) 1.68 (1.11–2.55) 1.66 (1.11–2.49) | 0.038 0.008 0.015 0.013 |
| MFS | rs246395 CC Regional recurrence | 2.79 (1.01–7.69) 5.56 (1.71–18.13) | 0.048 0.004 | Regional recurrence | 3.65 (1.62–8.22) | 0.002 | HPSCC Regional recurrence | 2.36 (1.13–4.89) 4.60 (2.37–8.92) | 0.021 6 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butkiewicz, D.; Gdowicz-Kłosok, A.; Krześniak, M.; Rutkowski, T.; Łasut-Szyszka, B.; Składowski, K. Germline Variants in Angiogenesis-Related Genes Contribute to Clinical Outcome in Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 1844. https://doi.org/10.3390/cancers14071844
Butkiewicz D, Gdowicz-Kłosok A, Krześniak M, Rutkowski T, Łasut-Szyszka B, Składowski K. Germline Variants in Angiogenesis-Related Genes Contribute to Clinical Outcome in Head and Neck Squamous Cell Carcinoma. Cancers. 2022; 14(7):1844. https://doi.org/10.3390/cancers14071844
Chicago/Turabian StyleButkiewicz, Dorota, Agnieszka Gdowicz-Kłosok, Małgorzata Krześniak, Tomasz Rutkowski, Barbara Łasut-Szyszka, and Krzysztof Składowski. 2022. "Germline Variants in Angiogenesis-Related Genes Contribute to Clinical Outcome in Head and Neck Squamous Cell Carcinoma" Cancers 14, no. 7: 1844. https://doi.org/10.3390/cancers14071844
APA StyleButkiewicz, D., Gdowicz-Kłosok, A., Krześniak, M., Rutkowski, T., Łasut-Szyszka, B., & Składowski, K. (2022). Germline Variants in Angiogenesis-Related Genes Contribute to Clinical Outcome in Head and Neck Squamous Cell Carcinoma. Cancers, 14(7), 1844. https://doi.org/10.3390/cancers14071844






