Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7
Abstract
Simple Summary
Abstract
1. Cancer Colorectal
1.1. Epidemiology
1.2. Molecular Definition of Colorectal Cancer
2. CXCL12 and Its Two Receptors CXCR4 and CXCR7
3. Physiological Roles of CXCL12 and Its Two Receptors CXCR4 and CXCR7
3.1. Chemokine CXCL12
3.2. CXCR4 Receptor
3.3. CXCR7 Receptor
4. CXCL12/CXCR4/CXCR7: Pathological Role in CRC
4.1. Receptor Expression
4.2. CXCL12 Expression
4.3. CXCL12/CXCR4/CXCR7 Axis in Cellular Interactions
5. Prognostic Value of CXCL121/CXCR4/CXCR7 Axis
5.1. CXCL12 as a Prognostic Factor
5.2. CXCR4 as a Prognostic Factor
5.3. CXCR4 as Stem Cell Marker
5.4. CXCR7 as a Prognostic Factor
| Authors | CXCL12 | CXCR4 | CXCR7 | References | |||
|---|---|---|---|---|---|---|---|
| Expression | Prognosis | Expression | Prognosis | Expression | Prognosis | ||
| Romain, 2017 | ↓; ↓ | If ↑; ↓ OS | [52] | ||||
| Fan (meta-analysis), 2018 | - | ↑ | ↓ OS; ↓ DFS | [81] | |||
| Romain, 2014 | - | ↑ | ↑ | [82] | |||
| Kim, 2005; 2006 | - | ↑ | ↓ OS | [84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121] | |||
| Yang, 2015 | - | ↑ | ↓ OS; ↓ DFS | [85] | |||
| Yang, 2015 | - | ↑ | ↓ OS; ↓ PFS | [85] | |||
| Xu, 2018 | - | ↑ | ↓ OS | [86] | |||
| Ingold, 2009 | ↑ | vascular | ↓ OS | [88] | |||
| Guillemot, 2012 | ↑ | ↑ | ↑ | [92] | |||
| Greijer, 2008 | ↑ | [93] | |||||
| Frick, 2011 | ↓ | ↑ | [94] | ||||
| Amara, 2015 | ↑ | ↓ OS | ↑ | ↓ OS | [95] | ||
| Yoshitake, 2008 | If ↑ | ↓ OS | If ↑ | ↓ OS | [96] | ||
| Akishima-Fukasawa, 2009 | If ↑ | ↓ OS | [97] | ||||
| Mousavi, 2018 | → | → | → | → | [98] | ||
| Wendt, 2006 | ↓ | [99] | |||||
| Mitchell, 2019 | ↑ | ↓ OS | ↑ | ↓ OS | ↑ | ↓ OS | [111] |
| Stanisavljević, 2016 | ↓; ↑ | ↓ DFS; ↑ DFS | ↑ | stage III, ↓ DFS | [112] | ||
| Li (meta-analysis), 2017 | ↑ | ↓ OS; ↓ DFS | ↑ | ↓ OS; ↓ DFS | [113] | ||
| Lalos, 2021 | ↑ | ↑ OS | [120] | ||||
| Schimanski, 2005 | - | If ↑ | ↓ OS | [122] | |||
| Lv, 2014 | - | ↑ | ↓ OS; ↓ DFS | [123] | |||
| Li, 2015 | ↑ | ↓ OS | [122] | ||||
| Jiang, 2019 | - | ↑ | ↓ OS | [125] | |||
| Ottaiano, 2020 | - | ↑ | ↓ OS | [124] | |||
| Yopp, 2012 | ↓; ↑ | → | If ↑ | ↓ OS; ↓ DFS | [127] | ||
| Nagasawa, 2021 | - | → | → | [128] | |||
| Jiao (CRC liver metastases), 2019 | → | → | → | → | [129] | ||
| Xu, 2007 | - | ↑ invasive border | ↓ OS | [132] | |||
| Kheirelseid, 2013 | - | If ↑ | ↑ OS | [144] | |||
6. Mechanisms of Expression Regulation
6.1. Regulation of CXCL12 Expression
6.2. Regulation of CXCR4 Expression
6.3. Regulation of CXCR7 Expression
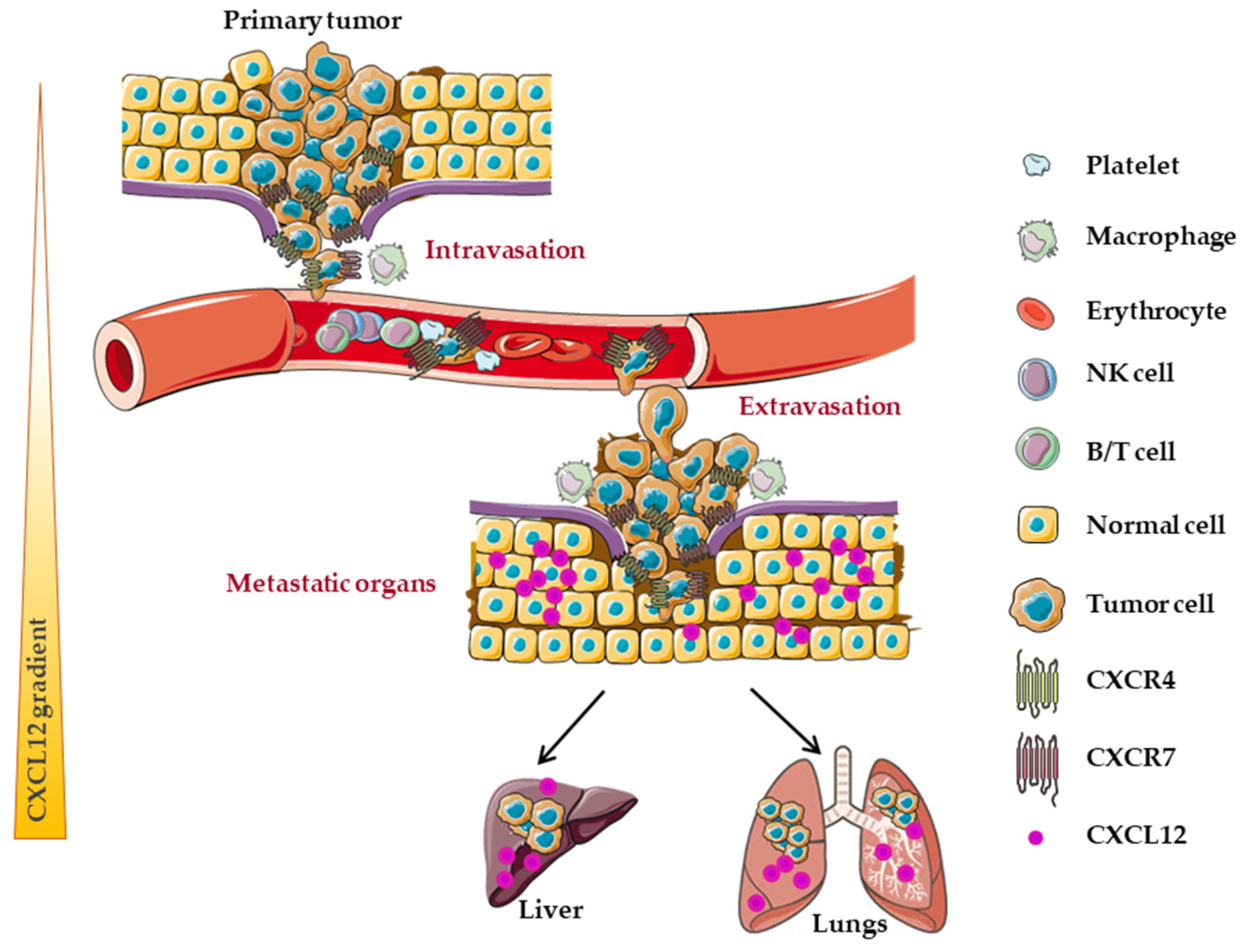
7. Implication of CXCL12/CXCR4/CXCR7 Axis in Metastatic Dissemination
7.1. CXCL12
7.2. CXCL12 and CXCR4
7.3. CXCL12 and CXCR7
7.4. CXCL12, CXCR4 and CXCR7
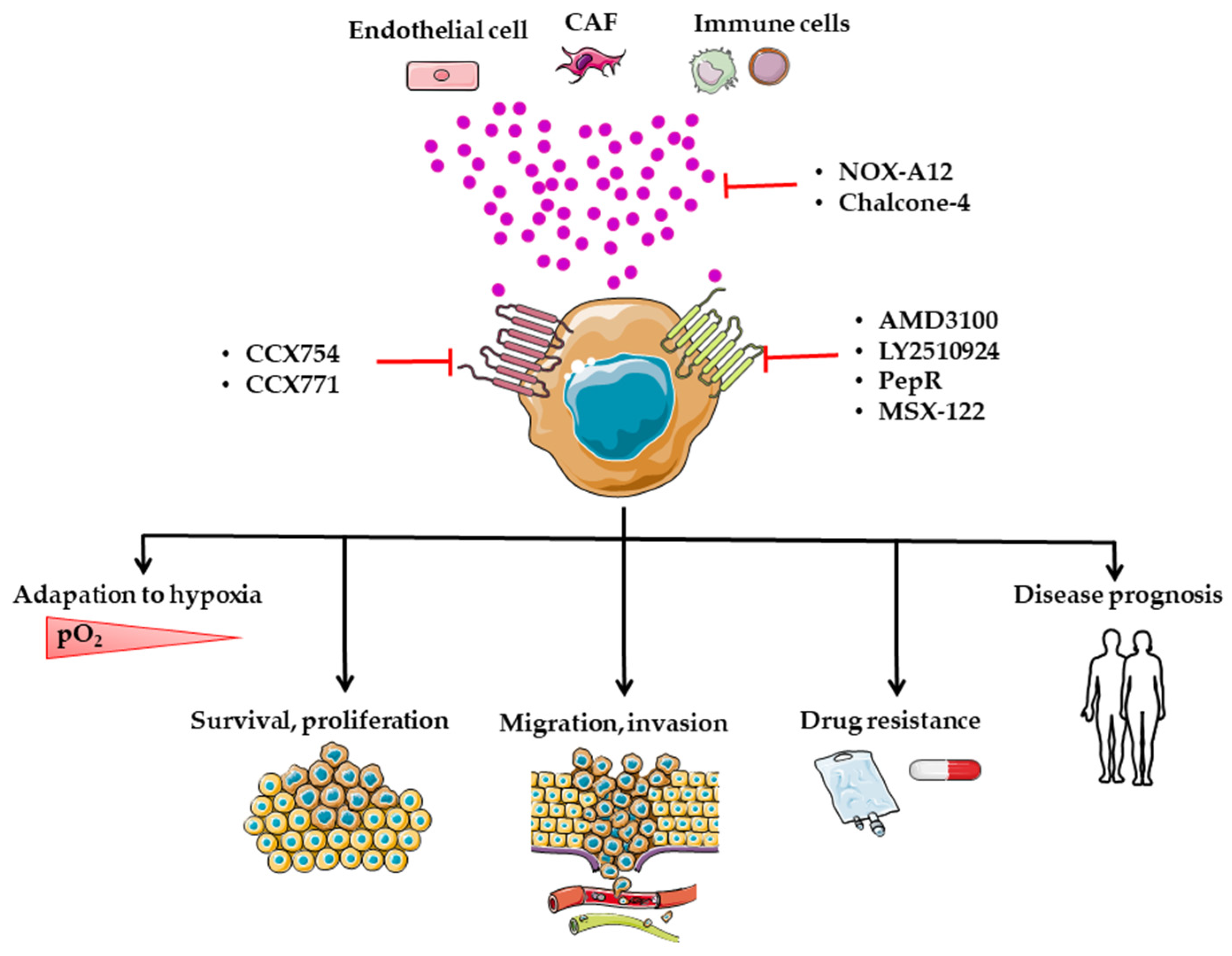
8. Targeting of the CXCL12/CXCR4/CXCR7 Axis in CRC
8.1. Preclinical Studies
8.2. AMD3100
8.3. LY2510924
8.4. PepR
8.5. MSX-122
8.6. CCX754 and CCX771
8.7. Chalcones
8.8. NOX-A12
| Inhibitor/Antagonist | Formula | IC50 | Target | References |
|---|---|---|---|---|
| AMD3100 | 1-[[4-(1,4,8,11 tetrazacyclotetradec-1-ylmethyl)phenyl]methyl]-1,4,8,11-tetrazacyclotetradecane | 37.5 nM | CXCR4 | [260] |
| LY2510924 | N(1)Phe-D-Tyr-Lys(iPr)-D-Arg-2Nal-Gly-D-Glu(1)-Lys(iPr)-NH2 | 0.079 nM | CXCR4 | [240] |
| PepR | (H-Arg-Ala-[Cys-Arg-Phe-Phe-Cys]-CO2H) | nd | CXCR4 | [242,243] |
| MSX-122 | N,N-9-(1,4-phenylenebis(methylene))dipyrimidin-2-amine | 10 nM | CXCR4 | [260] |
| CCX754 | nd | 5 nM | CXCR7 | [249] |
| CCX771 | nd | 4.1 nM | CXCR7 | [260] |
| Chalcone 4 | ((E)-1-(4′-chlorophenyl)-3-(4-hydroxy-3-metoxyphenyl) prop-2-en-1-one) | 150 nM | CXCL12 | [256] |
| NOX-A12 | nd | 5–200 nM | CXCL12 | [257] |
9. Clinical Trials
10. Resistance to Treatment
11. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ACKR3 | atypical chemokine receptor 3 |
| Akt | protein kinase B |
| cAMP | cyclic adenosine monophosphate |
| AOM | azoxymethane |
| APC | adenomatous polyposis coli |
| ARE | AU-rich element |
| ChIP | chromatin immunoprecipitation |
| CIN | chromosome instability |
| CIMP | CpG island methylator phenotype |
| CpG | cytosine-phosphate-guanine |
| CRC | colorectal cancer |
| CTLA-4 | cytotoxic T lymphocyte Antigen 4 |
| CXCL-10/12 | C-X-C motif ligand 10/12 |
| CXCR-3/4/7 | C-X-C motif receptor 3/4/7 |
| CAF | cancer-associated fibroblasts |
| DSS | dextran sodium sulfate |
| ECM | extracellular matrix |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial–mesenchymal transition |
| ERK | extracellular signal-regulated kinases |
| FAP | familial adenomatous polyposis |
| FGF | fibroblast growth factor |
| 5-FU | 5-fluorouracil |
| GAG | glycosaminoglycans |
| GEO | Gene Expression Omnibus |
| GRK2 | G-protein-coupled receptor kinase 2 |
| GTP | guanine–triphosphate |
| HDAC | histone deacetylases |
| HIC1 | hypermethylated in cancer 1 |
| HIF | hypoxia-inducible factor |
| HiRE | HIC1-responsive elements |
| HIV | human immunodeficiency virus |
| IBD | chronic inflammatory bowel disease |
| ICAM | intercellular adhesion molecule |
| ICI | immune checkpoint inhibitor |
| IGF | insulin-like growth factor |
| IL-2/6 | interleukin-2/6 |
| ITG | integrin |
| Jak | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| Lgr5 | leucine-rich repeat-containing G-protein coupled receptor 5 |
| MAPK | mitogen-activated protein kinases |
| MDSCs | myeloid-derived suppressor cells |
| MMP | matrix metalloproteinase |
| MMR | mismatch Repair |
| MSC | mesenchymal stromal cells |
| MSS | microsatellite stability |
| MSI | microsatellite instability |
| mTOR | mammalian target of rapamycin |
| NF-κB | nuclear factor-kappa B |
| PCAF | P300/CBP-associated factor |
| PDGF | platelet-derived growth factor |
| PD-1 | programmed cell death protein 1 |
| PD-L1 | programmed cell death protein ligand 1 |
| PI3K | phosphatidylinositol 3-kinase |
| PLC | Phospholipase C |
| PPPC1 | serine/threonine-protein phosphatase PP1-gamma catalytic |
| SDF-1 | stromal-cell-derived factor 1 |
| α-SMA | alpha-smooth muscle actin |
| STAT | signal transducer and activator of transcription |
| TAM | tumor associated macrophages |
| TCGA | the cancer genome atlas program |
| TGF | tumor growth factor |
| TIC | tumor-initiating cell |
| TIMP | tissue inhibitor of metalloproteinases |
| TNF | tumor necrosis factor |
| TNM | tumor, node, metastasis |
| TTP | tristetraprolin |
| UTR | untranslated transcribed region |
| VEGF | vascular endothelial growth factor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Ahmed, F.E. Effect of Diet, Life Style, and Other Environmental/Chemopreventive Factors on Colorectal Cancer Development, and Assessment of the Risks. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2004, 22, 91–147. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.N.; Giovannucci, E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Lukas, M. Inflammatory Bowel Disease as a Risk Factor for Colorectal Cancer. Dig. Dis. 2010, 28, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Morlote, D. Molecular Pathology of Colorectal Cancer. Adv. Anat. Pathol. 2020, 27, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Migliore, L.; Migheli, F.; Spisni, R.; Coppedè, F. Genetics, Cytogenetics, and Epigenetics of Colorectal Cancer. J. Biomed. Biotechnol. 2011, 2011, 792362. [Google Scholar] [CrossRef]
- Jass, J.R. Classification of Colorectal Cancer Based on Correlation of Clinical, Morphological and Molecular Features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Nakayama, M.; Oshima, M. Mutant P53 in Colon Cancer. J. Mol. Cell. Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Koumura, H.; Futamura, M.; Aoki, S.; Ymaguchi, K.; Kida, H.; Tanemura, H.; Shimokawa, K.; Saji, S. Somatic Alterations of the SMAD-2 Gene in Human Colorectal Cancers. Br. J. Cancer 1998, 78, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pernaute, A.; Pérez-Aguirre, E.; Cerdán, F.J.; Iniesta, P.; Díez Valladares, L.; de Juan, C.; Morán, A.; García-Botella, A.; García Aranda, C.; Benito, M.; et al. Overexpression of C-Myc and Loss of Heterozygosity on 2p, 3p, 5q, 17p and 18q in Sporadic Colorectal Carcinoma. Rev. Esp. Enferm. Dig. 2005, 97, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Watson, P.; Smyrk, T.C.; Lanspa, S.J.; Boman, B.M.; Boland, C.R.; Lynch, J.F.; Cavalieri, R.J.; Leppert, M.; White, R. Colon Cancer Genetics. Cancer 1992, 70, 1300–1312. [Google Scholar] [CrossRef][Green Version]
- Eshleman, J.R.; Markowitz, S.D. Mismatch Repair Defects in Human Carcinogenesis. Hum. Mol. Genet. 1996, 5, 1489–1494. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic Instability in Colorectal Cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef]
- Yamamoto, H.; Imai, K. Microsatellite Instability: An Update. Arch. Toxicol. 2015, 89, 899–921. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Smyrk, T. Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome). An Updated Review. Cancer 1996, 78, 1149–1167. [Google Scholar] [CrossRef]
- Samowitz, W.S.; Albertsen, H.; Herrick, J.; Levin, T.R.; Sweeney, C.; Murtaugh, M.A.; Wolff, R.K.; Slattery, M.L. Evaluation of a Large, Population-Based Sample Supports a CpG Island Methylator Phenotype in Colon Cancer. Gastroenterology 2005, 129, 837–845. [Google Scholar] [CrossRef]
- Lao, V.V.; Grady, W.M. Epigenetics and Colorectal Cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Gallois, C.; Laurent-Puig, P.; Taieb, J. Methylator Phenotype in Colorectal Cancer: A Prognostic Factor or Not? Crit. Rev. Oncol. Hematol. 2016, 99, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Watanabe, T.; Kanazawa, T.; Tanaka, J.; Tanaka, T.; Nagawa, H. Poorly Differentiated Colorectal Adenocarcinomas Show Higher Rates of Microsatellite Instability and Promoter Methylation of P16 and HMLH1: A Study Matched for T Classification and Tumor Location. J. Surg. Oncol. 2008, 97, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Popat, S.; Hubner, R.; Houlston, R.S. Systematic Review of Microsatellite Instability and Colorectal Cancer Prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef]
- Watanabe, T.; Kobunai, T.; Yamamoto, Y.; Matsuda, K.; Ishihara, S.; Nozawa, K.; Yamada, H.; Hayama, T.; Inoue, E.; Tamura, J.; et al. Chromosomal Instability (CIN) Phenotype, CIN High or CIN Low, Predicts Survival for Colorectal Cancer. J. Clin. Oncol. 2012, 30, 2256–2264. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Thiel, A.; Ristimäki, A. Toward a Molecular Classification of Colorectal Cancer: The Role of BRAF. Front. Oncol. 2013, 3, 281. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Lenz, H.-J.; Köhne, C.-H.; Heinemann, V.; Tejpar, S.; Melezínek, I.; Beier, F.; Stroh, C.; Rougier, P.; van Krieken, J.H.; et al. Fluorouracil, Leucovorin, and Irinotecan plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J. Clin. Oncol. 2015, 33, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; Van den Berg, J.G.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant Immunotherapy Leads to Pathological Responses in MMR-Proficient and MMR-Deficient Early-Stage Colon Cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Franke, A.J.; Skelton, W.P.; Starr, J.S.; Parekh, H.; Lee, J.J.; Overman, M.J.; Allegra, C.; George, T.J. Immunotherapy for Colorectal Cancer: A Review of Current and Novel Therapeutic Approaches. J. Natl. Cancer. Inst. 2019, 111, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Rollins, B.J. Chemokines. Blood 1997, 90, 909–928. [Google Scholar] [CrossRef]
- IUIS/WHO Subcommittee on Chemokine Nomenclature Chemokine/Chemokine Receptor Nomenclature. Cytokine 2003, 21, 48–49. [CrossRef]
- Murphy, P.M. Chemokine Receptors: Structure, Function and Role in Microbial Pathogenesis. Cytokine Growth Factor Rev. 1996, 7, 47–64. [Google Scholar] [CrossRef]
- Zou, Y.R.; Kottmann, A.H.; Kuroda, M.; Taniuchi, I.; Littman, D.R. Function of the Chemokine Receptor CXCR4 in Haematopoiesis and in Cerebellar Development. Nature 1998, 393, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, T.; Hirota, S.; Tachibana, K.; Takakura, N.; Nishikawa, S.; Kitamura, Y.; Yoshida, N.; Kikutani, H.; Kishimoto, T. Defects of B-Cell Lymphopoiesis and Bone-Marrow Myelopoiesis in Mice Lacking the CXC Chemokine PBSF/SDF-1. Nature 1996, 382, 635–638. [Google Scholar] [CrossRef]
- Yu, S.; Crawford, D.; Tsuchihashi, T.; Behrens, T.W.; Srivastava, D. The Chemokine Receptor CXCR7 Functions to Regulate Cardiac Valve Remodeling. Dev. Dyn. 2011, 240, 384–393. [Google Scholar] [CrossRef]
- Bleul, C.C.; Fuhlbrigge, R.C.; Casasnovas, J.M.; Aiuti, A.; Springer, T.A. A Highly Efficacious Lymphocyte Chemoattractant, Stromal Cell-Derived Factor 1 (SDF-1). J. Exp. Med. 1996, 184, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Ekbom, A.; Helmick, C.; Zack, M.; Adami, H.O. Ulcerative Colitis and Colorectal Cancer. A Population-Based Study. N. Engl. J. Med. 1990, 323, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Lewellis, S.W.; Knaut, H. Attractive Guidance: How the Chemokine SDF1/CXCL12 Guides Different Cells to Different Locations. Semin. Cell Dev. Biol. 2012, 23, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Petit, I.; Jin, D.; Rafii, S. The SDF-1-CXCR4 Signaling Pathway: A Molecular Hub Modulating Neo-Angiogenesis. Trends Immunol. 2007, 28, 299–307. [Google Scholar] [CrossRef]
- Siekmann, A.F.; Standley, C.; Fogarty, K.E.; Wolfe, S.A.; Lawson, N.D. Chemokine Signaling Guides Regional Patterning of the First Embryonic Artery. Genes Dev. 2009, 23, 2272–2277. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.; Willimann, K. Chemokines: Role in Inflammation and Immune Surveillance. Ann. Rheum. Dis. 2004, 63 (Suppl. 2), ii84–ii89. [Google Scholar] [CrossRef]
- White, G.E.; Iqbal, A.J.; Greaves, D.R. CC Chemokine Receptors and Chronic Inflammation--Therapeutic Opportunities and Pharmacological Challenges. Pharm. Rev. 2013, 65, 47–89. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F. Cancer and the Chemokine Network. Nat. Rev. Cancer 2004, 4, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A. Chemokines and Cancer. Int. J. Cancer 2006, 119, 2026–2029. [Google Scholar] [CrossRef] [PubMed]
- Raman, D.; Baugher, P.J.; Thu, Y.M.; Richmond, A. Role of Chemokines in Tumor Growth. Cancer Lett. 2007, 256, 137–165. [Google Scholar] [CrossRef]
- Nagasawa, T.; Kikutani, H.; Kishimoto, T. Molecular Cloning and Structure of a Pre-B-Cell Growth-Stimulating Factor. Proc. Natl. Acad. Sci. USA 1994, 91, 2305–2309. [Google Scholar] [CrossRef]
- Aiuti, A.; Webb, I.J.; Bleul, C.; Springer, T.; Gutierrez-Ramos, J.C. The Chemokine SDF-1 Is a Chemoattractant for Human CD34+ Hematopoietic Progenitor Cells and Provides a New Mechanism to Explain the Mobilization of CD34+ Progenitors to Peripheral Blood. J. Exp. Med. 1997, 185, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A. Involvement of Chemokine Receptors in Organ-Specific Metastasis. Contrib. Microbiol. 2006, 13, 191–199. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. Pathological Roles of the Homeostatic Chemokine CXCL12. Cytokine Growth Factor Rev.. 2018, 44, 51–68. [Google Scholar] [CrossRef]
- Roy, I.; Evans, D.B.; Dwinell, M.B. Chemokines and Chemokine Receptors: Update on Utility and Challenges for the Clinician. Surgery 2014, 155, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Shirozu, M.; Nakano, T.; Inazawa, J.; Tashiro, K.; Tada, H.; Shinohara, T.; Honjo, T. Structure and Chromosomal Localization of the Human Stromal Cell-Derived Factor 1 (SDF1) Gene. Genomics 1995, 28, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Cecil, J.; Peng, S.-B.; Schrementi, J.; Kovacevic, S.; Paul, D.; Su, E.W.; Wang, J. Identification and Expression of Novel Isoforms of Human Stromal Cell-Derived Factor 1. Gene 2006, 374, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Romain, B.; Benbrika-Nehmar, R.; Marisa, L.; Legrain, M.; Lobstein, V.; Oravecz, A.; Poidevin, L.; Bour, C.; Freund, J.-N.; Duluc, I.; et al. Histone Hypoacetylation Contributes to CXCL12 Downregulation in Colon Cancer: Impact on Tumor Growth and Cell Migration. Oncotarget 2017, 8, 38351–38366. [Google Scholar] [CrossRef]
- Smith, J.M.; Johanesen, P.A.; Wendt, M.K.; Binion, D.G.; Dwinell, M.B. CXCL12 Activation of CXCR4 Regulates Mucosal Host Defense through Stimulation of Epithelial Cell Migration and Promotion of Intestinal Barrier Integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G316–G326. [Google Scholar] [CrossRef] [PubMed]
- Bleul, C.C.; Farzan, M.; Choe, H.; Parolin, C.; Clark-Lewis, I.; Sodroski, J.; Springer, T.A. The Lymphocyte Chemoattractant SDF-1 Is a Ligand for LESTR/Fusin and Blocks HIV-1 Entry. Nature 1996, 382, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Loetscher, M.; Geiser, T.; O’Reilly, T.; Zwahlen, R.; Baggiolini, M.; Moser, B. Cloning of a Human Seven-Transmembrane Domain Receptor, LESTR, That Is Highly Expressed in Leukocytes. J. Biol. Chem. 1994, 269, 232–237. [Google Scholar] [CrossRef]
- Ma, Q.; Jones, D.; Borghesani, P.R.; Segal, R.A.; Nagasawa, T.; Kishimoto, T.; Bronson, R.T.; Springer, T.A. Impaired B-Lymphopoiesis, Myelopoiesis, and Derailed Cerebellar Neuron Migration in CXCR4- and SDF-1-Deficient Mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9448–9453. [Google Scholar] [CrossRef]
- Libert, F.; Parmentier, M.; Lefort, A.; Dumont, J.E.; Vassart, G. Complete Nucleotide Sequence of a Putative G Protein Coupled Receptor: RDC1. Nucleic Acids Res. 1990, 18, 1917. [Google Scholar] [CrossRef] [PubMed]
- Heesen, M.; Berman, M.A.; Charest, A.; Housman, D.; Gerard, C.; Dorf, M.E. Cloning and Chromosomal Mapping of an Orphan Chemokine Receptor: Mouse RDC1. Immunogenetics 1998, 47, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Mellado, M.; Rodríguez-Frade, J.M.; Mañes, S.; Martínez, A.C. Chemokine Signaling and Functional Responses: The Role of Receptor Dimerization and TK Pathway Activation. Annu. Rev. Immunol. 2001, 19, 397–421. [Google Scholar] [CrossRef] [PubMed]
- Walenkamp, A.M.E.; Lapa, C.; Herrmann, K.; Wester, H.-J. CXCR4 Ligands: The Next Big Hit? J. Nucl. Med. 2017, 58, 77S–82S. [Google Scholar] [CrossRef] [PubMed]
- Teixidó, J.; Martínez-Moreno, M.; Díaz-Martínez, M.; Sevilla-Movilla, S. The Good and Bad Faces of the CXCR4 Chemokine Receptor. Int. J. Biochem. Cell Biol. 2018, 95, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Balabanian, K.; Lagane, B.; Infantino, S.; Chow, K.Y.C.; Harriague, J.; Moepps, B.; Arenzana-Seisdedos, F.; Thelen, M.; Bachelerie, F. The Chemokine SDF-1/CXCL12 Binds to and Signals through the Orphan Receptor RDC1 in T Lymphocytes. J. Biol. Chem. 2005, 280, 35760–35766. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, S.; Kim, J.; Ahn, S.; Craig, S.; Lam, C.M.; Gerard, N.P.; Gerard, C.; Lefkowitz, R.J. Beta-Arrestin- but Not G Protein-Mediated Signaling by the “Decoy” Receptor CXCR7. Proc. Natl. Acad. Sci. USA 2010, 107, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Reyes-Alcaraz, A.; Yong, H.J.; Nguyen, L.P.; Park, H.-K.; Inoue, A.; Lee, C.S.; Seong, J.Y.; Hwang, J.-I. CXCR7: A β-Arrestin-Biased Receptor That Potentiates Cell Migration and Recruits β-Arrestin2 Exclusively through Gβγ Subunits and GRK2. Cell Biosci. 2020, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Sierro, F.; Biben, C.; Martínez-Muñoz, L.; Mellado, M.; Ransohoff, R.M.; Li, M.; Woehl, B.; Leung, H.; Groom, J.; Batten, M.; et al. Disrupted Cardiac Development but Normal Hematopoiesis in Mice Deficient in the Second CXCL12/SDF-1 Receptor, CXCR7. Proc. Natl. Acad. Sci. USA 2007, 104, 14759–14764. [Google Scholar] [CrossRef]
- Luker, K.; Gupta, M.; Luker, G. Bioluminescent CXCL12 Fusion Protein for Cellular Studies of CXCR4 and CXCR7. Biotechniques 2009, 47, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Levoye, A.; Balabanian, K.; Baleux, F.; Bachelerie, F.; Lagane, B. CXCR7 Heterodimerizes with CXCR4 and Regulates CXCL12-Mediated G Protein Signaling. Blood 2009, 113, 6085–6093. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, H.; van Ingen Schenau, D.S.; Bakker, N.E.C.; van Disseldorp, A.J.M.; Strik, A.; Hermens, L.S.; Koenen, T.B.; Krajnc-Franken, M.A.M.; Gossen, J.A. Early Postnatal Lethality and Cardiovascular Defects in CXCR7-Deficient Mice. Genesis 2008, 46, 235–245. [Google Scholar] [CrossRef]
- Sánchez-Alcañiz, J.A.; Haege, S.; Mueller, W.; Pla, R.; Mackay, F.; Schulz, S.; López-Bendito, G.; Stumm, R.; Marín, O. Cxcr7 Controls Neuronal Migration by Regulating Chemokine Responsiveness. Neuron 2011, 69, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Dambly-Chaudière, C.; Cubedo, N.; Ghysen, A. Control of Cell Migration in the Development of the Posterior Lateral Line: Antagonistic Interactions between the Chemokine Receptors CXCR4 and CXCR7/RDC1. BMC Dev. Biol. 2007, 7, 23. [Google Scholar] [CrossRef]
- Valentin, G.; Haas, P.; Gilmour, D. The Chemokine SDF1a Coordinates Tissue Migration through the Spatially Restricted Activation of Cxcr7 and Cxcr4b. Curr. Biol. 2007, 17, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Luker, K.E.; Steele, J.M.; Mihalko, L.A.; Ray, P.; Luker, G.D. Constitutive and Chemokine-Dependent Internalization and Recycling of CXCR7 in Breast Cancer Cells to Degrade Chemokine Ligands. Oncogene 2010, 29, 4599–4610. [Google Scholar] [CrossRef]
- Koenen, J.; Bachelerie, F.; Balabanian, K.; Schlecht-Louf, G.; Gallego, C. Atypical Chemokine Receptor 3 (ACKR3): A Comprehensive Overview of Its Expression and Potential Roles in the Immune System. Mol. Pharm. 2019, 96, 809–818. [Google Scholar] [CrossRef]
- Lau, S.; Feitzinger, A.; Venkiteswaran, G.; Wang, J.; Lewellis, S.W.; Koplinski, C.A.; Peterson, F.C.; Volkman, B.F.; Meier-Schellersheim, M.; Knaut, H. A Negative-Feedback Loop Maintains Optimal Chemokine Concentrations for Directional Cell Migration. Nat. Cell Biol. 2020, 22, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Quinn, K.E.; Mackie, D.I.; Caron, K.M. Emerging Roles of Atypical Chemokine Receptor 3 (ACKR3) in Normal Development and Physiology. Cytokine 2018, 109, 17–23. [Google Scholar] [CrossRef]
- Balkwill, F.R. The Chemokine System and Cancer. J. Pathol. 2012, 226, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Raman, D.; Sobolik-Delmaire, T.; Richmond, A. Chemokines in Health and Disease. Exp. Cell Res. 2011, 317, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of Chemokine Receptors in Breast Cancer Metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Hattermann, K.; Mentlein, R. An Infernal Trio: The Chemokine CXCL12 and Its Receptors CXCR4 and CXCR7 in Tumor Biology. Ann. Anat. 2013, 195, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guo, L.; Zhao, H.; Zhao, J.; Weng, H.; Zhao, B. CXCR4 Over-Expression and Survival in Cancer: A System Review and Meta-Analysis. Oncotarget 2015, 6, 5022–5040. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, W.; Yan, J.; Xiao, L.; Yang, L. Prognostic Significance of CXCR7 in Cancer Patients: A Meta-Analysis. Cancer Cell Int. 2018, 18, 212. [Google Scholar] [CrossRef] [PubMed]
- Romain, B.; Hachet-Haas, M.; Rohr, S.; Brigand, C.; Galzi, J.-L.; Gaub, M.-P.; Pencreach, E.; Guenot, D. Hypoxia Differentially Regulated CXCR4 and CXCR7 Signaling in Colon Cancer. Mol. Cancer 2014, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cheng, G.; Hao, M.; Zheng, J.; Zhou, X.; Zhang, J.; Taichman, R.S.; Pienta, K.J.; Wang, J. CXCL12/CXCR4/CXCR7 Chemokine Axis and Cancer Progression. Cancer Metastasis Rev. 2010, 29, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Takeuchi, H.; Lam, S.T.; Turner, R.R.; Wang, H.-J.; Kuo, C.; Foshag, L.; Bilchik, A.J.; Hoon, D.S.B. Chemokine Receptor CXCR4 Expression in Colorectal Cancer Patients Increases the Risk for Recurrence and for Poor Survival. J. Clin. Oncol. 2005, 23, 2744–2753. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Dai, T.; Xue, L.; Liu, X.; Wu, B.; Geng, J.; Mao, X.; Wang, R.; Chen, L.; Chu, X. Expression of Chemokine Receptor CXCR7 in Colorectal Carcinoma and Its Prognostic Significance. Int. J. Clin. Exp. Pathol. 2015, 8, 13051–13058. [Google Scholar] [PubMed]
- Xu, C.; Zheng, L.; Li, D.; Chen, G.; Gu, J.; Chen, J.; Yao, Q. CXCR4 Overexpression Is Correlated with Poor Prognosis in Colorectal Cancer. Life Sci. 2018, 208, 333–340. [Google Scholar] [CrossRef]
- Wani, N.; Nasser, M.W.; Ahirwar, D.K.; Zhao, H.; Miao, Z.; Shilo, K.; Ganju, R.K. C-X-C Motif Chemokine 12/C-X-C Chemokine Receptor Type 7 Signaling Regulates Breast Cancer Growth and Metastasis by Modulating the Tumor Microenvironment. Breast Cancer Res. 2014, 16, R54. [Google Scholar] [CrossRef]
- Ingold, B.; Schulz, S.; Budczies, J.; Neumann, U.; Ebert, M.P.A.; Weichert, W.; Röcken, C. The Role of Vascular CXCR4 Expression in Colorectal Carcinoma. Histopathology 2009, 55, 576–586. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Li, Z.; Zhang, Z.; Zhang, Y. Chemokine Receptor 7 Targets the Vascular Endothelial Growth Factor via the AKT/ERK Pathway to Regulate Angiogenesis in Colon Cancer. Cancer Med. 2019, 8, 5327–5340. [Google Scholar] [CrossRef]
- Chen, Y.; Teng, F.; Wang, G.; Nie, Z. Overexpression of CXCR7 Induces Angiogenic Capacity of Human Hepatocellular Carcinoma Cells via the AKT Signaling Pathway. Oncol. Rep. 2016, 36, 2275–2281. [Google Scholar] [CrossRef] [PubMed]
- Gentilini, A.; Caligiuri, A.; Raggi, C.; Rombouts, K.; Pinzani, M.; Lori, G.; Correnti, M.; Invernizzi, P.; Rovida, E.; Navari, N.; et al. CXCR7 Contributes to the Aggressive Phenotype of Cholangiocarcinoma Cells. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2246–2256. [Google Scholar] [CrossRef] [PubMed]
- Guillemot, E.; Karimdjee-Soilihi, B.; Pradelli, E.; Benchetrit, M.; Goguet-Surmenian, E.; Millet, M.-A.; Larbret, F.; Michiels, J.-F.; Birnbaum, D.; Alemanno, P.; et al. CXCR7 Receptors Facilitate the Progression of Colon Carcinoma within Lung Not within Liver. Br. J. Cancer 2012, 107, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Greijer, A.E.; Delis-van Diemen, P.M.; Fijneman, R.J.A.; Giles, R.H.; Voest, E.E.; van Hinsbergh, V.W.M.; Meijer, G.A. Presence of HIF-1 and Related Genes in Normal Mucosa, Adenomas and Carcinomas of the Colorectum. Virchows Arch. 2008, 452, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Frick, V.; Rubie, C.; Ghadjar, P.; Faust, S.K.; Wagner, M.; Gräber, S.; Schilling, M.K. Changes in CXCL12/CXCR4-Chemokine Expression during Onset of Colorectal Malignancies. Tumour Biol. 2011, 32, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Amara, S.; Chaar, I.; Khiari, M.; Ounissi, D.; Weslati, M.; Boughriba, R.; Hmida, A.B.; Bouraoui, S. Stromal Cell Derived Factor-1 and CXCR4 Expression in Colorectal Cancer Promote Liver Metastasis. Cancer Biomark 2015, 15, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, N.; Fukui, H.; Yamagishi, H.; Sekikawa, A.; Fujii, S.; Tomita, S.; Ichikawa, K.; Imura, J.; Hiraishi, H.; Fujimori, T. Expression of SDF-1 Alpha and Nuclear CXCR4 Predicts Lymph Node Metastasis in Colorectal Cancer. Br. J. Cancer 2008, 98, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Akishima-Fukasawa, Y.; Nakanishi, Y.; Ino, Y.; Moriya, Y.; Kanai, Y.; Hirohashi, S. Prognostic Significance of CXCL12 Expression in Patients with Colorectal Carcinoma. Am. J. Clin. Pathol. 2009, 132, 202–210; quiz 307. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, A.; Hashemzadeh, S.; Bahrami, T.; Estiar, M.A.; Feizi, M.A.H.; Pouladi, N.; Rostamizadeh, L.; Sakhinia, E. Expression Patterns of CXCL12 and Its Receptor in Colorectal Carcinoma. Clin. Lab. 2018, 64, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Wendt, M.K.; Johanesen, P.A.; Kang-Decker, N.; Binion, D.G.; Shah, V.; Dwinell, M.B. Silencing of Epithelial CXCL12 Expression by DNA Hypermethylation Promotes Colonic Carcinoma Metastasis. Oncogene 2006, 25, 4986–4997. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Bae, S.B.; Jeong, D.; Kim, E.S.; Kim, C.-N.; Park, D.-G.; Ahn, T.S.; Cho, S.W.; Shin, E.J.; Lee, M.S.; et al. Upregulation of Stromal Cell-Derived Factor 1α Expression Is Associated with the Resistance to Neoadjuvant Chemoradiotherapy of Locally Advanced Rectal Cancer: Angiogenic Markers of Neoadjuvant Chemoradiation. Oncol. Rep. 2014, 32, 2493–2500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tamas, K.; Domanska, U.M.; van Dijk, T.H.; Timmer-Bosscha, H.; Havenga, K.; Karrenbeld, A.; Sluiter, W.J.; Beukema, J.C.; van Vugt, M.A.T.M.; de Vries, E.G.E.; et al. CXCR4 and CXCL12 Expression in Rectal Tumors of Stage IV Patients Before and After Local Radiotherapy and Systemic Neoadjuvant Treatment. Curr. Pharm. Des. 2015, 21, 2276–2283. [Google Scholar] [CrossRef] [PubMed]
- Olaso, E.; Salado, C.; Egilegor, E.; Gutierrez, V.; Santisteban, A.; Sancho-Bru, P.; Friedman, S.L.; Vidal-Vanaclocha, F. Proangiogenic Role of Tumor-Activated Hepatic Stellate Cells in Experimental Melanoma Metastasis. Hepatology 2003, 37, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Wang, Y.; Liu, J.; Mok, S.C.; Xue, F.; Zhang, W. CXCL12/CXCR4: A Symbiotic Bridge Linking Cancer Cells and Their Stromal Neighbors in Oncogenic Communication Networks. Oncogene 2016, 35, 816–826. [Google Scholar] [CrossRef]
- Yu, P.F.; Huang, Y.; Xu, C.L.; Lin, L.Y.; Han, Y.Y.; Sun, W.H.; Hu, G.H.; Rabson, A.B.; Wang, Y.; Shi, Y.F. Downregulation of CXCL12 in Mesenchymal Stromal Cells by TGFβ Promotes Breast Cancer Metastasis. Oncogene 2017, 36, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Shinagawa, K.; Kitadai, Y.; Tanaka, M.; Sumida, T.; Kodama, M.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Mesenchymal Stem Cells Enhance Growth and Metastasis of Colon Cancer. Int. J. Cancer 2010, 127, 2323–2333. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.-C.; Sun, X.-W.; Su, H.; Chen, Q.; Guo, T.-K.; Li, Y.; Chen, X.-C.; Guo, J.; Gong, Z.-Q.; Zhao, X.-D.; et al. Fibroblast-Derived CXCL12/SDF-1α Promotes CXCL6 Secretion and Co-Operatively Enhances Metastatic Potential through the PI3K/Akt/MTOR Pathway in Colon Cancer. World J. Gastroenterol. 2017, 23, 5167–5178. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef]
- Cuiffo, B.G.; Karnoub, A.E. Mesenchymal Stem Cells in Tumor Development. Cell Adh. Migr. 2012, 6, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Erreni, M.; Bianchi, P.; Laghi, L.; Mirolo, M.; Fabbri, M.; Locati, M.; Mantovani, A.; Allavena, P. Expression of Chemokines and Chemokine Receptors in Human Colon Cancer. Meth. Enzymol. 2009, 460, 105–121. [Google Scholar] [CrossRef]
- Cheng, X.-S.; Li, Y.-F.; Tan, J.; Sun, B.; Xiao, Y.-C.; Fang, X.-B.; Zhang, X.-F.; Li, Q.; Dong, J.-H.; Li, M.; et al. CCL20 and CXCL8 Synergize to Promote Progression and Poor Survival Outcome in Patients with Colorectal Cancer by Collaborative Induction of the Epithelial-Mesenchymal Transition. Cancer Lett. 2014, 348, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Oladipo, O.; Conlon, S.; O’Grady, A.; Purcell, C.; Wilson, C.; Maxwell, P.J.; Johnston, P.G.; Stevenson, M.; Kay, E.W.; Wilson, R.H.; et al. The Expression and Prognostic Impact of CXC-Chemokines in Stage II and III Colorectal Cancer Epithelial and Stromal Tissue. Br. J. Cancer 2011, 104, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, S.; Teixeira, V.H.; Kalber, T.; Jose, R.J.; Floto, R.A.; Janes, S.M. Macrophage Migration Inhibitory Factor-CXCR4 Is the Dominant Chemotactic Axis in Human Mesenchymal Stem Cell Recruitment to Tumors. J. Immunol. 2015, 194, 3463–3474. [Google Scholar] [CrossRef] [PubMed]
- Cojoc, M.; Peitzsch, C.; Trautmann, F.; Polishchuk, L.; Telegeev, G.D.; Dubrovska, A. Emerging Targets in Cancer Management: Role of the CXCL12/CXCR4 Axis. Onco Targets 2013, 6, 1347–1361. [Google Scholar] [CrossRef]
- Mishra, P.; Banerjee, D.; Ben-Baruch, A. Chemokines at the Crossroads of Tumor-Fibroblast Interactions That Promote Malignancy. J. Leukoc. Biol. 2011, 89, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, P.; Haier, J.; Schlüter, K.; Domikowsky, B.; Wendel, C.; Wiesner, U.; Kubitza, R.; Engers, R.; Schneider, S.W.; Homey, B.; et al. CXCR4 Regulates the Early Extravasation of Metastatic Tumor Cells in Vivo. Neoplasia 2009, 11, 651–661. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.-N.; Pan, T.; Miao, R.-R.; Zhang, Y.-Y.; Wu, S.-H.; Qu, X.; Cui, S.-X. Atypical Chemokine Receptor 3 (ACKR3) Induces the Perturbation of RRNA Biogenesis in Colonic Cells: A Novel Mechanism of Colorectal Tumorigenesis. bioRxiv 2021. Posted 2 September 2021. [Google Scholar] [CrossRef]
- Miao, Z.; Luker, K.E.; Summers, B.C.; Berahovich, R.; Bhojani, M.S.; Rehemtulla, A.; Kleer, C.G.; Essner, J.J.; Nasevicius, A.; Luker, G.D.; et al. CXCR7 (RDC1) Promotes Breast and Lung Tumor Growth in Vivo and Is Expressed on Tumor-Associated Vasculature. Proc. Natl. Acad. Sci. USA 2007, 104, 15735–15740. [Google Scholar] [CrossRef]
- Salmaggi, A.; Maderna, E.; Calatozzolo, C.; Gaviani, P.; Canazza, A.; Milanesi, I.; Silvani, A.; DiMeco, F.; Carbone, A.; Pollo, B. CXCL12, CXCR4 and CXCR7 Expression in Brain Metastases. Cancer Biol. 2009, 8, 1608–1614. [Google Scholar] [CrossRef]
- Mitchell, A.; Hasanali, S.L.; Morera, D.S.; Baskar, R.; Wang, X.; Khan, R.; Talukder, A.; Li, C.S.; Manoharan, M.; Jordan, A.R.; et al. A Chemokine/Chemokine Receptor Signature Potentially Predicts Clinical Outcome in Colorectal Cancer Patients. Cancer Biomark 2019, 26, 291–301. [Google Scholar] [CrossRef]
- Stanisavljević, L.; Aßmus, J.; Storli, K.E.; Leh, S.M.; Dahl, O.; Myklebust, M.P. CXCR4, CXCL12 and the Relative CXCL12-CXCR4 Expression as Prognostic Factors in Colon Cancer. Tumour Biol. 2016, 37, 7441–7452. [Google Scholar] [CrossRef]
- Li, Y.-P.; Pang, J.; Gao, S.; Bai, P.-Y.; Wang, W.; Kong, P.; Cui, Y. Role of CXCR4 and SDF1 as Prognostic Factors for Survival and the Association with Clinicopathology in Colorectal Cancer: A Systematic Meta-Analysis. Tumour Biol. 2017, 39, 1010428317706206. [Google Scholar] [CrossRef] [PubMed]
- Marisa, L.; de Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.-C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, T.; O’Connor, T.P.; Crystal, R.G. Adenoviral Gene Transfer of Stromal Cell-Derived Factor-1 to Murine Tumors Induces the Accumulation of Dendritic Cells and Suppresses Tumor Growth. Cancer Res. 2006, 66, 3513–3522. [Google Scholar] [CrossRef][Green Version]
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Lugli, A.; Zlobec, I.; Hartmann, A.; Bifulco, C.; et al. Towards the Introduction of the “Immunoscore” in the Classification of Malignant Tumours. J. Pathol. 2014, 232, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Pagès, F.; Kirilovsky, A.; Mlecnik, B.; Asslaber, M.; Tosolini, M.; Bindea, G.; Lagorce, C.; Wind, P.; Marliot, F.; Bruneval, P.; et al. In Situ Cytotoxic and Memory T Cells Predict Outcome in Patients with Early-Stage Colorectal Cancer. J. Clin. Oncol. 2009, 27, 5944–5951. [Google Scholar] [CrossRef]
- Prall, F.; Dührkop, T.; Weirich, V.; Ostwald, C.; Lenz, P.; Nizze, H.; Barten, M. Prognostic Role of CD8+ Tumor-Infiltrating Lymphocytes in Stage III Colorectal Cancer with and without Microsatellite Instability. Hum. Pathol. 2004, 35, 808–816. [Google Scholar] [CrossRef]
- Chiba, T.; Ohtani, H.; Mizoi, T.; Naito, Y.; Sato, E.; Nagura, H.; Ohuchi, A.; Ohuchi, K.; Shiiba, K.; Kurokawa, Y.; et al. Intraepithelial CD8+ T-Cell-Count Becomes a Prognostic Factor after a Longer Follow-up Period in Human Colorectal Carcinoma: Possible Association with Suppression of Micrometastasis. Br. J. Cancer 2004, 91, 1711–1717. [Google Scholar] [CrossRef]
- Lalos, A.; Tülek, A.; Tosti, N.; Mechera, R.; Wilhelm, A.; Soysal, S.; Daester, S.; Kancherla, V.; Weixler, B.; Spagnoli, G.C.; et al. Prognostic Significance of CD8+ T-Cells Density in Stage III Colorectal Cancer Depends on SDF-1 Expression. Sci. Rep. 2021, 11, 775. [Google Scholar] [CrossRef]
- Lv, S.; Yang, Y.; Kwon, S.; Han, M.; Zhao, F.; Kang, H.; Dai, C.; Wang, R. The Association of CXCR4 Expression with Prognosis and Clinicopathological Indicators in Colorectal Carcinoma Patients: A Meta-Analysis. Histopathology 2014, 64, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-N.; Jiang, K.-T.; Tan, P.; Wang, A.-H.; Kong, Q.-Y.; Wang, C.-Y.; Lu, H.-R.; Wang, J. Prognosis and Clinicopathology of CXCR4 in Colorectal Cancer Patients: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2015, 16, 4077–4080. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Sun, Y.; Liu, X. CXCR4 as a Prognostic Biomarker in Gastrointestinal Cancer: A Meta-Analysis. Biomarkers 2019, 24, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Ottaiano, A.; Scala, S.; Normanno, N.; Botti, G.; Tatangelo, F.; Di Mauro, A.; Capozzi, M.; Facchini, S.; Tafuto, S.; Nasti, G. Prognostic and Predictive Role of CXC Chemokine Receptor 4 in Metastatic Colorectal Cancer Patients. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Yopp, A.C.; Shia, J.; Butte, J.M.; Allen, P.J.; Fong, Y.; Jarnagin, W.R.; DeMatteo, R.P.; D’Angelica, M.I. CXCR4 Expression Predicts Patient Outcome and Recurrence Patterns after Hepatic Resection for Colorectal Liver Metastases. Ann. Surg. Oncol. 2012, 19 (Suppl. S3), S339–S346. [Google Scholar] [CrossRef]
- D’Alterio, C.; Nasti, G.; Polimeno, M.; Ottaiano, A.; Conson, M.; Circelli, L.; Botti, G.; Scognamiglio, G.; Santagata, S.; De Divitiis, C.; et al. CXCR4-CXCL12-CXCR7, TLR2-TLR4, and PD-1/PD-L1 in Colorectal Cancer Liver Metastases from Neoadjuvant-Treated Patients. Oncoimmunology 2016, 5, e1254313. [Google Scholar] [CrossRef] [PubMed]
- Alamo, P.; Gallardo, A.; Di Nicolantonio, F.; Pavón, M.A.; Casanova, I.; Trias, M.; Mangues, M.A.; Lopez-Pousa, A.; Villaverde, A.; Vázquez, E.; et al. Higher Metastatic Efficiency of KRas G12V than KRas G13D in a Colorectal Cancer Model. FASEB J. 2015, 29, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, S.; Tsuchida, K.; Shiozawa, M.; Hiroshima, Y.; Kimura, Y.; Hashimoto, I.; Watanabe, H.; Kano, K.; Numata, M.; Aoyama, T.; et al. Clinical Significance of Chemokine Receptor CXCR4 and CCR7 MRNA Expression in Patients With Colorectal Cancer. Anticancer Res. 2021, 41, 4489–4495. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Shu, G.; Liu, H.; Zhang, Q.; Ma, Z.; Ren, C.; Guo, H.; Shi, J.; Liu, J.; Zhang, C.; et al. The Diagnostic Value of Chemokine/Chemokine Receptor Pairs in Hepatocellular Carcinoma and Colorectal Liver Metastasis. J. Histochem. Cytochem. 2019, 67, 299–308. [Google Scholar] [CrossRef]
- Xu, F.; Wang, F.; Di, M.; Huang, Q.; Wang, M.; Hu, H.; Jin, Y.; Dong, J.; Lai, M. Classification Based on the Combination of Molecular and Pathologic Predictors Is Superior to Molecular Classification on Prognosis in Colorectal Carcinoma. Clin. Cancer Res. 2007, 13, 5082–5088. [Google Scholar] [CrossRef][Green Version]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A Human Colon Cancer Cell Capable of Initiating Tumour Growth in Immunodeficient Mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and Expansion of Human Colon-Cancer-Initiating Cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; St Clair, R.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 Expression Is Not Restricted to Stem Cells, and Both CD133+ and CD133- Metastatic Colon Cancer Cells Initiate Tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, Y.; Yan, X.; Tian, Q.; Li, L.; Lin, E.H. CD133, Stem Cells, and Cancer Stem Cells: Myth or Reality? Curr. Colorectal Cancer Rep. 2011, 7, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Han, Z.; Jing, Y.; Tao, S.; Li, T.; Wang, H.; Wang, Y.; Li, R.; Yang, Y.; Zhao, X.; et al. CD133(+)CXCR4(+) Colon Cancer Cells Exhibit Metastatic Potential and Predict Poor Prognosis of Patients. BMC Med. 2012, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Silinsky, J.; Grimes, C.; Driscoll, T.; Green, H.; Cordova, J.; Davis, N.K.; Li, L.; Margolin, D.A. CD 133+ and CXCR4+ Colon Cancer Cells as a Marker for Lymph Node Metastasis. J. Surg. Res. 2013, 185, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cao, J.; Ji, Z.; Wang, J.; Jiang, T.; Ding, H. Co-Expression of Lgr5 and CXCR4 Characterizes Cancer Stem-like Cells of Colorectal Cancer. Oncotarget 2016, 7, 81144–81155. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Riese, D.J.; Shen, J. The Role of the CXCL12/CXCR4/CXCR7 Chemokine Axis in Cancer. Front. Pharm. 2020, 11, 574667. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, X.; Wei, M.; Wang, Z. The Role of CXCL12 Axis in Lung Metastasis of Colorectal Cancer. J. Cancer 2018, 9, 3898–3903. [Google Scholar] [CrossRef]
- Yamada, S.; Shimada, M.; Utsunomiya, T.; Morine, Y.; Imura, S.; Ikemoto, T.; Mori, H.; Arakawa, Y.; Kanamoto, M.; Iwahashi, S.; et al. CXC Receptor 4 and Stromal Cell-Derived Factor 1 in Primary Tumors and Liver Metastases of Colorectal Cancer. J. Surg. Res. 2014, 187, 107–112. [Google Scholar] [CrossRef]
- Rubie, C.; Kollmar, O.; Frick, V.O.; Wagner, M.; Brittner, B.; Gräber, S.; Schilling, M.K. Differential CXC Receptor Expression in Colorectal Carcinomas. Scand. J. Immunol. 2008, 68, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Sherif, M.F.; Ismail, I.M.; Ata, S.M.S. Expression of CXCR7 in Colorectal Adenoma and Adenocarcinoma: Correlation with Clinicopathological Parameters. Ann. Diagn. Pathol. 2020, 49, 151621. [Google Scholar] [CrossRef] [PubMed]
- Kheirelseid, E.A.H.; Miller, N.; Chang, K.H.; Nugent, M.; Kerin, M.J. Clinical Applications of Gene Expression in Colorectal Cancer. J. Gastrointest. Oncol. 2013, 4, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, U.M.; Ortega, S.B.; Hu, R.; Gilchrist, R.; Kong, X.; Partin, A.; Plautz, E.J.; Klein, R.S.; Gidday, J.M.; Stowe, A.M. Preconditioning-Induced CXCL12 Upregulation Minimizes Leukocyte Infiltration after Stroke in Ischemia-Tolerant Mice. J. Cereb. Blood Flow Metab. 2017, 37, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Ceradini, D.J.; Kulkarni, A.R.; Callaghan, M.J.; Tepper, O.M.; Bastidas, N.; Kleinman, M.E.; Capla, J.M.; Galiano, R.D.; Levine, J.P.; Gurtner, G.C. Progenitor Cell Trafficking Is Regulated by Hypoxic Gradients through HIF-1 Induction of SDF-1. Nat. Med. 2004, 10, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Karshovska, E.; Zernecke, A.; Sevilmis, G.; Millet, A.; Hristov, M.; Cohen, C.D.; Schmid, H.; Krotz, F.; Sohn, H.-Y.; Klauss, V.; et al. Expression of HIF-1alpha in Injured Arteries Controls SDF-1alpha Mediated Neointima Formation in Apolipoprotein E Deficient Mice. Arter. Thromb Vasc. Biol. 2007, 27, 2540–2547. [Google Scholar] [CrossRef] [PubMed]
- Hitchon, C.; Wong, K.; Ma, G.; Reed, J.; Lyttle, D.; El-Gabalawy, H. Hypoxia-Induced Production of Stromal Cell-Derived Factor 1 (CXCL12) and Vascular Endothelial Growth Factor by Synovial Fibroblasts. Arthritis Rheum. 2002, 46, 2587–2597. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Prasad, S.B.; Prasad, C.B.; Pandey, L.K.; Pradhan, S.; Singh, S.; Narayan, G. CXCL12 Is a Key Regulator in Tumor Microenvironment of Cervical Cancer: An in Vitro Study. Clin. Exp. Metastasis 2016, 33, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Kim, Y.-O.; Jo, I. Differential Expression of Stromal Cell-Derived Factor 1 in Human Brain Microvascular Endothelial Cells and Pericytes Involves Histone Modifications. Biochem. Biophys. Res. Commun. 2009, 382, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Wendt, M.K.; Drury, L.J.; Vongsa, R.A.; Dwinell, M.B. Constitutive CXCL12 Expression Induces Anoikis in Colorectal Carcinoma Cells. Gastroenterology 2008, 135, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Halees, A.S.; El-Badrawi, R.; Khabar, K.S.A. ARED Organism: Expansion of ARED Reveals AU-Rich Element Cluster Variations between Human and Mouse. Nucleic Acids Res. 2008, 36, D137–D140. [Google Scholar] [CrossRef]
- Al-Souhibani, N.; Al-Ghamdi, M.; Al-Ahmadi, W.; Khabar, K.S.A. Posttranscriptional Control of the Chemokine Receptor CXCR4 Expression in Cancer Cells. Carcinogenesis 2014, 35, 1983–1992. [Google Scholar] [CrossRef]
- Semenza, G.L. Life with Oxygen. Science 2007, 318, 62–64. [Google Scholar] [CrossRef]
- Zong, S.; Li, W.; Li, H.; Han, S.; Liu, S.; Shi, Q.; Hou, F. Identification of Hypoxia-Regulated Angiogenic Genes in Colorectal Cancer. Biochem. Biophys. Res. Commun. 2017, 493, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; De Marzo, A.M.; Laughner, E.; Lim, M.; Hilton, D.A.; Zagzag, D.; Buechler, P.; Isaacs, W.B.; Semenza, G.L.; Simons, J.W. Overexpression of Hypoxia-Inducible Factor 1alpha in Common Human Cancers and Their Metastases. Cancer Res. 1999, 59, 5830–5835. [Google Scholar] [PubMed]
- Harris, A.L. Hypoxia--a Key Regulatory Factor in Tumour Growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, M.; Xu, H.; Shimin, Z.; He, S.; Wang, L.; Zhang, Y. Clinicopathologic Significance of HIF-1α, CXCR4, and VEGF Expression in Colon Cancer. Clin. Dev. Immunol. 2010, 2010, 537531. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Simińska, D.; Gąssowska-Dobrowolska, M.; Listos, J.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation through HIF-1 and NF-ΚB Activation: A Review of the Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 10701. [Google Scholar] [CrossRef]
- Schioppa, T.; Uranchimeg, B.; Saccani, A.; Biswas, S.K.; Doni, A.; Rapisarda, A.; Bernasconi, S.; Saccani, S.; Nebuloni, M.; Vago, L.; et al. Regulation of the Chemokine Receptor CXCR4 by Hypoxia. J. Exp. Med. 2003, 198, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Staller, P.; Sulitkova, J.; Lisztwan, J.; Moch, H.; Oakeley, E.J.; Krek, W. Chemokine Receptor CXCR4 Downregulated by von Hippel-Lindau Tumour Suppressor PVHL. Nature 2003, 425, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Fertala, J.; Vresilovic, E.J.; Albert, T.J.; Shapiro, I.M. Nucleus Pulposus Cells Upregulate PI3K/Akt and MEK/ERK Signaling Pathways under Hypoxic Conditions and Resist Apoptosis Induced by Serum Withdrawal. Spine 2005, 30, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Hu, Y.; Zheng, H. Hypoxia Increases Expression of CXC Chemokine Receptor 4 via Activation of PI3K/Akt Leading to Enhanced Migration of Endothelial Progenitor Cells. Eur. Rev. Med. Pharm. Sci. 2017, 21, 1820–1827. [Google Scholar]
- Treins, C.; Giorgetti-Peraldi, S.; Murdaca, J.; Semenza, G.L.; Van Obberghen, E. Insulin Stimulates Hypoxia-Inducible Factor 1 through a Phosphatidylinositol 3-Kinase/Target of Rapamycin-Dependent Signaling Pathway. J. Biol. Chem. 2002, 277, 27975–27981. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.-C.; Han, N.; Ping, G.-F.; Zheng, P.-F.; Feng, H.-L.; Qin, L.; He, P. MicroRNA-9 Functions as a Tumor Suppressor in Colorectal Cancer by Targeting CXCR4. Int. J. Clin. Exp. Pathol. 2018, 11, 526–536. [Google Scholar] [PubMed]
- Liu, Y.; Zhou, Y.; Feng, X.; An, P.; Quan, X.; Wang, H.; Ye, S.; Yu, C.; He, Y.; Luo, H. MicroRNA-126 Functions as a Tumor Suppressor in Colorectal Cancer Cells by Targeting CXCR4 via the AKT and ERK1/2 Signaling Pathways. Int. J. Oncol. 2014, 44, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Feng, X.; Yang, P.; Yang, J.; An, P.; Wang, H.; Ye, S.; Yu, C.; He, Y.; et al. Low Expression of MicroRNA-126 Is Associated with Poor Prognosis in Colorectal Cancer. Genes Chromosomes Cancer 2014, 53, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Sun, B.; Li, Z.; Chen, Z.; Xiang, J. MiR-622 Inhibited Colorectal Cancer Occurrence and Metastasis by Suppressing K-Ras. Mol. Carcinog. 2016, 55, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Sun, B.; Wang, J.; Wang, Y. MiR-622 Inhibits Angiogenesis by Suppressing the CXCR4-VEGFA Axis in Colorectal Cancer. Gene 2019, 699, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.-T.; Qian, F.; Fang, K.; Lin, K.-Y.; Wang, W.-T.; Chen, Y.-Q. MiR-133b, a Muscle-Specific MicroRNA, Is a Novel Prognostic Marker That Participates in the Progression of Human Colorectal Cancer via Regulation of CXCR4 Expression. Mol. Cancer 2013, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Fu, H.-J.; Xie, Q.-S.; Wang, L.; Zhang, R.; Guo, Z.-Y.; Zhao, J.; Meng, Y.-L.; Ren, X.-L.; Wang, T.; et al. HER2 Interacts with CD44 to Up-Regulate CXCR4 via Epigenetic Silencing of MicroRNA-139 in Gastric Cancer Cells. Gastroenterology 2011, 141, 2076–2087.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, J.-Y.; Zhang, J.; Long, Y.-X.; Li, Y.-J.; Guo, X.-D.; Wei, M.-N.; Liu, W.-J. Role of Visfatin in Promoting Proliferation and Invasion of Colorectal Cancer Cells by Downregulating SDF-1/CXCR4-Mediated MiR-140-3p Expression. Eur. Rev. Med. Pharm. Sci. 2020, 24, 5367–5377. [Google Scholar] [CrossRef]
- Yu, X.; Shi, W.; Zhang, Y.; Wang, X.; Sun, S.; Song, Z.; Liu, M.; Zeng, Q.; Cui, S.; Qu, X. CXCL12/CXCR4 Axis Induced MiR-125b Promotes Invasion and Confers 5-Fluorouracil Resistance through Enhancing Autophagy in Colorectal Cancer. Sci. Rep. 2017, 7, 42226. [Google Scholar] [CrossRef] [PubMed]
- Stuckel, A.J.; Zhang, W.; Zhang, X.; Zeng, S.; Dougherty, U.; Mustafi, R.; Zhang, Q.; Perreand, E.; Khare, T.; Joshi, T.; et al. Enhanced CXCR4 Expression Associates with Increased Gene Body 5-Hydroxymethylcytosine Modification but Not Decreased Promoter Methylation in Colorectal Cancer. Cancers 2020, 12, 539. [Google Scholar] [CrossRef]
- Branco, M.R.; Ficz, G.; Reik, W. Uncovering the Role of 5-Hydroxymethylcytosine in the Epigenome. Nat. Rev. Genet. 2011, 13, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. 5-Hydroxymethylcytosine Signatures in Circulating Cell-Free DNA as Diagnostic Biomarkers for Human Cancers. Cell Res. 2017, 27, 1243–1257. [Google Scholar] [CrossRef]
- Krook, M.A.; Nicholls, L.A.; Scannell, C.A.; Chugh, R.; Thomas, D.G.; Lawlor, E.R. Stress-Induced CXCR4 Promotes Migration and Invasion of Ewing Sarcoma. Mol. Cancer Res. 2014, 12, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Krook, M.A.; Hawkins, A.G.; Patel, R.M.; Lucas, D.R.; Van Noord, R.; Chugh, R.; Lawlor, E.R. A Bivalent Promoter Contributes to Stress-Induced Plasticity of CXCR4 in Ewing Sarcoma. Oncotarget 2016, 7, 61775–61788. [Google Scholar] [CrossRef]
- Urosevic, J.; Blasco, M.T.; Llorente, A.; Bellmunt, A.; Berenguer-Llergo, A.; Guiu, M.; Cañellas, A.; Fernandez, E.; Burkov, I.; Clapés, M.; et al. ERK1/2 Signaling Induces Upregulation of ANGPT2 and CXCR4 to Mediate Liver Metastasis in Colon Cancer. Cancer Res. 2020, 80, 4668–4680. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Huang, Q.; Wei, G.-H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Huang, W.; Chen, J.; Qiao, C.; Liu, D.; Ji, X.; Xie, M.; Zhang, T.; Wang, Y.; Sun, M.; et al. CXCL12-Mediated HOXB5 Overexpression Facilitates Colorectal Cancer Metastasis through Transactivating CXCR4 and ITGB3. Theranostics 2021, 11, 2612–2633. [Google Scholar] [CrossRef] [PubMed]
- Esencay, M.; Sarfraz, Y.; Zagzag, D. CXCR7 Is Induced by Hypoxia and Mediates Glioma Cell Migration towards SDF-1α. BMC Cancer 2013, 13, 347. [Google Scholar] [CrossRef] [PubMed]
- Nosho, K.; Kure, S.; Irahara, N.; Shima, K.; Baba, Y.; Spiegelman, D.; Meyerhardt, J.A.; Giovannucci, E.L.; Fuchs, C.S.; Ogino, S. A Prospective Cohort Study Shows Unique Epigenetic, Genetic, and Prognostic Features of Synchronous Colorectal Cancers. Gastroenterology 2009, 137, 1609–1620.e1-3. [Google Scholar] [CrossRef]
- Bagci, B.; Sari, M.; Karadayi, K.; Turan, M.; Ozdemir, O.; Bagci, G. KRAS, BRAF Oncogene Mutations and Tissue Specific Promoter Hypermethylation of Tumor Suppressor SFRP2, DAPK1, MGMT, HIC1 and P16 Genes in Colorectal Cancer Patients. Cancer Biomark. 2016, 17, 133–143. [Google Scholar] [CrossRef]
- Van Rechem, C.; Rood, B.R.; Touka, M.; Pinte, S.; Jenal, M.; Guérardel, C.; Ramsey, K.; Monté, D.; Bégue, A.; Tschan, M.P.; et al. Scavenger Chemokine (CXC Motif) Receptor 7 (CXCR7) Is a Direct Target Gene of HIC1 (Hypermethylated in Cancer 1). J. Biol. Chem. 2009, 284, 20927–20935. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-M.; Zhang, F.; Chen, X.-B.; Jun, C.-M.; Jing, X.; Wei, D.-X.; Xia, Y.; Zhou, Y.-B.; Xiao, X.-Q.; Jia, R.-Q.; et al. MiR-100 Suppresses the Proliferation and Tumor Growth of Esophageal Squamous Cancer Cells via Targeting CXCR7. Oncol. Rep. 2016, 35, 3453–3459. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, X.; Zhu, X.; Zhong, Z.; Xu, R.; Wang, Z.; Cao, J.; Hou, Y. Decreased Expression of MiR-430 Promotes the Development of Bladder Cancer via the Upregulation of CXCR7. Mol. Med. Rep. 2013, 8, 140–146. [Google Scholar] [CrossRef]
- Ge, Y.; Shu, J.; Shi, G.; Yan, F.; Li, Y.; Ding, H. MiR-100 Suppresses the Proliferation, Invasion, and Migration of Hepatocellular Carcinoma Cells via Targeting CXCR7. J. Immunol. Res. 2021, 2021, 9920786. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Song, J.; Ge, J.; Song, Z.; Chen, J.; Wu, C. MicroRNA-100 Suppresses Human Gastric Cancer Cell Proliferation by Targeting CXCR7. Oncol. Lett. 2018, 15, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, O.; Rupertus, K.; Scheuer, C.; Nickels, R.M.; Haberl, G.C.Y.; Tilton, B.; Menger, M.D.; Schilling, M.K. CXCR4 and CXCR7 Regulate Angiogenesis and CT26.WT Tumor Growth Independent from SDF-1. Int. J. Cancer. 2010, 126, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, O.; Rupertus, K.; Scheuer, C.; Junker, B.; Tilton, B.; Schilling, M.K.; Menger, M.D. Stromal Cell-Derived Factor-1 Promotes Cell Migration and Tumor Growth of Colorectal Metastasis. Neoplasia 2007, 9, 862–870. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Picardo, A.; Karpoff, H.M.; Ng, B.; Lee, J.; Brennan, M.F.; Fong, Y. Partial Hepatectomy Accelerates Local Tumor Growth: Potential Roles of Local Cytokine Activation. Surgery 1998, 124, 57–64. [Google Scholar] [CrossRef]
- Rashidi, B.; An, Z.; Sun, F.X.; Sasson, A.; Gamagammi, R.; Moossa, A.R.; Hoffman, R.M. Minimal Liver Resection Strongly Stimulates the Growth of Human Colon Cancer in the Liver of Nude Mice. Clin. Exp. Metastasis 1999, 17, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Roy, I.; Zimmerman, N.P.; Mackinnon, A.C.; Tsai, S.; Evans, D.B.; Dwinell, M.B. CXCL12 Chemokine Expression Suppresses Human Pancreatic Cancer Growth and Metastasis. PLoS ONE 2014, 9, e90400. [Google Scholar] [CrossRef] [PubMed]
- Brand, S.; Dambacher, J.; Beigel, F.; Olszak, T.; Diebold, J.; Otte, J.-M.; Göke, B.; Eichhorst, S.T. CXCR4 and CXCL12 Are Inversely Expressed in Colorectal Cancer Cells and Modulate Cancer Cell Migration, Invasion and MMP-9 Activation. Exp. Cell Res. 2005, 310, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Hongo, K.; Tsuno, N.H.; Kawai, K.; Sasaki, K.; Kaneko, M.; Hiyoshi, M.; Murono, K.; Tada, N.; Nirei, T.; Sunami, E.; et al. Hypoxia Enhances Colon Cancer Migration and Invasion through Promotion of Epithelial-Mesenchymal Transition. J. Surg. Res. 2013, 182, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-Y.; Gao, Z.-H.; Chu, J.-H.; Han, X.-Z.; Qu, X.-J. Downregulation of the CXCR4/CXCL12 Axis Blocks the Activation of the Wnt/β-Catenin Pathway in Human Colon Cancer Cells. Biomed. Pharm. 2015, 71, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-Y.; Wang, F.; Cui, S.-X.; Qu, X.-J. Knockdown of CXCR4 Inhibits CXCL12-Induced Angiogenesis in HUVECs through Downregulation of the MAPK/ERK and PI3K/AKT and the Wnt/β-Catenin Pathways. Cancer Investig. 2018, 36, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Zhang, Z.; Flamini, V.; Jiang, W.G.; Cui, Y. The Axis of CXCR4/SDF-1 Plays a Role in Colon Cancer Cell Adhesion Through Regulation of the AKT and IGF1R Signalling Pathways. Anticancer Res. 2017, 37, 4361–4369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Li, C.-L.; Wang, L.; Yu, W.-B.; Yin, H.-P.; Zhang, G.-Y.; Zhang, L.-F.; Li, S.; Hu, S.-Y. Influence of CXCR4/SDF-1 Axis on E-Cadherin/β-Catenin Complex Expression in HT29 Colon Cancer Cells. World J. Gastroenterol. 2011, 17, 625–632. [Google Scholar] [CrossRef]
- Tung, S.-Y.; Chang, S.-F.; Chou, M.-H.; Huang, W.-S.; Hsieh, Y.-Y.; Shen, C.-H.; Kuo, H.-C.; Chen, C.-N. CXC Chemokine Ligand 12/Stromal Cell-Derived Factor-1 Regulates Cell Adhesion in Human Colon Cancer Cells by Induction of Intercellular Adhesion Molecule-1. J. Biomed. Sci. 2012, 19, 91. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kang, S.-M.; Sawada, T.; Nishiguchi, Y.; Yashiro, M.; Ogawa, Y.; Ohira, M.; Ishikawa, T.; Hirakawa-YS Chung, K. Expression of Intercellular Adhesion Molecule-1 and Prognosis in Colorectal Cancer. Oncol. Rep. 2002, 9, 511–514. [Google Scholar] [CrossRef]
- Tan, H.-X.; Gong, W.-Z.; Zhou, K.; Xiao, Z.-G.; Hou, F.-T.; Huang, T.; Zhang, L.; Dong, H.-Y.; Zhang, W.-L.; Liu, Y.; et al. CXCR4/TGF-Β1 Mediated Hepatic Stellate Cells Differentiation into Carcinoma-Associated Fibroblasts and Promoted Liver Metastasis of Colon Cancer. Cancer Biol. 2020, 21, 258–268. [Google Scholar] [CrossRef]
- Guleng, B.; Tateishi, K.; Ohta, M.; Kanai, F.; Jazag, A.; Ijichi, H.; Tanaka, Y.; Washida, M.; Morikane, K.; Fukushima, Y.; et al. Blockade of the Stromal Cell-Derived Factor-1/CXCR4 Axis Attenuates in Vivo Tumor Growth by Inhibiting Angiogenesis in a Vascular Endothelial Growth Factor-Independent Manner. Cancer Res. 2005, 65, 5864–5871. [Google Scholar] [CrossRef]
- Shin, H.-N.; Moon, H.-H.; Ku, J.-L. Stromal Cell-Derived Factor-1α and Macrophage Migration-Inhibitory Factor Induce Metastatic Behavior in CXCR4-Expressing Colon Cancer Cells. Int. J. Mol. Med. 2012, 30, 1537–1543. [Google Scholar] [CrossRef]
- Gouveia-Fernandes, S. Colorectal Cancer Aggressiveness Is Related to Fibronectin Over Expression, Driving the Activation of SDF-1:CXCR4 Axis. Int. J. Cancer Clin. Res. 2016, 3, 1–9. [Google Scholar] [CrossRef]
- Zeelenberg, I.S.; Ruuls-Van Stalle, L.; Roos, E. The Chemokine Receptor CXCR4 Is Required for Outgrowth of Colon Carcinoma Micrometastases. Cancer Res. 2003, 63, 3833–3839. [Google Scholar] [PubMed]
- Matsusue, R.; Kubo, H.; Hisamori, S.; Okoshi, K.; Takagi, H.; Hida, K.; Nakano, K.; Itami, A.; Kawada, K.; Nagayama, S.; et al. Hepatic Stellate Cells Promote Liver Metastasis of Colon Cancer Cells by the Action of SDF-1/CXCR4 Axis. Ann. Surg. Oncol. 2009, 16, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tao, L.; Qi, K.E.; Zhang, H.; Feng, D.; Wei, W.; Kong, H.; Chen, T.; Lin, Q.; Chen, D. CXCR7 Functions in Colon Cancer Cell Survival and Migration. Exp. Med. 2015, 10, 1720–1724. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Kawada, K.; Iwamoto, M.; Akagami, M.; Hida, K.; Nakanishi, Y.; Kanda, K.; Kawada, M.; Seno, H.; Taketo, M.M.; et al. The Role of CXCR3 and CXCR4 in Colorectal Cancer Metastasis. Int. J. Cancer 2013, 132, 276–287. [Google Scholar] [CrossRef]
- Li, X.-X.; Zheng, H.-T.; Huang, L.-Y.; Shi, D.-B.; Peng, J.-J.; Liang, L.; Cai, S.-J. Silencing of CXCR7 Gene Represses Growth and Invasion and Induces Apoptosis in Colorectal Cancer through ERK and β-Arrestin Pathways. Int. J. Oncol. 2014, 45, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wu, Q.; Dang, S.; Jin, M.; Xu, J.; Cheng, Y.; Pan, M.; Wu, Y.; Zhang, C.; Zhang, Y. Alteration of CXCR7 Expression Mediated by TLR4 Promotes Tumor Cell Proliferation and Migration in Human Colorectal Carcinoma. PLoS ONE 2011, 6, e27399. [Google Scholar] [CrossRef] [PubMed]
- Zabel, B.A.; Lewén, S.; Berahovich, R.D.; Jaén, J.C.; Schall, T.J. The Novel Chemokine Receptor CXCR7 Regulates Trans-Endothelial Migration of Cancer Cells. Mol. Cancer 2011, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Würth, R.; Bajetto, A.; Harrison, J.K.; Barbieri, F.; Florio, T. CXCL12 Modulation of CXCR4 and CXCR7 Activity in Human Glioblastoma Stem-like Cells and Regulation of the Tumor Microenvironment. Front. Cell. Neurosci. 2014, 8, 144. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-Y.; Wang, F.; Cui, S.-X.; Gao, Z.-H.; Qu, X.-J. CXCR7/CXCR4 Heterodimer-Induced Histone Demethylation: A New Mechanism of Colorectal Tumorigenesis. Oncogene 2019, 38, 1560–1575. [Google Scholar] [CrossRef]
- Mc Donnell, S.; Chaudhry, V.; Mansilla-Soto, J.; Zeng, Z.S.; Shu, W.P.; Guillem, J.G. Metastatic and Non-Metastatic Colorectal Cancer (CRC) Cells Induce Host Metalloproteinase Production in Vivo. Clin. Exp. Metastasis 1999, 17, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Parmo-Cabañas, M.; Molina-Ortiz, I.; Matías-Román, S.; García-Bernal, D.; Carvajal-Vergara, X.; Valle, I.; Pandiella, A.; Arroyo, A.G.; Teixidó, J. Role of Metalloproteinases MMP-9 and MT1-MMP in CXCL12-Promoted Myeloma Cell Invasion across Basement Membranes. J. Pathol. 2006, 208, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Turunen, S.P.; Tatti-Bugaeva, O.; Lehti, K. Membrane-Type Matrix Metalloproteases as Diverse Effectors of Cancer Progression. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1974–1988. [Google Scholar] [CrossRef] [PubMed]
- Halama, N.; Pruefer, U.; Frömming, A.; Beyer, D.; Eulberg, D.; Jungnelius, J.U.B.; Mangasarian, A. Experience with CXCL12 Inhibitor NOX-A12 plus Pembrolizumab in Patients with Microsatellite-Stable, Metastatic Colorectal or Pancreatic Cancer. JCO 2019, 37, e14143. [Google Scholar] [CrossRef]
- Micallef, I.N.; Stiff, P.J.; Nademanee, A.P.; Maziarz, R.T.; Horwitz, M.E.; Stadtmauer, E.A.; Kaufman, J.L.; McCarty, J.M.; Vargo, R.; Cheverton, P.D.; et al. Plerixafor Plus Granulocyte Colony-Stimulating Factor for Patients with Non-Hodgkin Lymphoma and Multiple Myeloma: Long-Term Follow-Up Report. Biol. Blood Marrow Transpl. 2018, 24, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Micallef, I.N.; Stiff, P.J.; DiPersio, J.F.; Maziarz, R.T.; McCarty, J.M.; Bridger, G.; Calandra, G. Successful Stem Cell Remobilization Using Plerixafor (Mozobil) plus Granulocyte Colony-Stimulating Factor in Patients with Non-Hodgkin Lymphoma: Results from the Plerixafor NHL Phase 3 Study Rescue Protocol. Biol. Blood Marrow Transpl. 2009, 15, 1578–1586. [Google Scholar] [CrossRef]
- De Clercq, E. The Bicyclam AMD3100 Story. Nat. Rev. Drug Discov. 2003, 2, 581–587. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. AMD3100/CXCR4 Inhibitor. Front. Immunol. 2015, 6, 276. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tannous, B.A.; Poznansky, M.C.; Chen, H. CXCR4 Antagonist AMD3100 (Plerixafor): From an Impurity to a Therapeutic Agent. Pharm. Res. 2020, 159, 105010. [Google Scholar] [CrossRef]
- Benedicto, A.; Romayor, I.; Arteta, B. CXCR4 Receptor Blockage Reduces the Contribution of Tumor and Stromal Cells to the Metastatic Growth in the Liver. Oncol. Rep. 2018, 39, 2022–2030. [Google Scholar] [CrossRef]
- Kalatskaya, I.; Berchiche, Y.A.; Gravel, S.; Limberg, B.J.; Rosenbaum, J.S.; Heveker, N. AMD3100 Is a CXCR7 Ligand with Allosteric Agonist Properties. Mol. Pharmacol. 2009, 75, 1240–1247. [Google Scholar] [CrossRef]
- Li, J.-K.; Yu, L.; Shen, Y.; Zhou, L.-S.; Wang, Y.-C.; Zhang, J.-H. Inhibition of CXCR4 Activity with AMD3100 Decreases Invasion of Human Colorectal Cancer Cells in Vitro. World J. Gastroenterol. 2008, 14, 2308–2313. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Zhai, B.; Yu, Y.; Kiyotsugu, Y.; Raschle, T.; Etzkorn, M.; Seo, H.-C.; Nagiec, M.; Luna, R.E.; Reinherz, E.L.; et al. Quantitative Phosphoproteomic Analysis Reveals System-Wide Signaling Pathways Downstream of SDF-1/CXCR4 in Breast Cancer Stem Cells. Proc. Natl. Acad. Sci. USA 2014, 111, E2182–E2190. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-G.; Go, R.-E.; Hwang, K.-A.; Choi, K.-C. Resveratrol Inhibits DHT-Induced Progression of Prostate Cancer Cell Line through Interfering with the AR and CXCR4 Pathway. J. Steroid Biochem. Mol. Biol. 2019, 192, 105406. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chien, E.Y.T.; Mol, C.D.; Fenalti, G.; Liu, W.; Katritch, V.; Abagyan, R.; Brooun, A.; Wells, P.; Bi, F.C.; et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science 2010, 330, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-B.; Zhang, X.; Paul, D.; Kays, L.M.; Gough, W.; Stewart, J.; Uhlik, M.T.; Chen, Q.; Hui, Y.-H.; Zamek-Gliszczynski, M.J.; et al. Identification of LY2510924, a Novel Cyclic Peptide CXCR4 Antagonist That Exhibits Antitumor Activities in Solid Tumor and Breast Cancer Metastatic Models. Mol. Cancer 2015, 14, 480–490. [Google Scholar] [CrossRef]
- Levine, D.; Hellmers, L.; Maresh, G.; Zhang, X.; Moret, R.; Green, H.; Margolin, D.; Li, L. Evaluating the Inhibitory Effects of LY2510924, a Cyclic Peptide CXCR4 Antagonist, in Human Colon Cancer Metastasis Using an Orthotopic Xenograft Model. J. Am. Coll. Surg. 2018, 227, S64–S65. [Google Scholar] [CrossRef]
- D’Alterio, C.; Zannetti, A.; Trotta, A.M.; Ieranò, C.; Napolitano, M.; Rea, G.; Greco, A.; Maiolino, P.; Albanese, S.; Scognamiglio, G.; et al. New CXCR4 Antagonist Peptide R (Pep R) Improves Standard Therapy in Colorectal Cancer. Cancers 2020, 12, 1952. [Google Scholar] [CrossRef]
- D’Alterio, C.; Buoncervello, M.; Ieranò, C.; Napolitano, M.; Portella, L.; Rea, G.; Barbieri, A.; Luciano, A.; Scognamiglio, G.; Tatangelo, F.; et al. Targeting CXCR4 Potentiates Anti-PD-1 Efficacy Modifying the Tumor Microenvironment and Inhibiting Neoplastic PD-1. J. Exp. Clin. Cancer Res. 2019, 38, 432. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Fearon, D.T. T Cell Exclusion, Immune Privilege, and the Tumor Microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Lesokhin, A.M.; Hohl, T.M.; Kitano, S.; Cortez, C.; Hirschhorn-Cymerman, D.; Avogadri, F.; Rizzuto, G.A.; Lazarus, J.J.; Pamer, E.G.; Houghton, A.N.; et al. Monocytic CCR2(+) Myeloid-Derived Suppressor Cells Promote Immune Escape by Limiting Activated CD8 T-Cell Infiltration into the Tumor Microenvironment. Cancer Res. 2012, 72, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhan, W.; Zhu, A.; Yoon, Y.; Lin, S.; Sasaki, M.; Klapproth, J.-M.A.; Yang, H.; Grossniklaus, H.E.; Xu, J.; et al. Development of a Unique Small Molecule Modulator of CXCR4. PLoS ONE 2012, 7, e34038. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, B.; Dougherty, U.; Mustafi, R.; Haider, H.; Joseph, L.; Souris, J.; Hart, J.; Pewkow, J.; LI, Y. CXCR4 Inhibitor, MSX-122 Suppresses AOM-induced Colon Cancer in Apc+/Min Mouse. FASEB J. 2018, 32, 677.4. [Google Scholar] [CrossRef]
- Suzui, M.; Okuno, M.; Tanaka, T.; Nakagama, H.; Moriwaki, H. Enhanced Colon Carcinogenesis Induced by Azoxymethane in Min Mice Occurs via a Mechanism Independent of Beta-Catenin Mutation. Cancer Lett. 2002, 183, 31–41. [Google Scholar] [CrossRef]
- Burns, J.M.; Summers, B.C.; Wang, Y.; Melikian, A.; Berahovich, R.; Miao, Z.; Penfold, M.E.T.; Sunshine, M.J.; Littman, D.R.; Kuo, C.J.; et al. A Novel Chemokine Receptor for SDF-1 and I-TAC Involved in Cell Survival, Cell Adhesion, and Tumor Development. J. Exp. Med. 2006, 203, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Zabel, B.A.; Wang, Y.; Lewén, S.; Berahovich, R.D.; Penfold, M.E.T.; Zhang, P.; Powers, J.; Summers, B.C.; Miao, Z.; Zhao, B.; et al. Elucidation of CXCR7-Mediated Signaling Events and Inhibition of CXCR4-Mediated Tumor Cell Transendothelial Migration by CXCR7 Ligands. J. Immunol. 2009, 183, 3204–3211. [Google Scholar] [CrossRef] [PubMed]
- Saita, Y.; Kodama, E.; Orita, M.; Kondo, M.; Miyazaki, T.; Sudo, K.; Kajiwara, K.; Matsuoka, M.; Shimizu, Y. Structural Basis for the Interaction of CCR5 with a Small Molecule, Functionally Selective CCR5 Agonist. J. Immunol. 2006, 177, 3116–3122. [Google Scholar] [CrossRef]
- Ali, S.; O’Boyle, G.; Mellor, P.; Kirby, J.A. An Apparent Paradox: Chemokine Receptor Agonists Can Be Used for Anti-Inflammatory Therapy. Mol. Immunol. 2007, 44, 1477–1482. [Google Scholar] [CrossRef] [PubMed]
- Lounsbury, N. Advances in CXCR7 Modulators. Pharmaceuticals 2020, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Gravel, S.; Malouf, C.; Boulais, P.E.; Berchiche, Y.A.; Oishi, S.; Fujii, N.; Leduc, R.; Sinnett, D.; Heveker, N. The Peptidomimetic CXCR4 Antagonist TC14012 Recruits Beta-Arrestin to CXCR7: Roles of Receptor Domains. J. Biol. Chem. 2010, 285, 37939–37943. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Orengo, L.; Holman, D.W.; Dorsey, D.; Zhou, L.; Zhang, P.; Wright, M.; McCandless, E.E.; Patel, J.R.; Luker, G.D.; Littman, D.R.; et al. CXCR7 Influences Leukocyte Entry into the CNS Parenchyma by Controlling Abluminal CXCL12 Abundance during Autoimmunity. J. Exp. Med. 2011, 208, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Hachet-Haas, M.; Balabanian, K.; Rohmer, F.; Pons, F.; Franchet, C.; Lecat, S.; Chow, K.Y.C.; Dagher, R.; Gizzi, P.; Didier, B.; et al. Small Neutralizing Molecules to Inhibit Actions of the Chemokine CXCL12. J. Biol. Chem. 2008, 283, 23189–23199. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Sacco, A.; Purschke, W.G.; Moschetta, M.; Buchner, K.; Maasch, C.; Zboralski, D.; Zöllner, S.; Vonhoff, S.; Mishima, Y.; et al. SDF-1 Inhibition Targets the Bone Marrow Niche for Cancer Therapy. Cell Rep. 2014, 9, 118–128. [Google Scholar] [CrossRef]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a Novel CXCL12 Inhibitor, Interferes with Chronic Lymphocytic Leukemia Cell Motility and Causes Chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Zboralski, D.; Hoehlig, K.; Eulberg, D.; Frömming, A.; Vater, A. Increasing Tumor-Infiltrating T Cells through Inhibition of CXCL12 with NOX-A12 Synergizes with PD-1 Blockade. Cancer Immunol. Res. 2017, 5, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Adlere, I.; Caspar, B.; Arimont, M.; Dekkers, S.; Visser, K.; Stuijt, J.; de Graaf, C.; Stocks, M.; Kellam, B.; Briddon, S.; et al. Modulators of CXCR4 and CXCR7/ACKR3 Function. Mol. Pharm. 2019, 96, 737–752. [Google Scholar] [CrossRef] [PubMed]
- Gómez-España, M.A.; Gallego, J.; González-Flores, E.; Maurel, J.; Páez, D.; Sastre, J.; Aparicio, J.; Benavides, M.; Feliu, J.; Vera, R. SEOM Clinical Guidelines for Diagnosis and Treatment of Metastatic Colorectal Cancer (2018). Clin. Transl. Oncol. 2019, 21, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Formica, V.; Roselli, M. Targeted Therapy in First Line Treatment of RAS Wild Type Colorectal Cancer. World J. Gastroenterol. 2015, 21, 2871–2874. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Bissonnette, M.; Khare, S. CXCL12-CXCR4/CXCR7 Axis in Colorectal Cancer: Therapeutic Target in Preclinical and Clinical Studies. Int. J. Mol. Sci. 2021, 22, 7371. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Mayer, I.A.; Walenkamp, A.M.E.; Lapa, C.; Andreeff, M.; Bobirca, A. At the Bedside: Profiling and Treating Patients with CXCR4-Expressing Cancers. J. Leukoc. Biol. 2021, 109, 953–967. [Google Scholar] [CrossRef] [PubMed]
- Santagata, S.; Ieranò, C.; Trotta, A.M.; Capiluongo, A.; Auletta, F.; Guardascione, G.; Scala, S. CXCR4 and CXCR7 Signaling Pathways: A Focus on the Cross-Talk Between Cancer Cells and Tumor Microenvironment. Front. Oncol. 2021, 11, 591386. [Google Scholar] [CrossRef] [PubMed]
- Galsky, M.D.; Vogelzang, N.J.; Conkling, P.; Raddad, E.; Polzer, J.; Roberson, S.; Stille, J.R.; Saleh, M.; Thornton, D. A Phase I Trial of LY2510924, a CXCR4 Peptide Antagonist, in Patients with Advanced Cancer. Clin. Cancer Res. 2014, 20, 3581–3588. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Carmona, M.; Williams, A.; Schreiber, J.; Hohmann, N.; Pruefer, U.; Krauss, J.; Jäger, D.; Frömming, A.; Beyer, D.; Eulberg, D.; et al. Combined Inhibition of CXCL12 and PD-1 in MSS Colorectal and Pancreatic Cancer: Modulation of the Microenvironment and Clinical Effects. J. Immunother. Cancer 2021, 9, e002505. [Google Scholar] [CrossRef] [PubMed]
- Biasci, D.; Smoragiewicz, M.; Connell, C.M.; Wang, Z.; Gao, Y.; Thaventhiran, J.E.D.; Basu, B.; Magiera, L.; Johnson, T.I.; Bax, L.; et al. CXCR4 Inhibition in Human Pancreatic and Colorectal Cancers Induces an Integrated Immune Response. Proc. Natl. Acad. Sci. USA 2020, 117, 28960–28970. [Google Scholar] [CrossRef]
- Shimizu, S.; Brown, M.; Sengupta, R.; Penfold, M.E.; Meucci, O. CXCR7 Protein Expression in Human Adult Brain and Differentiated Neurons. PLoS ONE 2011, 6, e20680. [Google Scholar] [CrossRef] [PubMed]
- Lubner, S.J.; Uboha, N.V.; Deming, D.A. Primary and Acquired Resistance to Biologic Therapies in Gastrointestinal Cancers. J. Gastrointest Oncol. 2017, 8, 499–512. [Google Scholar] [CrossRef]
- Gottesman, M.M. Mechanisms of Cancer Drug Resistance. Annu. Rev. Med. 2002, 53, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem Cells, Cancer, and Cancer Stem Cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Margolin, D.A.; Silinsky, J.; Grimes, C.; Spencer, N.; Aycock, M.; Green, H.; Cordova, J.; Davis, N.K.; Driscoll, T.; Li, L. Lymph Node Stromal Cells Enhance Drug-Resistant Colon Cancer Cell Tumor Formation through SDF-1α/CXCR4 Paracrine Signaling. Neoplasia 2011, 13, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Margolin, D.A.; Myers, T.; Zhang, X.; Bertoni, D.M.; Reuter, B.A.; Obokhare, I.; Borgovan, T.; Grimes, C.; Green, H.; Driscoll, T.; et al. The Critical Roles of Tumor-Initiating Cells and the Lymph Node Stromal Microenvironment in Human Colorectal Cancer Extranodal Metastasis Using a Unique Humanized Orthotopic Mouse Model. FASEB J. 2015, 29, 3571–3581. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Ji, H.; Jia, C.; Brockmeier, U.; Hermann, D.M.; Metzen, E.; Zhu, Y.; Chi, B. Synergistic Antitumor Effects of Endostar in Combination with Oxaliplatin via Inhibition of HIF and CXCR4 in the Colorectal Cell Line SW1116. PLoS ONE 2012, 7, e47161. [Google Scholar] [CrossRef] [PubMed]
- Heckmann, D.; Maier, P.; Laufs, S.; Wenz, F.; Zeller, W.J.; Fruehauf, S.; Allgayer, H. CXCR4 Expression and Treatment with SDF-1α or Plerixafor Modulate Proliferation and Chemosensitivity of Colon Cancer Cells. Transl. Oncol. 2013, 6, 124–132. [Google Scholar] [CrossRef]
- Heckmann, D.; Maier, P.; Laufs, S.; Li, L.; Sleeman, J.P.; Trunk, M.J.; Leupold, J.H.; Wenz, F.; Zeller, W.J.; Fruehauf, S.; et al. The Disparate Twins: A Comparative Study of CXCR4 and CXCR7 in SDF-1α-Induced Gene Expression, Invasion and Chemosensitivity of Colon Cancer. Clin. Cancer Res. 2014, 20, 604–616. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goïta, A.A.; Guenot, D. Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7. Cancers 2022, 14, 1810. https://doi.org/10.3390/cancers14071810
Goïta AA, Guenot D. Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7. Cancers. 2022; 14(7):1810. https://doi.org/10.3390/cancers14071810
Chicago/Turabian StyleGoïta, Aïssata Aimée, and Dominique Guenot. 2022. "Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7" Cancers 14, no. 7: 1810. https://doi.org/10.3390/cancers14071810
APA StyleGoïta, A. A., & Guenot, D. (2022). Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7. Cancers, 14(7), 1810. https://doi.org/10.3390/cancers14071810





