The Diagnostic Utility of Cell-Free DNA from Ex Vivo Bronchoalveolar Lavage Fluid in Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
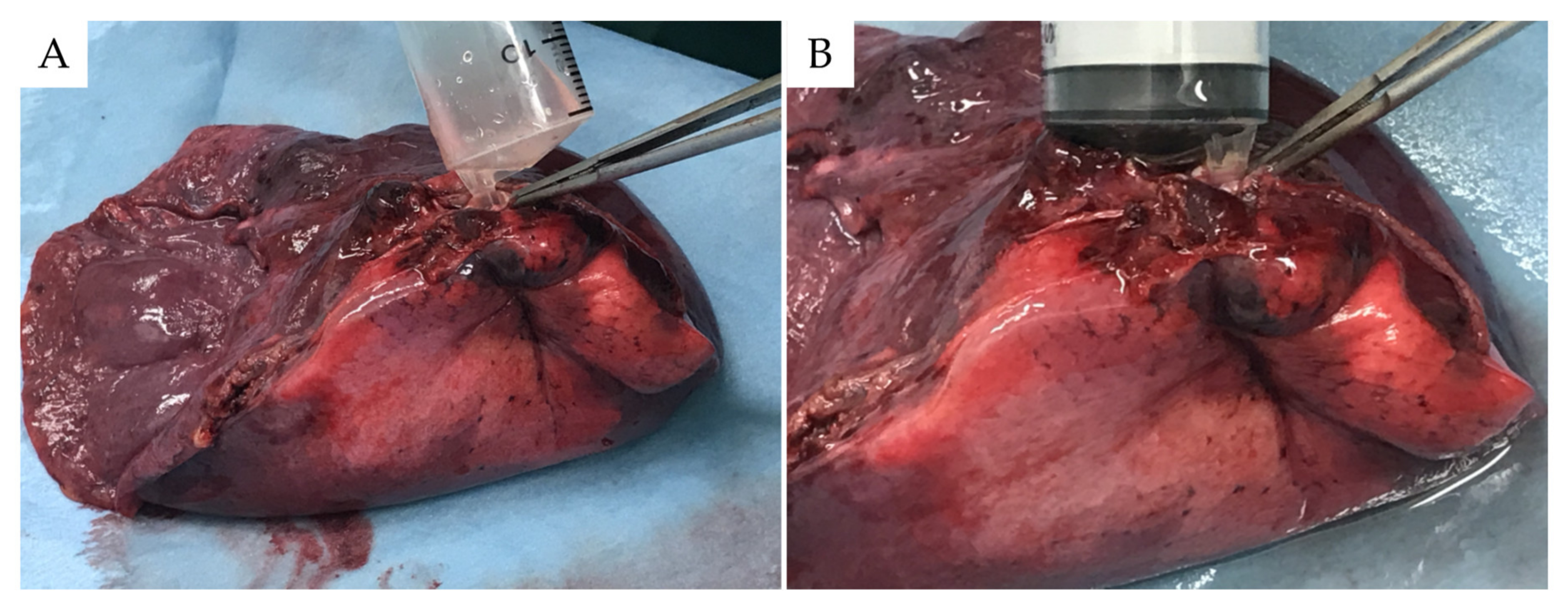
2.2. Ex Vivo BAL Model
2.3. Isolation and Purification of Genomic DNA Samples from Specimens
2.4. Quality Assessment of DNA Samples by Quantitative Real-Time PCR
2.5. Deep Sequencing for the Detection of Targeted Lung Cancer-Related Gene Mutations and Data Analysis
2.6. OncomineTarget Dx
2.7. Statistical Analyses
3. Results
3.1. Patient Characteristics
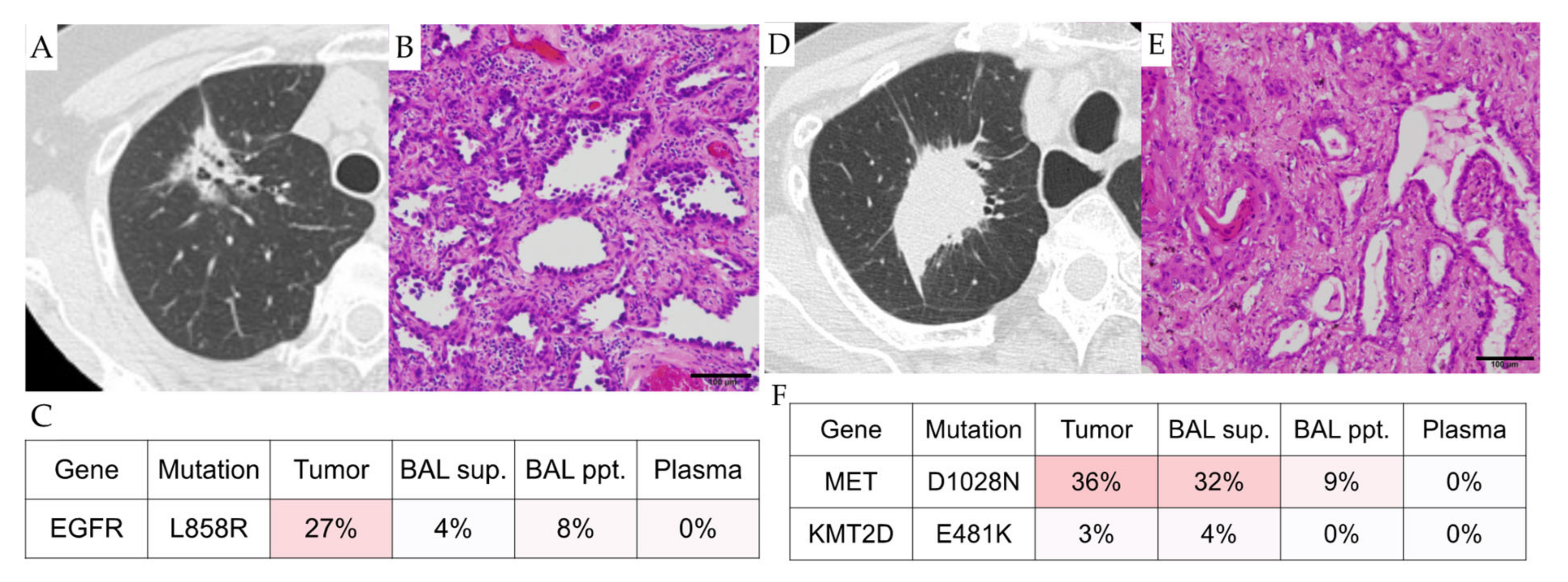
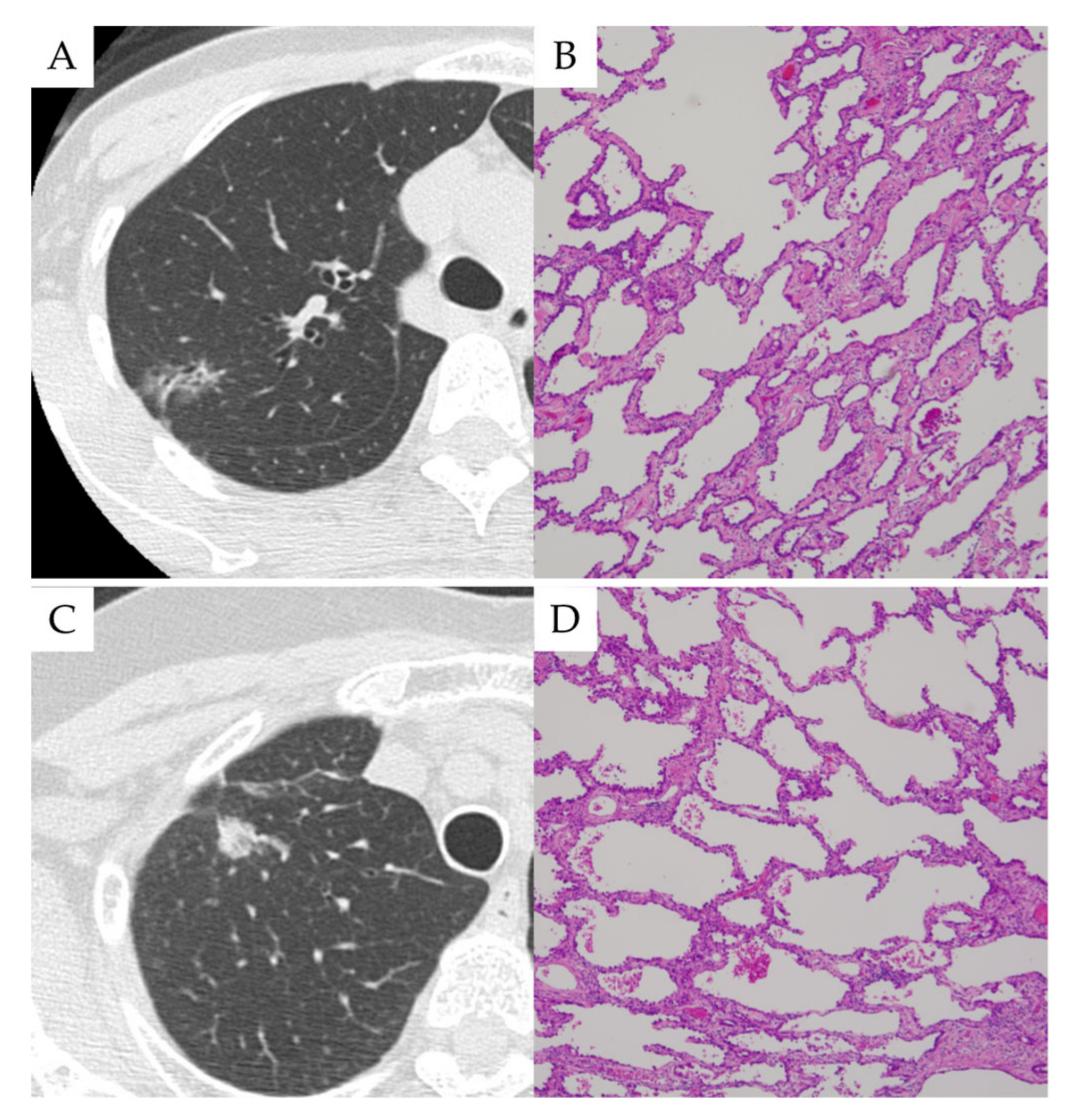
3.2. Case Presentations
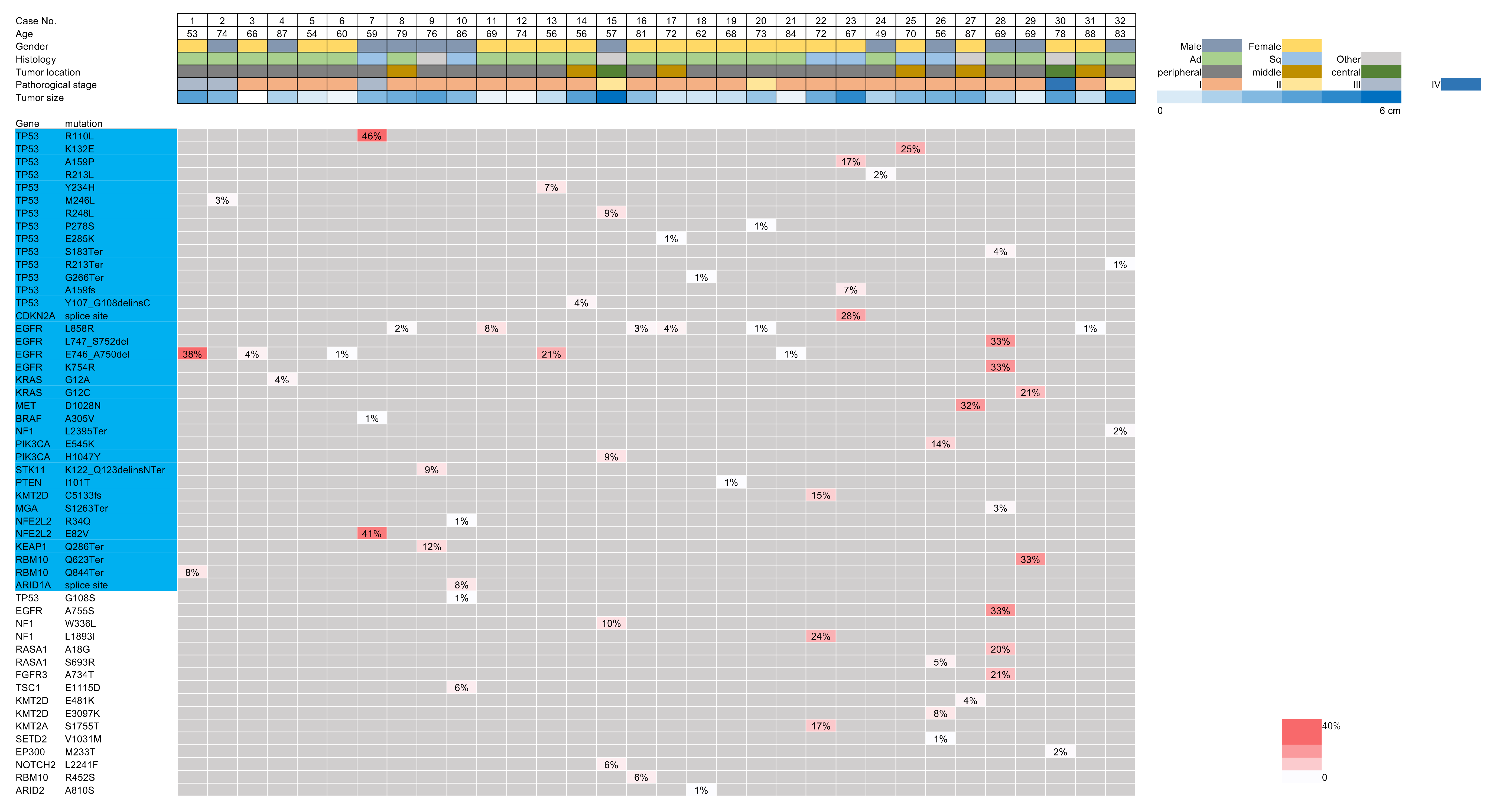
3.3. Targeted Sequencing Identified Shared Mutations in the Ex Vivo BAL Fluid and Lung Cancers
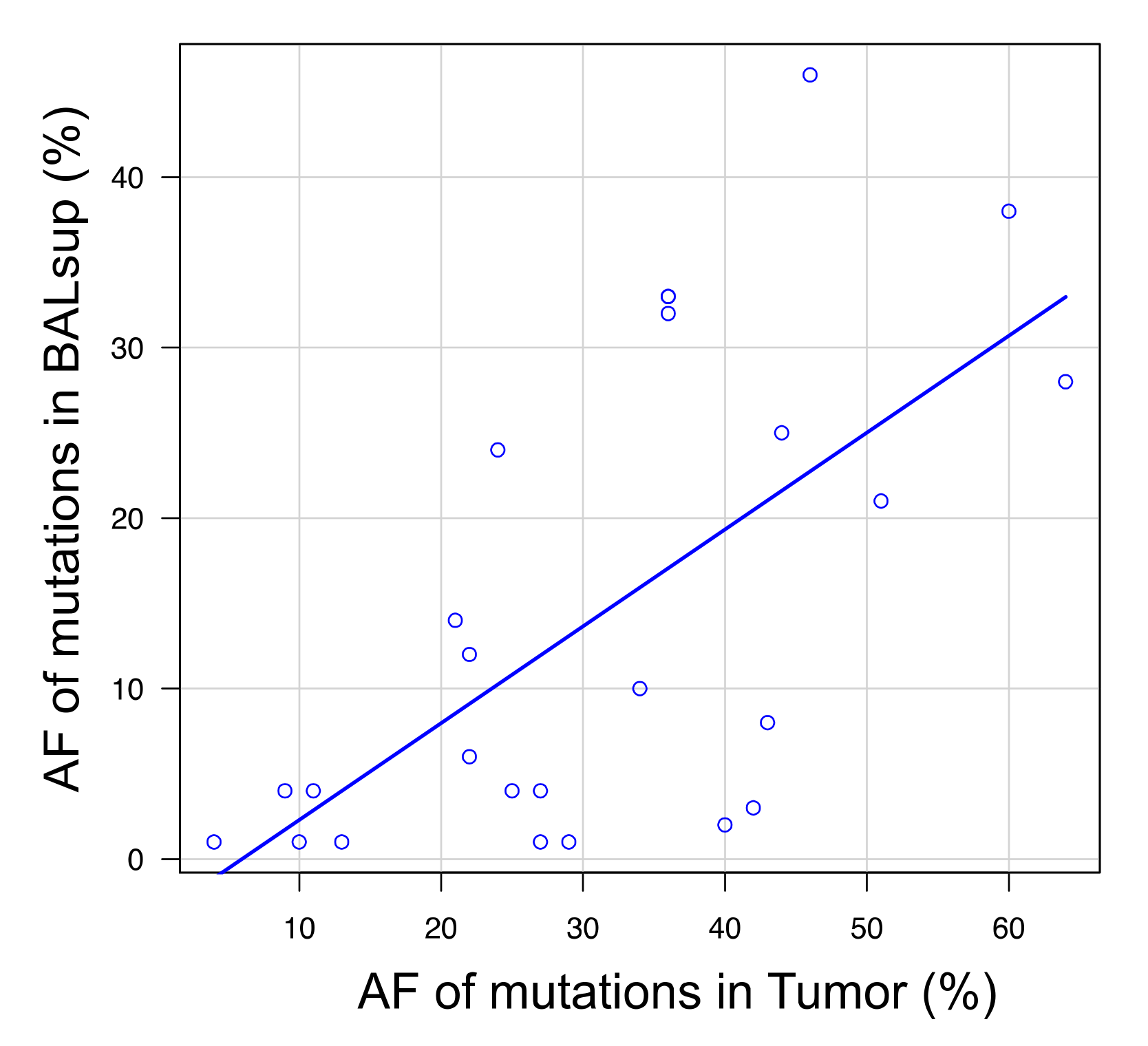
3.4. Analysis of the Mutational Overlap across Different Sample Types
3.5. Microscopic Pathological Examination vs. Genomic DNA Sequencing Approach for Bronchoscopic Specimens
3.6. Comparison between dtDNA Positive and Negative Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goto, T. Effect of the coronavirus disease pandemic on bronchoscopic diagnosis of lung cancer in a provincial city in Japan. J. Cardiothorac. Surg. 2021, 16, 115. [Google Scholar] [CrossRef]
- Wang Memoli, J.S.; Nietert, P.J.; Silvestri, G.A. Meta-analysis of guided bronchoscopy for the evaluation of the pulmonary nodule. Chest 2012, 142, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Goto, T. Robotic bronchoscopy: Is it classic? J. Thorac. Dis. 2021, 13, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Goto, T. Is tomosynthesis an ingenious scheme for bronchoscopic diagnosis of lung nodules? Respirology 2021, 26, 125. [Google Scholar] [CrossRef]
- Yutaka, Y.; Sato, T.; Isowa, M.; Murata, Y.; Tanaka, S.; Yamada, Y.; Ohsumi, A.; Nakajima, D.; Hamaji, M.; Menju, T.; et al. Electromagnetic navigation bronchoscopy versus virtual bronchoscopy navigation for improving the diagnosis of peripheral lung lesions: Analysis of the predictors of successful diagnosis. Surg. Today 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Anan, K.; Oshima, Y.; Ogura, T.; Tanabe, Y.; Higashi, A.; Iwashita, Y.; Fujita, K.; Yoshida, T.; Ando, K.; Okamori, S.; et al. Safety and harms of bronchoalveolar lavage for acute respiratory failure: A systematic review and meta-analysis. Respir. Investig. 2021, 60, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Hirotsu, Y.; Nakagomi, T.; Shikata, D.; Yokoyama, Y.; Amemiya, K.; Tsutsui, T.; Kakizaki, Y.; Oyama, T.; Mochizuki, H.; et al. Detection of tumor-derived DNA dispersed in the airway improves the diagnostic accuracy of bronchoscopy for lung cancer. Oncotarget 2017, 8, 79404–79413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hattori, A.; Takamochi, K.; Oh, S.; Suzuki, K. New revisions and current issues in the eighth edition of the TNM classification for non-small cell lung cancer. Jpn. J. Clin. Oncol. 2019, 49, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.W.; Erasmus, J.J. IDKD Springer Series, Current Concepts in the Diagnosis and Staging of Lung Cancer. In Diseases of the Chest, Breast, Heart and Vessels 2019–2022: Diagnostic and Interventional Imaging; Hodler, J., Kubik-Huch, R.A., von Schulthess, G.K., Eds.; Springer: Cham, Switzerland, 2019; pp. 79–93. [Google Scholar]
- Higuchi, R.; Goto, T.; Hirotsu, Y.; Otake, S.; Oyama, T.; Amemiya, K.; Mochizuki, H.; Omata, M. Streptococcus australis and Ralstonia pickettii as Major Microbiota in Mesotheliomas. J. Pers. Med. 2021, 11, 297. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Goto, T.; Hirotsu, Y.; Otake, S.; Oyama, T.; Amemiya, K.; Ohyama, H.; Mochizuki, H.; Omata, M. Sphingomonas and Phenylobacterium as Major Microbiota in Thymic Epithelial Tumors. J. Pers. Med. 2021, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Goto, T.; Nakagomi, T.; Hirotsu, Y.; Oyama, T.; Amemiya, K.; Mochizuki, H.; Omata, M. Discrimination Between Primary Lung Cancer and Lung Metastases by Genomic Profiling. JTO Clin. Res. Rep. 2021, 2, 100255. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, K.; Hirotsu, Y.; Goto, T.; Nakagomi, H.; Mochizuki, H.; Oyama, T.; Omata, M. Touch imprint cytology with massively parallel sequencing (TIC-seq): A simple and rapid method to snapshot genetic alterations in tumors. Cancer Med. 2016, 5, 3426–3436. [Google Scholar] [CrossRef]
- Nakagomi, T.; Goto, T.; Hirotsu, Y.; Shikata, D.; Yokoyama, Y.; Higuchi, R.; Amemiya, K.; Okimoto, K.; Oyama, T.; Mochizuki, H.; et al. New therapeutic targets for pulmonary sarcomatoid carcinomas based on their genomic and phylogenetic profiles. Oncotarget 2018, 9, 10635–10649. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Goto, T.; Hirotsu, Y.; Oyama, T.; Amemiya, K.; Omata, M. Analysis of tumor-derived DNA in plasma and bone marrow fluid in lung cancer patients. Med. Oncol. 2016, 33, 29. [Google Scholar] [CrossRef]
- Goto, T.; Kunimasa, K.; Hirotsu, Y.; Nakagomi, T.; Yokoyama, Y.; Higuchi, R.; Otake, S.; Oyama, T.; Amemiya, K.; Mochizuki, H.; et al. Association of Mutation Profiles with Postoperative Survival in Patients with Non-Small Cell Lung Cancer. Cancers 2020, 12, 3472. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Nakagomi, T.; Goto, T.; Hirotsu, Y.; Shikata, D.; Yokoyama, Y.; Otake, S.; Amemiya, K.; Oyama, T.; Mochizuki, H.; et al. Identification of Clonality through Genomic Profile Analysis in Multiple Lung Cancers. J. Clin. Med. 2020, 9, 573. [Google Scholar] [CrossRef] [PubMed]
- Iijima, Y.; Hirotsu, Y.; Amemiya, K.; Ooka, Y.; Mochizuki, H.; Oyama, T.; Nakagomi, T.; Uchida, Y.; Kobayashi, Y.; Tsutsui, T.; et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. Eur. J. Cancer 2017, 86, 349–357. [Google Scholar] [CrossRef]
- Kunimasa, K.; Hirotsu, Y.; Amemiya, K.; Nagakubo, Y.; Goto, T.; Miyashita, Y.; Kakizaki, Y.; Tsutsui, T.; Otake, S.; Kobayashi, H.; et al. Genome analysis of peeling archival cytology samples detects driver mutations in lung cancer. Cancer Med. 2020, 9, 4501–4511. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Kojima, Y.; Okimoto, K.; Amemiya, K.; Mochizuki, H.; Omata, M. Comparison between two amplicon-based sequencing panels of different scales in the detection of somatic mutations associated with gastric cancer. BMC Genom. 2016, 17, 833. [Google Scholar] [CrossRef] [PubMed]
- Nakagomi, T.; Goto, T.; Hirotsu, Y.; Shikata, D.; Amemiya, K.; Oyama, T.; Mochizuki, H.; Omata, M. Elucidation of radiation-resistant clones by a serial study of intratumor heterogeneity before and after stereotactic radiotherapy in lung cancer. J. Thorac. Dis. 2017, 9, e598–e604. [Google Scholar] [CrossRef] [PubMed]
- Nakagomi, T.; Goto, T.; Hirotsu, Y.; Shikata, D.; Yokoyama, Y.; Higuchi, R.; Otake, S.; Amemiya, K.; Oyama, T.; Mochizuki, H.; et al. Genomic Characteristics of Invasive Mucinous Adenocarcinomas of the Lung and Potential Therapeutic Targets of B7-H3. Cancers 2018, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Nakagomi, T.; Hirotsu, Y.; Goto, T.; Shikata, D.; Yokoyama, Y.; Higuchi, R.; Otake, S.; Amemiya, K.; Oyama, T.; Mochizuki, H.; et al. Clinical Implications of Noncoding Indels in the Surfactant-Encoding Genes in Lung Cancer. Cancers 2019, 11, 552. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Rogers, M.F.; Shihab, H.A.; Mort, M.; Cooper, D.N.; Gaunt, T.R.; Campbell, C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2017, 34, 511–513. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Amemiya, K.; Hirotsu, Y.; Nagakubo, Y.; Mochizuki, H.; Higuchi, R.; Tsutsui, T.; Kakizaki, Y.; Miyashita, Y.; Oyama, T.; Omata, M. Actionable driver DNA variants and fusion genes can be detected in archived cytological specimens with the Oncomine Dx Target Test Multi-CDx system in lung cancer. Cancer Cytopathol. 2021, 129, 729–738. [Google Scholar] [CrossRef]
- Paasinen-Sohns, A.; Koelzer, V.H.; Frank, A.; Schafroth, J.; Gisler, A.; Sachs, M.; Graber, A.; Rothschild, S.I.; Wicki, A.; Cathomas, G.; et al. Single-Center Experience with a Targeted Next Generation Sequencing Assay for Assessment of Relevant Somatic Alterations in Solid Tumors. Neoplasia 2017, 19, 196–206. [Google Scholar] [CrossRef]
- Silvestri, G.A.; Vachani, A.; Whitney, D.; Elashoff, M.; Porta Smith, K.; Ferguson, J.S.; Parsons, E.; Mitra, N.; Brody, J.; Lenburg, M.E.; et al. A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. N. Engl. J. Med. 2015, 373, 243–251. [Google Scholar] [CrossRef]
- Whitney, D.H.; Elashoff, M.R.; Porta-Smith, K.; Gower, A.C.; Vachani, A.; Ferguson, J.S.; Silvestri, G.A.; Brody, J.S.; Lenburg, M.E.; Spira, A. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med. Genom. 2015, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Hirotsu, Y.; Amemiya, K.; Nakagomi, T.; Shikata, D.; Yokoyama, Y.; Okimoto, K.; Oyama, T.; Mochizuki, H.; Omata, M. Distribution of circulating tumor DNA in lung cancer: Analysis of the primary lung and bone marrow along with the pulmonary venous and peripheral blood. Oncotarget 2017, 8, 59268–59281. [Google Scholar] [CrossRef] [PubMed]
- Ohira, T.; Sakai, K.; Matsubayashi, J.; Kajiwara, N.; Kakihana, M.; Hagiwara, M.; Hibi, M.; Yoshida, K.; Maeda, J.; Ohtani, K.; et al. Tumor volume determines the feasibility of cell-free DNA sequencing for mutation detection in non-small cell lung cancer. Cancer Sci. 2016, 107, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Bick, A.G.; Weinstock, J.S.; Nandakumar, S.K.; Fulco, C.P.; Bao, E.L.; Zekavat, S.M.; Szeto, M.D.; Liao, X.; Leventhal, M.J.; Nasser, J.; et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020, 586, 763–768. [Google Scholar] [CrossRef]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Goto, T. Cell-Free DNA From Nontumor Tissue in Patients With Non-small Cell Lung Cancer. Chest 2021, 160, e374–e375. [Google Scholar] [CrossRef] [PubMed]
- Van der Leest, P.; Hiddinga, B.; Miedema, A.; Aguirre Azpurua, M.L.; Rifaela, N.; Ter Elst, A.; Timens, W.; Groen, H.J.M.; van Kempen, L.C.; Hiltermann, T.J.N.; et al. Circulating tumor DNA as a biomarker for monitoring early treatment responses of patients with advanced lung adenocarcinoma receiving immune checkpoint inhibitors. Mol. Oncol. 2021, 15, 2910–2922. [Google Scholar] [CrossRef]
- Stitz, R.; Buder, A.; Silye, R.; Baumgartner, B.; Pühringer, F.; Filipits, M.; Oberndorfer, E.; Heitzer, E. Validation of a next-generation sequencing assay for the detection of EGFR mutations in cell-free circulating tumor DNA. Exp. Mol. Pathol. 2021, 123, 104685. [Google Scholar] [CrossRef] [PubMed]
- Olmedillas-López, S.; Olivera-Salazar, R.; García-Arranz, M.; García-Olmo, D. Current and Emerging Applications of Droplet Digital PCR in Oncology: An Updated Review. Mol. Diagn. Ther. 2022, 26, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Huang, Q.; Yin, W.; Tan, S.; Chen, C.; Liu, W.; Tang, J.; Wang, X.; Zhang, B.; Zou, M.; et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020, 10, 561598. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016, 7, 11815. [Google Scholar] [CrossRef] [PubMed]
- Bratman, S.V.; Newman, A.M.; Alizadeh, A.A.; Diehn, M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev. Mol. Diagn. 2015, 15, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Gowers, K.H.C.; Lee-Six, H.; Chandrasekharan, D.P.; Coorens, T.; Maughan, E.F.; Beal, K.; Menzies, A.; Millar, F.R.; Anderson, E.; et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 2020, 578, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Tian, B.; Geng, X.; Zhou, H.; Xu, Z.; Lai, T.; Wu, Y.; Bao, Z.; Chen, Z.; Li, W.; et al. IL-17-Mediated Inflammation Promotes Cigarette Smoke-Induced Genomic Instability. Cells 2021, 10, 1173. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Fowler, J.C.; Wabik, A.; Lawson, A.R.J.; Abascal, F.; Hall, M.W.J.; Cagan, A.; Murai, K.; Mahbubani, K.; Stratton, M.R.; et al. Somatic mutant clones colonize the human esophagus with age. Science 2018, 362, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Lee-Six, H.; Olafsson, S.; Ellis, P.; Osborne, R.J.; Sanders, M.A.; Moore, L.; Georgakopoulos, N.; Torrente, F.; Noorani, A.; Goddard, M.; et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019, 574, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, R.; Lupini, L.; Miotto, E.; Saccenti, E.; Mascetti, S.; Morandi, L.; Bassi, C.; Rasio, D.; Callegari, E.; Conti, V.; et al. Molecular testing on bronchial washings for the diagnosis and predictive assessment of lung cancer. Mol. Oncol. 2020, 14, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Hirotsu, Y.; Amemiya, K.; Mochizuki, H.; Omata, M. Understanding Intratumor Heterogeneity and Evolution in NSCLC and Potential New Therapeutic Approach. Cancers 2018, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.S.; Lim, J.H.; Lee, M.K.; Lee, S.J.; Kim, H.J.; Kim, M.J.; Park, M.H.; Kim, J.S.; Nam, H.S.; Park, N.; et al. Feasibility of Bronchial Washing Fluid-Based Approach to Early-Stage Lung Cancer Diagnosis. Oncologist 2019, 24, e603–e606. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Hirotsu, Y.; Mochizuki, H.; Nakagomi, T.; Oyama, T.; Amemiya, K.; Omata, M. Stepwise addition of genetic changes correlated with histological change from “well-differentiated” to “sarcomatoid” phenotypes: A case report. BMC Cancer 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]


| Parameters | Variables | Number |
|---|---|---|
| Total number | 32 | |
| Age, years | Mean ± SD | 70.0 ± 11.5 |
| Sex | Male | 12 (37.5%) |
| Female | 20 (62.5%) | |
| Smoking history | Smoker | 19 (59.4%) |
| Non-smoker | 13 (40.6%) | |
| Tumor size, mm | Mean ± SD | 24.3 ± 14.3 |
| Tumor location | Central | 1 (3.1%) |
| Middle | 7 (21.9%) | |
| Peripheral | 24 (75%) | |
| Histology | Adenocarcinoma | 22 (68.8%) |
| Squamous cell carcinoma | 6 (18.8%) | |
| Pleomorphic carcinoma | 2 (6.3%) | |
| Adenosquamous cell carcinoma | 1 (3.1%) | |
| Large cell neuroendocrine carcinoma | 1 (3.1%) | |
| Pathological stage | I | 25 (78.1%) |
| II | 3 (9.4%) | |
| III | 3 (9.4%) | |
| IV | 1 (3.1%) |
| Case | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
| Microscopic Dx | ||||||||||||||||
| Cytology | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Genomic Dx | ||||||||||||||||
| BALF sup. | + | + | − | + | − | − | + | + | + | + | + | − | + | + | + | + |
| BALF ppt. | + | − | + | − | − | + | − | + | + | − | + | − | − | − | − | + |
| Plasma | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Case | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 |
| Microscopic Dx | ||||||||||||||||
| Cytology | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Genomic Dx | ||||||||||||||||
| BALF sup. | + | + | + | + | − | + | + | + | + | + | + | + | + | − | + | − |
| BALF ppt. | − | − | − | − | + | + | + | − | − | − | + | + | − | + | − | + |
| Plasma | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otake, S.; Goto, T.; Higuchi, R.; Nakagomi, T.; Hirotsu, Y.; Amemiya, K.; Oyama, T.; Mochizuki, H.; Omata, M. The Diagnostic Utility of Cell-Free DNA from Ex Vivo Bronchoalveolar Lavage Fluid in Lung Cancer. Cancers 2022, 14, 1764. https://doi.org/10.3390/cancers14071764
Otake S, Goto T, Higuchi R, Nakagomi T, Hirotsu Y, Amemiya K, Oyama T, Mochizuki H, Omata M. The Diagnostic Utility of Cell-Free DNA from Ex Vivo Bronchoalveolar Lavage Fluid in Lung Cancer. Cancers. 2022; 14(7):1764. https://doi.org/10.3390/cancers14071764
Chicago/Turabian StyleOtake, Sotaro, Taichiro Goto, Rumi Higuchi, Takahiro Nakagomi, Yosuke Hirotsu, Kenji Amemiya, Toshio Oyama, Hitoshi Mochizuki, and Masao Omata. 2022. "The Diagnostic Utility of Cell-Free DNA from Ex Vivo Bronchoalveolar Lavage Fluid in Lung Cancer" Cancers 14, no. 7: 1764. https://doi.org/10.3390/cancers14071764
APA StyleOtake, S., Goto, T., Higuchi, R., Nakagomi, T., Hirotsu, Y., Amemiya, K., Oyama, T., Mochizuki, H., & Omata, M. (2022). The Diagnostic Utility of Cell-Free DNA from Ex Vivo Bronchoalveolar Lavage Fluid in Lung Cancer. Cancers, 14(7), 1764. https://doi.org/10.3390/cancers14071764






