Transforming Growth Factor-Beta Orchestrates Tumour and Bystander Cells in B-Cell Non-Hodgkin Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
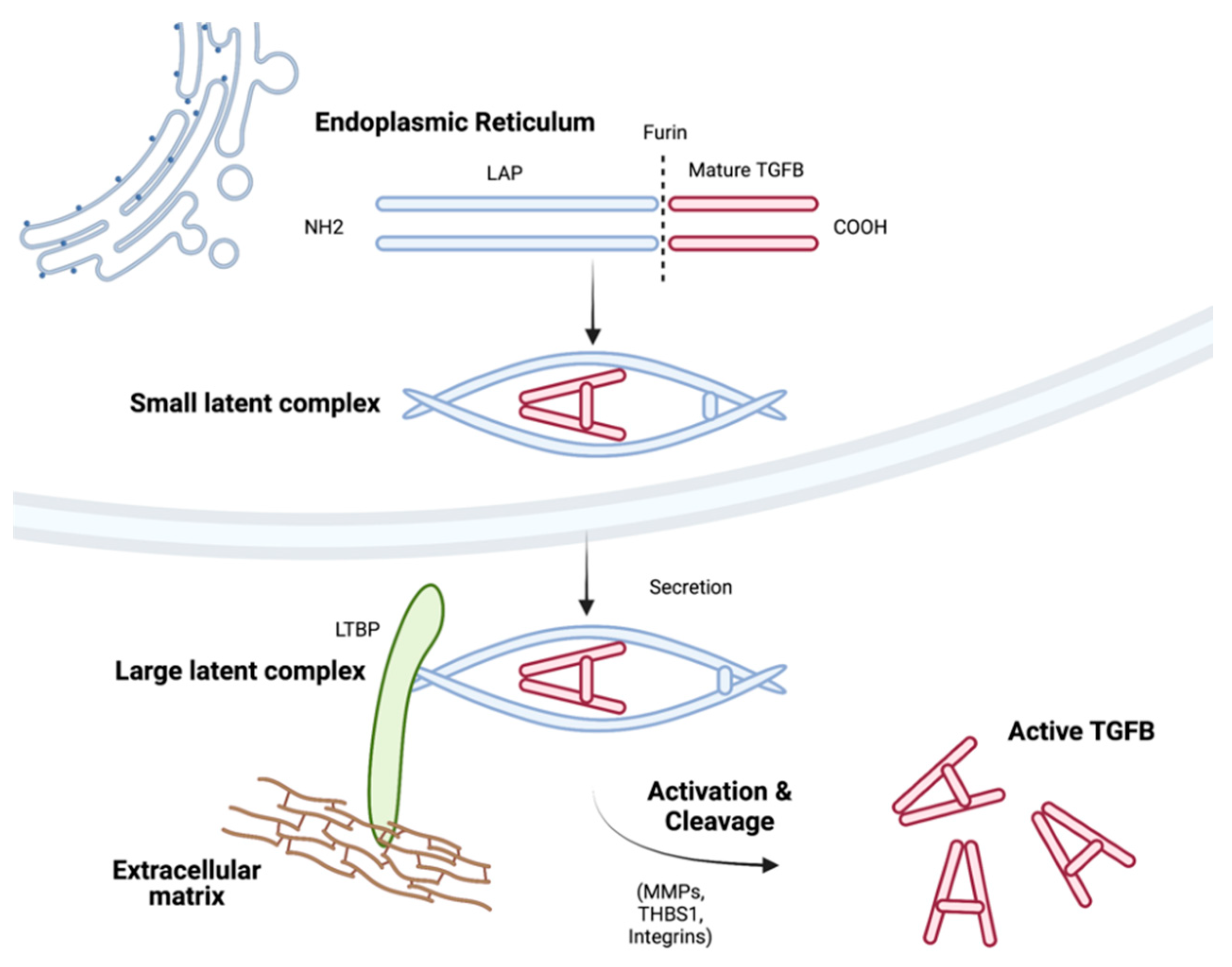
2. TGFB Isoforms, Secretion, and Activation
3. Canonical and Non-Canonical TGFB Signalling
4. TGFB and Haematopoiesis
5. TGFB and the Bone Marrow Niche
6. TGFB and Normal B-Cells
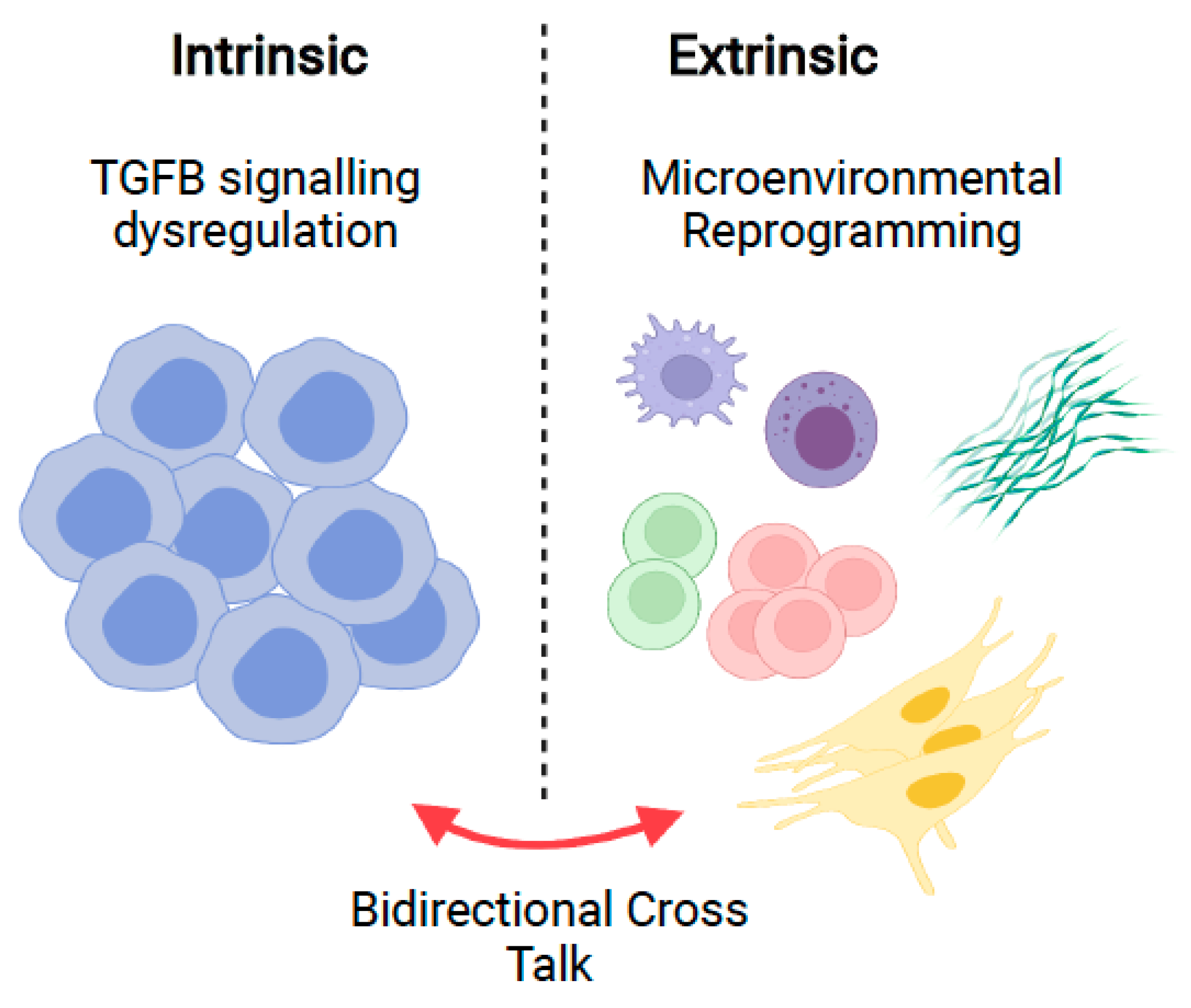
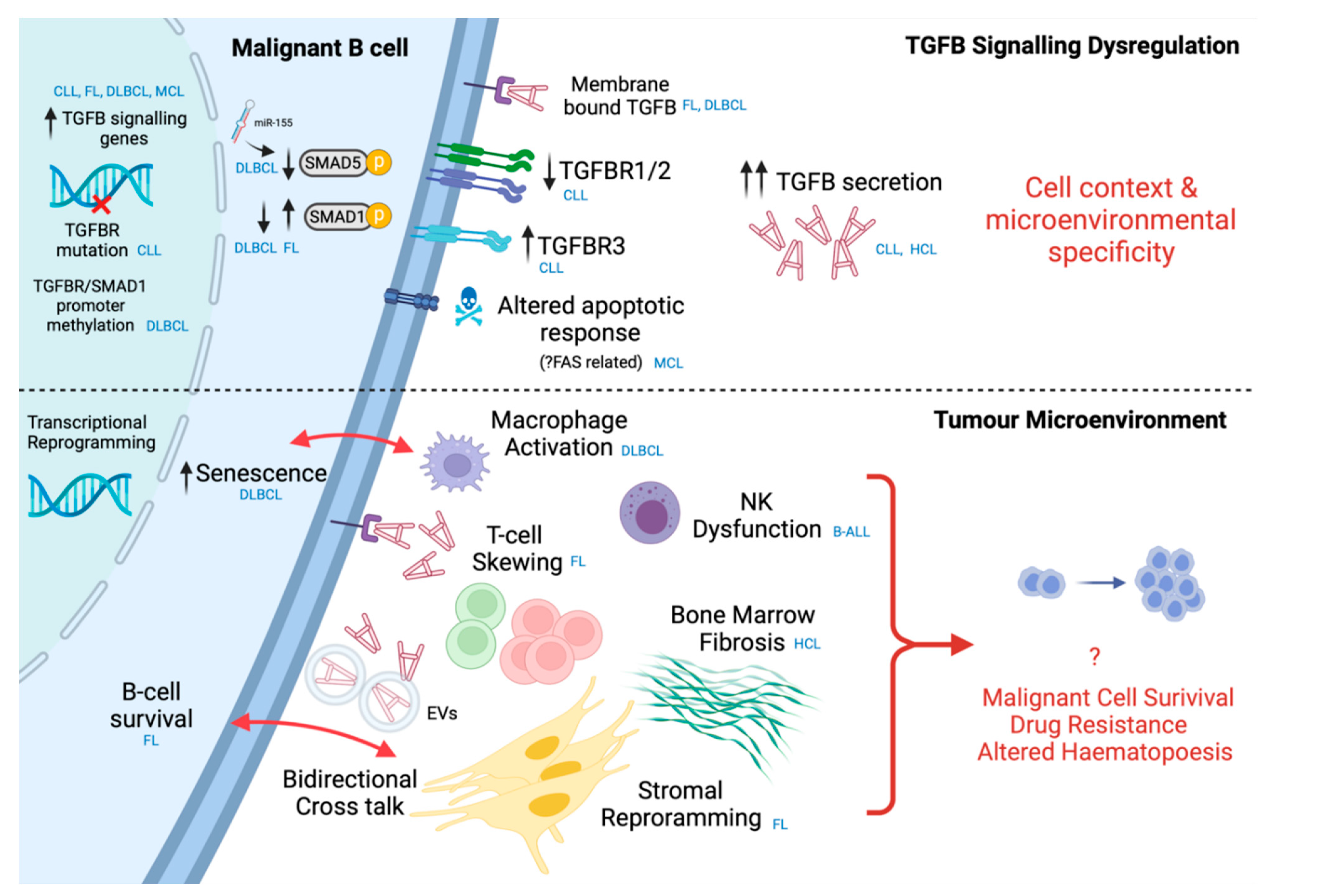
7. Intrinsic Dysregulation in Malignant B-Cells
8. Microenvironmental TGFB in B-Cell Malignancies
8.1. T-Cells
8.2. Stromal Cells
8.3. Innate Immune Cells
9. Therapeutic Perspective
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Meads, M.B.; Gatenby, R.A.; Dalton, W.S. Environment-mediated drug resistance: A major contributor to minimal residual disease. Nat. Cancer 2009, 9, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.X.; Müller, S.; Keerthivasan, S.; Koeppen, H.; Hung, J.; Gierke, S.; Breart, B.; Foreman, O.; Bainbridge, T.W.; Castiglioni, A.; et al. Single-Cell RNA Sequencing Reveals Stromal Evolution into LRRC15+ Myofibroblasts as a Determinant of Patient Response to Cancer Immunotherapy. Cancer Discov. 2020, 10, 232–253. [Google Scholar] [CrossRef] [PubMed]
- Grauel, A.L.; Nguyen, B.; Ruddy, D.; Laszewski, T.; Schwartz, S.; Chang, J.; Chen, J.; Piquet, M.; Pelletier, M.; Yan, Z.; et al. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat. Commun. 2020, 11, 6315. [Google Scholar] [CrossRef] [PubMed]
- Travis, M.A.; Sheppard, D. TGF-β Activation and Function in Immunity. Annu. Rev. Immunol. 2014, 32, 51–82. [Google Scholar] [CrossRef]
- Batlle, E.; Massagué, J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Dubois, C.M.; Laprise, M.-H.; Blanchette, F.; Gentry, L.E.; Leduc, R. Processing of Transforming Growth Factor β1 Precursor by Human Furin Convertase. J. Biol. Chem. 1995, 270, 10618–10624. [Google Scholar] [CrossRef]
- Proetzel, G.; Pawlowski, S.A.; Wiles, M.V.; Yin, M.; Boivin, G.P.; Howles, P.N.; Ding, J.; Ferguson, M.W.; Doetschman, T. Transforming growth factor–β3 is required for secondary palate fusion. Nat. Genet. 1995, 11, 409–414. [Google Scholar] [CrossRef]
- Shull, M.M.; Ormsby, I.; Kier, A.B.; Pawlowski, S.; Diebold, R.J.; Yin, M.Y.; Allen, R.; Sidman, C.; Proetzel, G.; Calvin, D.; et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 1992, 359, 693–699. [Google Scholar] [CrossRef]
- Dickson, M.C.; Martin, J.S.; Cousins, F.M.; Kulkarni, A.B.; Karlsson, S.; Akhurst, R.J. Defective haematopoiesis and vasculogenesis in transforming growth factor-beta 1 knock out mice. Development 1995, 121, 1845–1854. [Google Scholar] [CrossRef]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef]
- Millan, F.A.; Denhez, F.; Kondaiah, P.; Akhurst, R.J. Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development 1991, 111, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.S.; Huang, X.; Kawakatsu, H.; Griffiths, M.J.; Dalton, S.L.; Wu, J.; Pittet, J.-F.; Kaminski, N.; Garat, C.; Matthay, M.A. A mechanism for regulating pulmonary inflammation and fibrosis: The integrin αvβ6 binds and activates latent TGF β1. Cell 1999, 96, 319–328. [Google Scholar] [CrossRef]
- Schultz-Cherry, S.; Ribeiro, S.; Gentry, L.; Murphy-Ullrich, J.E. Thrombospondin binds and activates the small and large forms of latent transforming growth factor-beta in a chemically defined system. J. Biol. Chem. 1994, 269, 26775–26782. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, D.; Spinetti, G.; Zhang, J.; Jiang, L.-Q.; Pintus, G.; Monticone, R.; Lakatta, E.G. Matrix metalloproteinase 2 activation of transforming growth factor-β1 (TGF-β1) and TGF-β1–type II receptor signaling within the aged arterial wall. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1503–1509. [Google Scholar] [CrossRef]
- Karsdal, M.A.; Larsen, L.; Engsig, M.T.; Lou, H.; Ferreras, M.; Lochter, A.; Delaissé, J.-M.; Foged, N.T. Matrix metalloproteinase-dependent activation of latent transforming growth factor-β controls the conversion of osteoblasts into osteocytes by blocking osteoblast apoptosis. J. Biol. Chem. 2002, 277, 44061–44067. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 2000, 14, 163–176. [Google Scholar] [CrossRef]
- López-Casillas, F.; Wrana, J.L.; Massagué, J. Betaglycan presents ligand to the TGFβ signaling receptor. Cell 1993, 73, 1435–1444. [Google Scholar] [CrossRef]
- Cheifetz, S.; Bellón, T.; Calés, C.; Vera, S.; Bernabeu, C.; Massague, J.; Letarte, M. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J. Biol. Chem. 1992, 267, 19027–19030. [Google Scholar] [CrossRef]
- Schmierer, B.; Hill, C.S. TGFβ–SMAD signal transduction: Molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007, 8, 970–982. [Google Scholar] [CrossRef]
- Daly, A.C.; Randall, R.A.; Hill, C.S. Transforming growth factor β-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol. Cell. Biol. 2008, 28, 6889–6902. [Google Scholar] [CrossRef] [PubMed]
- Munoz, O.; Fend, F.; De Beaumont, R.; Husson, H.; Astier, A.; Freedman, A. TGF β-mediated activation of Smad1 in B-cell non-Hodgkin’s lymphoma and effect on cell proliferation. Leukemia 2004, 18, 2015–2025. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.; Lin, X.; Yu, P.; Feng, X.-H. Transforming Growth Factor β Can Stimulate Smad1 Phosphorylation Independently of Bone Morphogenic Protein Receptors. J. Biol. Chem. 2009, 284, 9755–9763. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Vizan, P.; Das, D.; Chakravarty, P.; Vogt, J.; Rogers, K.W.; Müller, P.; Hinck, A.P.; Sapkota, G.P.; Hill, C.S. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife 2018, 7, e31756. [Google Scholar] [CrossRef] [PubMed]
- Flinn, I.W.; Hillmen, P.; Montillo, M.; Nagy, Z.; Illés, A.; Etienne, G.; Delgado, J.; Kuss, B.J.; Tam, C.S.; Gasztonyi, Z.; et al. The phase 3 DUO trial: Duvelisib vs. ofatumumab in relapsed and refractory CLL/SLL. Blood 2018, 132, 2446–2455. [Google Scholar] [CrossRef]
- Hayashi, H.; Abdollah, S.; Qiu, Y.; Cai, J.; Xu, Y.-Y.; Grinnell, B.W.; Richardson, M.A.; Topper, J.N.; Gimbrone, M.A., Jr.; Wrana, J.L. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 1997, 89, 1165–1173. [Google Scholar] [CrossRef]
- Shi, W.; Sun, C.; He, B.; Xiong, W.; Shi, X.; Yao, D.; Cao, X. GADD34–PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J. Cell Biol. 2004, 164, 291–300. [Google Scholar] [CrossRef]
- Nakao, A.; Afrakhte, M.; Morn, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.-E.; Heldin, C.-H.; et al. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 1997, 389, 631–635. [Google Scholar] [CrossRef]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 Binds to Smurf2 to Form an E3 Ubiquitin Ligase that Targets the TGFβ Receptor for Degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef]
- Imamura, T.; Takase, M.; Nishihara, A.; Oeda, E.; Hanai, J.-I.; Kawabata, M.; Miyazono, K. Smad6 inhibits signalling by the TGF-β superfamily. Nature 1997, 389, 622–626. [Google Scholar] [CrossRef]
- Di Guglielmo, G.M.; LE Roy, C.; Goodfellow, A.F.; Wrana, J.L. Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat. Cell Biol. 2003, 5, 410–421. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Ottmann, O.; Pelus, L.M. Differential proliferative effects of transforming growth factor-beta on human hematopoietic progenitor cells. J. Immunol. 1988, 140, 2661–2665. [Google Scholar] [PubMed]
- Sing, G.K.; Keller, J.R.; Ellingsworth, L.R.; Ruscetti, F.W. Transforming growth factor beta selectively inhibits normal and leukemic human bone marrow cell growth in vitro. Blood 1988, 72, 1504–1511. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.; Blank, U.; Helgadottir, H.; Björnsson, J.M.; Ehinger, M.; Goumans, M.-J.; Fan, X.; Levéen, P.; Karlsson, S. TGF-β signaling–deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood 2003, 102, 3129–3135. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Blobe, G.C. Role of transforming growth factor-β in hematologic malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef] [PubMed]
- Langer, J.C.; Henckaerts, E.J.V.; Orenstein, J.; Snoeck, H.-W. Quantitative Trait Analysis Reveals Transforming Growth Factor-β2 as a Positive Regulator of Early Hematopoietic Progenitor and Stem Cell Function. J. Exp. Med. 2004, 199, 5–14. [Google Scholar] [CrossRef]
- Jacobsen, S.; Keller, J.; Ruscetti, F.; Kondaiah, P.; Roberts, A.; Falk, L. Bidirectional effects of transforming growth factor beta (TGF-beta) on colony-stimulating factor-induced human myelopoiesis in vitro: Differential effects of distinct TGF-beta isoforms. Blood 1991, 78, 2239–2247. [Google Scholar] [CrossRef]
- Santoro, A.; Andrei, C.; Bryant, C.; Calderbank, E.; Wray, A.; Baxter, J.E.; Godfrey, A.; Laurenti, E.; Ringshausen, I. Chronic lymphocytic leukemia increases the pool of peripheral blood hematopoietic stem cells and skews differentiation. Blood Adv. 2020, 4, 6310–6314. [Google Scholar] [CrossRef]
- Challen, G.A.; Boles, N.C.; Chambers, S.M.; Goodell, M.A. Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1. Cell Stem Cell 2010, 6, 265–278. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ema, H.; Karlsson, G.; Yamaguchi, T.; Miyoshi, H.; Shioda, S.; Taketo, M.M.; Karlsson, S.; Iwama, A.; Nakauchi, H. Nonmyelinating Schwann Cells Maintain Hematopoietic Stem Cell Hibernation in the Bone Marrow Niche. Cell 2011, 147, 1146–1158. [Google Scholar] [CrossRef]
- Zhao, M.; Perry, J.M.; Marshall, H.; Venkatraman, A.; Qian, P.; He, X.C.; Ahamed, J.; Li, L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 2014, 20, 1321–1326. [Google Scholar] [CrossRef] [PubMed]
- Kay, N.E.; Shanafelt, T.D.; Strege, A.K.; Lee, Y.K.; Bone, N.D.; Raza, A. Bone biopsy derived marrow stromal elements rescue chronic lymphocytic leukemia B-cells from spontaneous and drug induced cell death and facilitates an “angiogenic switch”. Leuk. Res. 2007, 31, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Panayiotidis, P.; Jones, D.; Ganeshaguru, K.; Foroni, L.; Hoffbrand, A.V. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cellsin vitro. Br. J. Haematol. 1996, 92, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Burger, M.; Kipps, T.J. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood 1999, 94, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Kurtova, A.V.; Balakrishnan, K.; Chen, R.; Ding, W.; Schnabl, S.; Quiroga, M.P.; Sivina, M.; Wierda, W.G.; Estrov, Z.; Keating, M.J.; et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: Development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood 2009, 114, 4441–4450. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Tang, Y.; Wu, X.; Lei, W.; Pang, L.; Wan, C.; Shi, Z.; Zhao, L.; Nagy, T.R.; Peng, X.; Hu, J.; et al. TGF-β1–induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009, 15, 757–765. [Google Scholar] [CrossRef]
- Zhao, L.; Hantash, B.M. TGF-β1 Regulates Differentiation of Bone Marrow Mesenchymal Stem Cells. Vitam. Horm. 2011, 87, 127–141. [Google Scholar] [CrossRef]
- Sime, P.J.; Xing, Z.; Graham, F.L.; Csaky, K.G.; Gauldie, J. Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J. Clin. Investig. 1997, 100, 768–776. [Google Scholar] [CrossRef]
- Desmoulière, A.; Geinoz, A.; Gabbiani, F.; Gabiani, G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J. Cell Biol. 1993, 122, 103–111. [Google Scholar] [CrossRef]
- Hawinkels, L.J.A.C.; Paauwe, M.; Verspaget, H.W.; Wiercinska, E.; Van Der Zon, J.M.; Van Der Ploeg, K.; Koelink, P.J.; Lindeman, J.H.N.; Mesker, W.; Dijke, P.T.; et al. Interaction with colon cancer cells hyperactivates TGF-β signaling in cancer-associated fibroblasts. Oncogene 2014, 33, 97–107. [Google Scholar] [CrossRef]
- Shangguan, L.; Ti, X.; Krause, U.; Hai, B.; Zhao, Y.; Yang, Z.; Liu, F. Inhibition of TGF-β/Smad Signaling by BAMBI Blocks Differentiation of Human Mesenchymal Stem Cells to Carcinoma-Associated Fibroblasts and Abolishes their Protumor Effects. Stem Cells 2012, 30, 2810–2819. [Google Scholar] [CrossRef] [PubMed]
- Franses, J.W.; Baker, A.B.; Chitalia, V.C.; Edelman, E.R. Stromal Endothelial Cells Directly Influence Cancer Progression. Sci. Transl. Med. 2011, 3, 66ra5. [Google Scholar] [CrossRef]
- Solimando, A.G.; De Summa, S.; Vacca, A.; Ribatti, D. Cancer-Associated Angiogenesis: The Endothelial Cell as a Checkpoint for Immunological Patrolling. Cancers 2020, 12, 3380. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Annese, T.; Tamma, R.; Ingravallo, G.; Maiorano, E.; Vacca, A.; Specchia, G.; Ribatti, D. New Insights into Diffuse Large B-Cell Lymphoma Pathobiology. Cancers 2020, 12, 1869. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Bhowmick, N.A.; Chytil, A.; Gorksa, A.E.; Brown, K.A.; Muraoka, R.; Arteaga, C.L.; Neilson, E.G.; Hayward, S.W.; Moses, H.L. Loss of TGF-β type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-α-, MSP-and HGF-mediated signaling networks. Oncogene 2005, 24, 5053–5068. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.E.; Jiang, M.; Strand, D.W.; Peacock, J.; Fernandez, S.; Jackson, R.S.; Revelo, M.P.; Bhowmick, N.A.; Hayward, S.W. Altered TGF-β Signaling in a Subpopulation of Human Stromal Cells Promotes Prostatic Carcinogenesis. Cancer Res. 2011, 71, 1272–1281. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Chytil, A.; Plieth, D.; Gorska, A.E.; Dumont, N.; Shappell, S.; Washington, M.K.; Neilson, E.G.; Moses, H.L. TGF-ß Signaling in Fibroblasts Modulates the Oncogenic Potential of Adjacent Epithelia. Science 2004, 303, 848–851. [Google Scholar] [CrossRef]
- Gong, D.; Shi, W.; Yi, S.-J.; Chen, H.; Groffen, J.; Heisterkamp, N. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012, 13, 31. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Marcoe, J.P.; Lim, J.R.; Schaubert, K.L.; Fodil-Cornu, N.; Matka, M.; McCubbrey, A.L.; Farr, A.R.; Vidal, S.M.; Laouar, Y. TGF-β is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat. Immunol. 2012, 13, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Marie, J.; Liggitt, D.; Rudensky, A.Y. Cellular Mechanisms of Fatal Early-Onset Autoimmunity in Mice with the T Cell-Specific Targeting of Transforming Growth Factor-β Receptor. Immunity 2006, 25, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Li, M.O.; Flavell, R.A. TGF-β: A master of all T cell trades. Cell 2008, 134, 392–404. [Google Scholar] [CrossRef]
- Arron, J.R.; Choi, Y. Bone versus immune system. Nature 2000, 408, 535–536. [Google Scholar] [CrossRef]
- Gnoni, A.; Brunetti, O.; Longo, V.; Calabrese, A.; Argentiero, A.-l.; Calbi, R.; Antonio, G.S.; Licchetta, A. Immune system and bone microenvironment: Rationale for targeted cancer therapies. Oncotarget 2020, 11, 480. [Google Scholar] [PubMed]
- Baryawno, N.; Przybylski, D.; Kowalczyk, M.S.; Kfoury, Y.; Severe, N.; Gustafsson, K.; Kokkaliaris, K.D.; Mercier, F.; Tabaka, M.; Hofree, M.; et al. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 2019, 177, 1915–1932.e16. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Ellingsworth, L.R.; Gillis, S.; Wall, R.; Kincade, P.W. Beta transforming growth factors are potential regulators of B lymphopoiesis. J. Exp. Med. 1987, 166, 1290–1299. [Google Scholar] [CrossRef]
- Rehmann, J.A.; LeBien, T.W. Transforming growth factor-β regulates normal human pre-B cell differentiation. Int. Immunol. 1994, 6, 315–322. [Google Scholar] [CrossRef]
- Albright, A.R.; Kabat, J.; Li, M.; Raso, F.; Reboldi, A.; Muppidi, J.R. TGFβ signaling in germinal center B cells promotes the transition from light zone to dark zone. J. Exp. Med. 2019, 216, 2531–2545. [Google Scholar] [CrossRef]
- Stelling, A.; Hashwah, H.; Bertram, K.; Manz, M.G.; Tzankov, A.; Müller, A. The tumor suppressive TGF-β/SMAD1/S1PR2 signaling axis is recurrently inactivated in diffuse large B-cell lymphoma. Blood 2018, 131, 2235–2246. [Google Scholar] [CrossRef]
- Li, R.; Rosendahl, A.; Brodin, G.; Cheng, A.M.; Ahgren, A.; Sundquist, C.; Kulkarni, S.; Pawson, T.; Heldin, C.-H.; Heuchel, R. Deletion of Exon I of SMAD7 in Mice Results in Altered B Cell Responses. J. Immunol. 2006, 176, 6777–6784. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Ju, W.; Heyer, J.; Wittek, B.; Haneke, T.; Knaus, P.; Kucherlapati, R.; Böttinger, E.P.; Nitschke, L.; Kneitz, B. B Cell-Specific Deficiency for Smad2 In Vivo Leads to Defects in TGF-β-Directed IgA Switching and Changes in B Cell Fate. J. Immunol. 2006, 176, 2389–2396. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Sanjo, H.; Takeda, K.; Ninomiya-Tsuji, J.; Yamamoto, M.; Kawai, T.; Matsumoto, K.; Takeuchi, O.; Akira, S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005, 6, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Lømo, J.; Blomhoff, H.K.; Beiske, K.; Stokke, T.; Smeland, E.B. TGF-beta 1 and cyclic AMP promote apoptosis in resting human B lymphocytes. J. Immunol. 1995, 154, 1634–1643. [Google Scholar]
- Saltzman, A.; Munro, R.; Searfoss, G.; Franks, C.; Jaye, M.; Ivashchenko, Y. Transforming Growth Factor-β-Mediated Apoptosis in the Ramos B-Lymphoma Cell Line Is Accompanied by Caspase Activation and Bcl-XLDownregulation. Exp. Cell Res. 1998, 242, 244–254. [Google Scholar] [CrossRef]
- Wildey, G.M.; Patil, S.; Howe, P.H. Smad3 Potentiates Transforming Growth Factor β (TGFβ)-induced Apoptosis and Expression of the BH3-only Protein Bim in WEHI 231 B Lymphocytes. J. Biol. Chem. 2003, 278, 18069–18077. [Google Scholar] [CrossRef]
- Spender, L.; O’Brien, D.; Simpson, D.; Dutt, D.; Gregory, C.; Allday, M.; Clark, L.; Inman, G. TGF-β induces apoptosis in human B cells by transcriptional regulation of BIK and BCL-X L. Cell Death Differ. 2009, 16, 593–602. [Google Scholar] [CrossRef]
- Cazac, B.B.; Roes, J. TGF-β Receptor Controls B Cell Responsiveness and Induction of IgA In Vivo. Immunity 2000, 13, 443–451. [Google Scholar] [CrossRef]
- Tamayo, E.; Alvarez, P.; Merino, R. TGFβ Superfamily Members as Regulators of B Cell Development and Function—Implications for Autoimmunity. Int. J. Mol. Sci. 2018, 19, 3928. [Google Scholar] [CrossRef]
- Bataller, A.; Montalban-Bravo, G.; Soltysiak, K.A.; Garcia-Manero, G. The role of TGFβ in hematopoiesis and myeloid disorders. Leukemia 2019, 33, 1076–1089. [Google Scholar] [CrossRef]
- Rizzatti, E.G.; Falcão, R.P.; Panepucci, R.A.; Proto-Siqueira, R.; Anselmo-Lima, W.T.; Okamoto, O.K.; Zago, M.A. Gene expression profiling of mantle cell lymphoma cells reveals aberrant expression of genes from the PI3K-AKT, WNT and TGFβ signalling pathways. Br. J. Haematol. 2005, 130, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Dumontet, E.; Pangault, C.; Roulois, D.; Desoteux, M.; Léonard, S.; Marchand, T.; Latour, M.; Legoix, P.; Loew, D.; Dingli, F.; et al. Extracellular vesicles shed by follicular lymphoma B cells promote polarization of the bone marrow stromal cell niche. Blood 2021, 138, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Husson, H.; Carideo, E.G.; Neuberg, D.; Schultze, J.; Munoz, O.; Marks, P.W.; Donovan, J.W.; Chillemi, A.C.; O’Connell, P.; Freedman, A.S. Gene expression profiling of follicular lymphoma and normal germinal center B cells using cDNA arrays. Blood 2002, 99, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Schwarzmeier, J.D.; Hilgarth, M.; Hubmann, R.; Duechler, M.; Gisslinger, H. TGF-β1 induces bone marrow reticulin fibrosis in hairy cell leukemia. J. Clin. Investig. 2004, 113, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Lagneaux, L.; Delforge, A.; Bron, D.; Massy, M.; Bernier, M.; Stryckmans, P. Heterogenous response of B lymphocytes to transforming growth factor-beta in B-cell chronic lymphocytic leukaemia: Correlation with the expression of TGF-β receptors. Br. J. Haematol. 1997, 97, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Urashima, M.; Ogata, A.; Chauhan, D.; Hatziyanni, M.; Vidriales, M.B.; Dedera, D.A.; Schlossman, R.L.; Anderson, K.C. Transforming growth factor-beta1: Differential effects on multiple myeloma versus normal B cells. Blood 1996, 87, 1928–1938. [Google Scholar] [CrossRef]
- Lagneaux, L.; Delforge, A.; Bron, D.; Bosmans, E.; Stryckmans, P. Comparative Analysis of Cytokines Released by Bone Marrow Stromal Cells from Normal Donors and B-cell Chronic Lymphocytic Leukemic Patients. Leuk. Lymphoma 1995, 17, 127–133. [Google Scholar] [CrossRef]
- Kremer, J.-P.; Reisbach, G.; Nerl, C.; Döurmer, P. B-cell chronic lymphocytic leukaemia cells express and release transforming growth factor-β. Br. J. Haematol. 1992, 80, 480–487. [Google Scholar] [CrossRef]
- Klein, U.; Tu, Y.; Stolovitzky, G.A.; Mattioli, M.; Cattoretti, G.; Husson, H.; Freedman, A.; Inghirami, G.; Cro, L.M.; Baldini, L.; et al. Gene Expression Profiling of B Cell Chronic Lymphocytic Leukemia Reveals a Homogeneous Phenotype Related to Memory B Cells. J. Exp. Med. 2001, 194, 1625–1638. [Google Scholar] [CrossRef]
- Lagneaux, L.; Delforge, A.; Dorval, C.; Bron, D.; Stryckmans, P. Excessive production of transforming growth factor-beta by bone marrow stromal cells in B-cell chronic lymphocytic leukemia inhibits growth of hematopoietic precursors and interleukin-6 production. Blood 1993, 82, 2379–2385. [Google Scholar] [CrossRef]
- DeCoteau, J.F.; Knaus, P.I.; Yankelev, H.; Reis, M.D.; Lowsky, R.; Lodish, H.F.; Kadin, M.E. Loss of functional cell surface transforming growth factor β (TGF-β) type 1 receptor correlates with insensitivity to TGF-β in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 1997, 94, 5877–5881. [Google Scholar] [CrossRef] [PubMed]
- Lagneaux, L.; Delforge, A.; Bernier, M.; Stryckmans, P.; Bron, D. TGF-β Activity and Expression of its Receptors in B-Cell Chronic Lymphocytic Leukemia. Leuk. Lymphoma 1998, 31, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Douglas, R.S.; Capocasale, R.J.; Lamb, R.J.; Nowell, P.C.; Moore, J.S. Chronic Lymphocytic Leukemia B Cells Are Resistant to the Apoptotic Effects of Transforming Growth Factor-β. Blood 1997, 89, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Friedenberg, W.R.; Salzman, S.A.; Phan, S.M.; Burmester, J.K. Transforming growth factor-β and multidrug resistance in chronic lymphocytic leukemia. Med. Oncol. 1999, 16, 110–118. [Google Scholar] [CrossRef]
- Kumar, A.; Rogers, T.; Maizel, A.; Sharma, S. Loss of transforming growth factor beta 1 receptors and its effects on the growth of EBV-transformed human B cells. J. Immunol. 1991, 147, 998–1006. [Google Scholar]
- Blomhoff, H.K.; Smeland, E.; Mustafa, A.S.; Godal, T.; Ohlsson, R. Epstein-Barr virus mediates a switch in responsiveness to transforming growth factor, type beta, in cells of the B cell lineage. Eur. J. Immunol. 1987, 17, 299–301. [Google Scholar] [CrossRef]
- Inman, G.; Allday, M.J. Resistance to TGF-β1 correlates with a reduction of TGF-β type II receptor expression in Burkitt’s lymphoma and Epstein–Barr virus-transformed B lymphoblastoid cell lines. J. Gen. Virol. 2000, 81, 1567–1578. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Monti, S.; Savage, K.J.; Kutok, J.L.; Feuerhake, F.; Kurtin, P.; Mihm, M.; Wu, B.; Pasqualucci, L.; Neuberg, D.D.; Aguiar, R.C.; et al. Molecular Profiling of Diffuse Large B-Cell Lymphoma Identifies Robust Subtypes Including One Characterized by Host Inflammatory Response. Blood 2005, 105, 1851–1861. [Google Scholar] [CrossRef]
- Schiemann, W.P.; Rotzer, D.; Pfeifer, W.M.; Levi, E.; Rai, K.R.; Knaus, P.; Kadin, M.E. Transforming growth factor-β (TGF-β)-resistant B cells from chronic lymphocytic leukemia patients contain recurrent mutations in the signal sequence of the type I TGF-β receptor. Cancer Detect. Prev. 2004, 28, 57–64. [Google Scholar] [CrossRef]
- Chen, G.; Ghosh, P.; Osawa, H.; Sasaki, C.Y.; Rezanka, L.; Yang, J.; O’Farrell, T.J.; Longo, D.L. Resistance to TGF-β1 correlates with aberrant expression of TGF-β receptor II in human B-cell lymphoma cell lines. Blood 2007, 109, 5301–5307. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beckwith, M.; Ruscetti, F.; Sing, G.; Urba, W.; Longo, D. Anti-IgM induces transforming growth factor-beta sensitivity in a human B-lymphoma cell line: Inhibition of growth is associated with a downregulation of mutant p53. Blood 1995, 85, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Kim, J.-Y.; Park, J.-H. TGF-β1 inhibits Fas-mediated apoptosis by regulating surface Fas and cFLIPL expression in human leukaemia/lymphoma cells. Int. J. Mol. Med. 2004, 13, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Maesako, Y.; Uchiyama, T.; Ohno, H. Comparison of gene expression profiles of lymphoma cell lines from transformed follicular lymphoma, Burkitt’s lymphoma and de novo diffuse large B-cell lymphoma. Cancer Sci. 2003, 94, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Goumans, M.J.; Valdimarsdottir, G.; Itoh, S.; Rosendahl, A.; Sideras, P.; ten Dijke, P. Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J. 2002, 21, 1743–1753. [Google Scholar] [CrossRef] [PubMed]
- Stelling, A.; Wu, C.-T.; Bertram, K.; Hashwah, H.; Theocharides, A.; Manz, M.; Tzankov, A.; Müller, A. Pharmacological DNA demethylation restores SMAD1 expression and tumor suppressive signaling in diffuse large B-cell lymphoma. Blood Adv. 2019, 3, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Clozel, T.; Yang, S.; Elstrom, R.L.; Tam, W.; Martin, P.; Kormaksson, M.; Banerjee, S.; VasanthaKumar, A.; Culjkovic, B.; Scott, D.W.; et al. Mechanism-Based Epigenetic Chemosensitization Therapy of Diffuse Large B-Cell Lymphoma. Cancer Discov. 2013, 3, 1002–1019. [Google Scholar] [CrossRef]
- Rai, D.; Kim, S.-W.; McKeller, M.R.; Dahia, P.L.M.; Aguiar, R.C.T. Targeting of SMAD5 links microRNA-155 to the TGF-β pathway and lymphomagenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 3111–3116. [Google Scholar] [CrossRef]
- Rai, D.; Karanti, S.; Jung, I.; Dahia, P.L.; Aguiar, R.C. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 2008, 181, 8–15. [Google Scholar] [CrossRef]
- Ahmadvand, M.; Eskandari, M.; Pashaiefar, H.; Yaghmaie, M.; Manoochehrabadi, S.; Khakpour, G.; Sheikhsaran, F.; Zohour, M.M. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leuk. Res. 2018, 70, 45–48. [Google Scholar] [CrossRef]
- Lutzny, G.; Kocher, T.; Schmidt-Supprian, M.; Rudelius, M.; Klein-Hitpass, L.; Finch, A.J.; Dürig, J.; Wagner, M.; Haferlach, C.; Kohlmann, A. Protein kinase c-β-dependent activation of NF-κB in stromal cells is indispensable for the survival of chronic lymphocytic leukemia B cells in vivo. Cancer Cell 2013, 23, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Streubel, B.; Chott, A.; Huber, D.; Exner, M.; Jäger, U.; Wagner, O.; Schwarzinger, I. Lymphoma-Specific Genetic Aberrations in Microvascular Endothelial Cells in B-Cell Lymphomas. N. Engl. J. Med. 2004, 351, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef]
- Yang, Z.-Z.; Grote, D.M.; Ziesmer, S.C.; Xiu, B.; Yates, N.R.; Secreto, F.J.; Hodge, L.S.; Witzig, T.E.; Novak, A.J.; Ansell, S.M. Soluble and Membrane-Bound TGF-β-Mediated Regulation of Intratumoral T Cell Differentiation and Function in B-Cell Non-Hodgkin Lymphoma. PLoS ONE 2013, 8, e59456. [Google Scholar] [CrossRef]
- Yang, Z.-Z.; Grote, D.M.; Xiu, B.; Ziesmer, S.C.; Price-Troska, T.L.; Hodge, L.S.; Yates, D.M.; Novak, A.J.; Ansell, S.M. TGF-β upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia 2014, 28, 1872–1884. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Kitani, A.; Strober, W. Cell Contact–Dependent Immunosuppression by Cd4+Cd25+Regulatory T Cells Is Mediated by Cell Surface–Bound Transforming Growth Factor beta. J. Exp. Med. 2001, 194, 629–644. [Google Scholar] [CrossRef]
- Baker, K.; Raut, P.; Jass, J.R. Colorectal cancer cells express functional cell surface-bound TGFβ. Int. J. Cancer 2007, 122, 1695–1700. [Google Scholar] [CrossRef]
- Chung, J.-S.; Shiue, L.H.; Duvic, M.; Pandya, A.; Cruz Jr, P.D.; Ariizumi, K. Sézary syndrome cells overexpress syndecan-4 bearing distinct heparan sulfate moieties that suppress T-cell activation by binding DC-HIL and trapping TGF-β on the cell surface. Blood 2011, 117, 3382–3390. [Google Scholar] [CrossRef]
- Mourcin, F.; Verdière, L.; Roulois, D.; Amin, R.; Lamaison, C.; Sibut, V.; Thamphya, B.; Pangault, C.; Monvoisin, C.; Huet, S. Follicular lymphoma triggers phenotypic and functional remodeling of the human lymphoid stromal cell landscape. Immunity 2021, 54, 1788–1806. [Google Scholar] [CrossRef]
- Park, E.; Chen, J.; Moore, A.; Mangolini, M.; Santoro, A.; Boyd, J.R.; Schjerven, H.; Ecker, V.; Buchner, M.; Williamson, J.C. Stromal cell protein kinase C-β inhibition enhances chemosensitivity in B cell malignancies and overcomes drug resistance. Sci. Transl. Med. 2020, 12, eaax9340. [Google Scholar] [CrossRef]
- Kotlov, N.; Bagaev, A.; Revuelta, M.V.; Phillip, J.M.; Cacciapuoti, M.T.; Antysheva, Z.; Svekolkin, V.; Tikhonova, E.; Miheecheva, N.; Kuzkina, N.; et al. Clinical and Biological Subtypes of B-cell Lymphoma Revealed by Microenvironmental Signatures. Cancer Discov. 2021, 11, 1468–1489. [Google Scholar] [CrossRef] [PubMed]
- Rouce, R.H.; Shaim, H.; Sekine, T.; Weber, G.; Ballard, B.; Ku, S.; Barese, C.; Murali, V.; Wu, M.-F.; Liu, H.; et al. The TGF-β/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia 2016, 30, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Reimann, M.; Lee, S.; Loddenkemper, C.; Dörr, J.R.; Tabor, V.; Aichele, P.; Stein, H.; Dörken, B.; Jenuwein, T.; Schmitt, C.A. Tumor Stroma-Derived TGF-β Limits Myc-Driven Lymphomagenesis via Suv39h1-Dependent Senescence. Cancer Cell 2010, 17, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Däbritz, J.H.M.; Zhao, Z.; Yu, Y.; Dörr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Niemann, C.; Herman, S.E.M.; Maric, I.; Gomez-Rodriguez, J.; Biancotto, A.; Chang, B.Y.; Martyr, S.; Stetler-Stevenson, M.; Yuan, C.M.; Calvo, K.R.; et al. Disruption of in vivo Chronic Lymphocytic Leukemia Tumor–Microenvironment Interactions by Ibrutinib—Findings from an Investigator-Initiated Phase II Study. Clin. Cancer Res. 2016, 22, 1572–1582. [Google Scholar] [CrossRef]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel III, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Kelley, R.K.; Gane, E.; Assenat, E.; Siebler, J.; Galle, P.R.; Merle, P.; Hourmand, I.O.; Cleverly, A.; Zhao, Y.; Gueorguieva, I.; et al. A Phase 2 Study of Galunisertib (TGF-β1 Receptor Type I Inhibitor) and Sorafenib in Patients with Advanced Hepatocellular Carcinoma. Clin. Transl. Gastroenterol. 2019, 10, e00056. [Google Scholar] [CrossRef]
- Melisi, D.; Garcia-Carbonero, R.; Macarulla, T.; Pezet, D.; Deplanque, G.; Fuchs, M.; Trojan, J.; Oettle, H.; Kozloff, M.; Cleverly, A.; et al. Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br. J. Cancer 2018, 119, 1208–1214. [Google Scholar] [CrossRef]
- Teixeira, A.F.; Ten Dijke, P.; Zhu, H.-J. On-target anti-TGF-β therapies are not succeeding in clinical cancer treatments: What are remaining challenges? Front. Cell Dev. Biol. 2020, 8, 605. [Google Scholar] [CrossRef]
- Lu, A.; Pallero, M.A.; Lei, W.; Hong, H.; Yang, Y.; Suto, M.J.; Murphy-Ullrich, J.E. Inhibition of Transforming Growth Factor-β Activation Diminishes Tumor Progression and Osteolytic Bone Disease in Mouse Models of Multiple Myeloma. Am. J. Pathol. 2016, 186, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Bollard, C.M.; Rössig, C.; Calonge, M.J.; Huls, M.H.; Wagner, H.-J.; Massague, J.; Brenner, M.K.; Heslop, H.E.; Rooney, C.M. Adapting a transforming growth factor β–related tumor protection strategy to enhance antitumor immunity. Blood 2002, 99, 3179–3187. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.E.; Dotti, G.; Lu, A.; Khalil, M.; Brenner, M.K.; Heslop, H.E.; Rooney, C.M.; Bollard, C.M. Antitumor Activity of EBV-specific T Lymphocytes Transduced with a Dominant Negative TGF-β Receptor. J. Immunother. 2008, 31, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Kloss, C.C.; Lee, J.; Zhang, A.; Chen, F.; Melenhorst, J.J.; Lacey, S.F.; Maus, M.V.; Fraietta, J.A.; Zhao, Y.; June, C.H. Dominant-Negative TGF-β Receptor Enhances PSMA-Targeted Human CAR T Cell Proliferation and Augments Prostate Cancer Eradication. Mol. Ther. 2018, 26, 1855–1866. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timmins, M.A.; Ringshausen, I. Transforming Growth Factor-Beta Orchestrates Tumour and Bystander Cells in B-Cell Non-Hodgkin Lymphoma. Cancers 2022, 14, 1772. https://doi.org/10.3390/cancers14071772
Timmins MA, Ringshausen I. Transforming Growth Factor-Beta Orchestrates Tumour and Bystander Cells in B-Cell Non-Hodgkin Lymphoma. Cancers. 2022; 14(7):1772. https://doi.org/10.3390/cancers14071772
Chicago/Turabian StyleTimmins, Matthew A., and Ingo Ringshausen. 2022. "Transforming Growth Factor-Beta Orchestrates Tumour and Bystander Cells in B-Cell Non-Hodgkin Lymphoma" Cancers 14, no. 7: 1772. https://doi.org/10.3390/cancers14071772
APA StyleTimmins, M. A., & Ringshausen, I. (2022). Transforming Growth Factor-Beta Orchestrates Tumour and Bystander Cells in B-Cell Non-Hodgkin Lymphoma. Cancers, 14(7), 1772. https://doi.org/10.3390/cancers14071772





