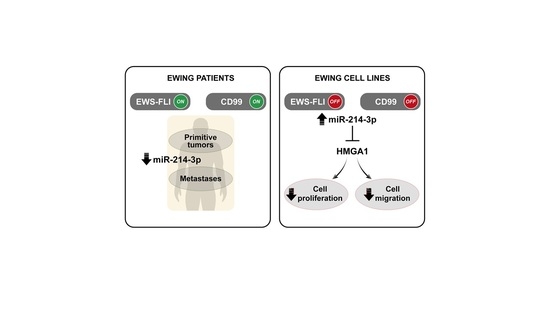
miR-214-3p Is Commonly Downregulated by EWS-FLI1 and by CD99 and Its Restoration Limits Ewing Sarcoma Aggressiveness
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Lines and Primary Cultures
2.2. Anti-CD99 Antibody Treatment
2.3. Stable and Transient Transfection
2.4. Clinical Samples
2.5. RNA Extraction and Quantitative PCR
2.6. Cell Growth Assays
2.7. Migration Assay
2.8. Western Blotting
2.9. Statistical Analyses
2.10. Software Analysis
3. Results
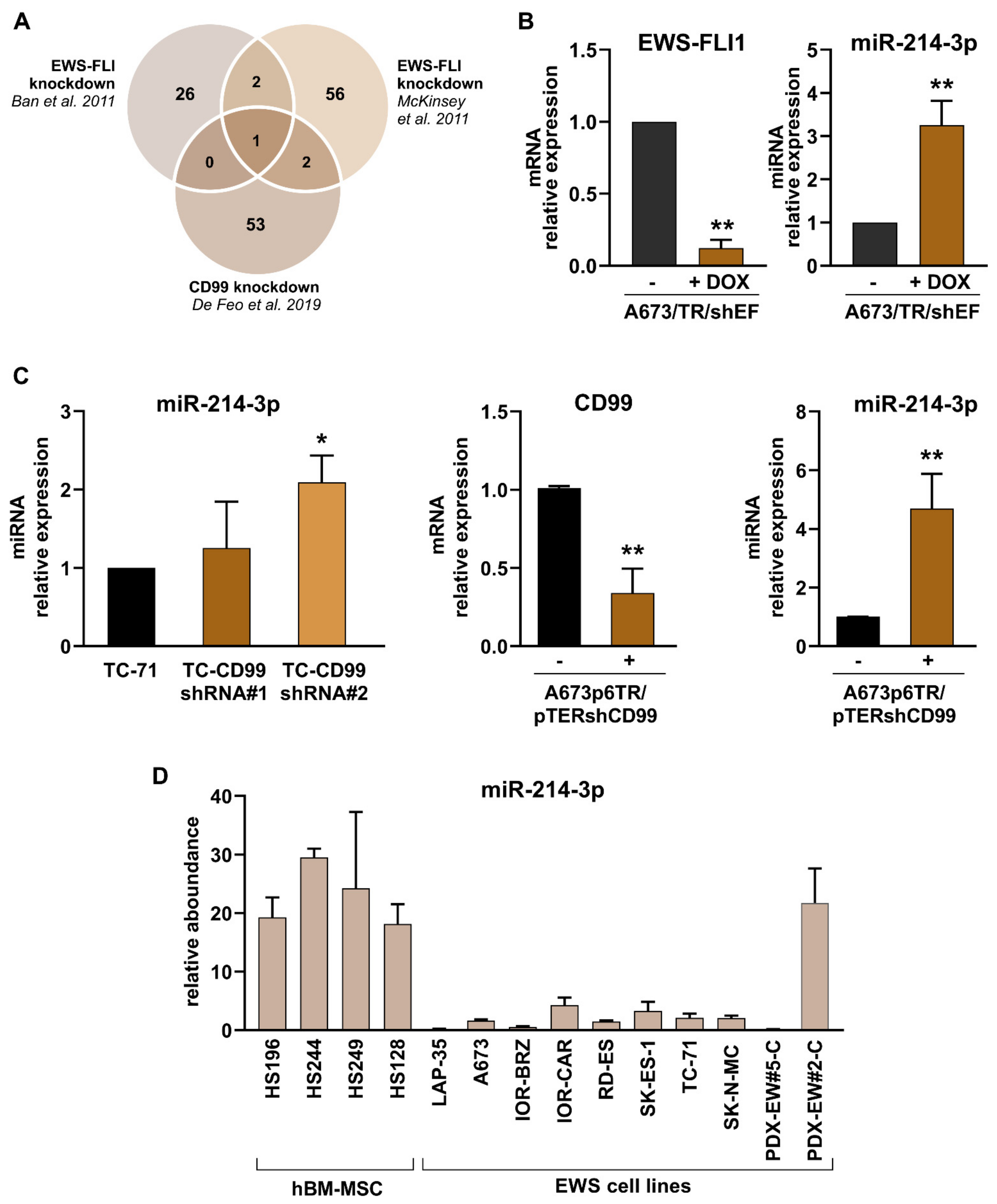
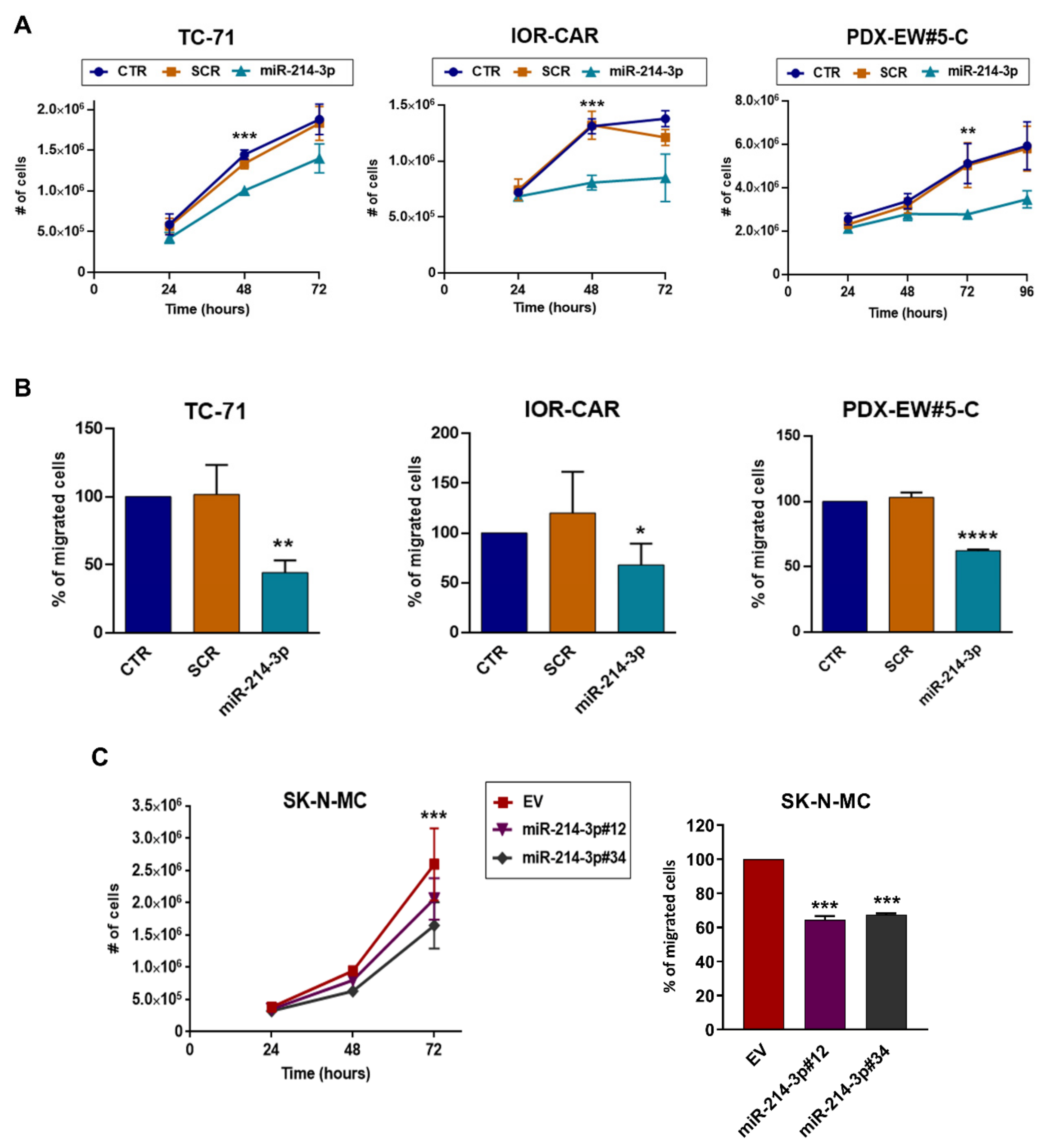
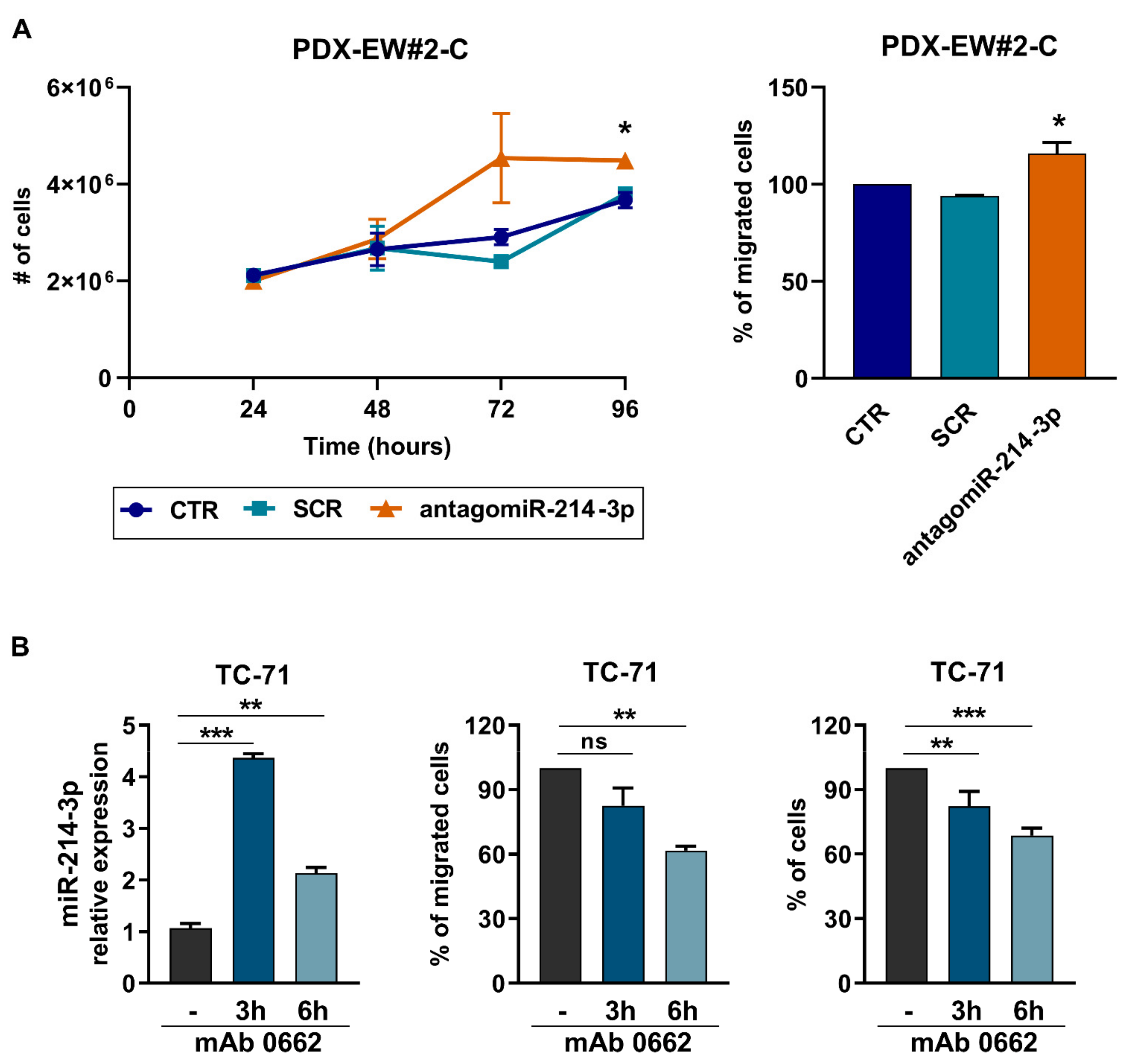
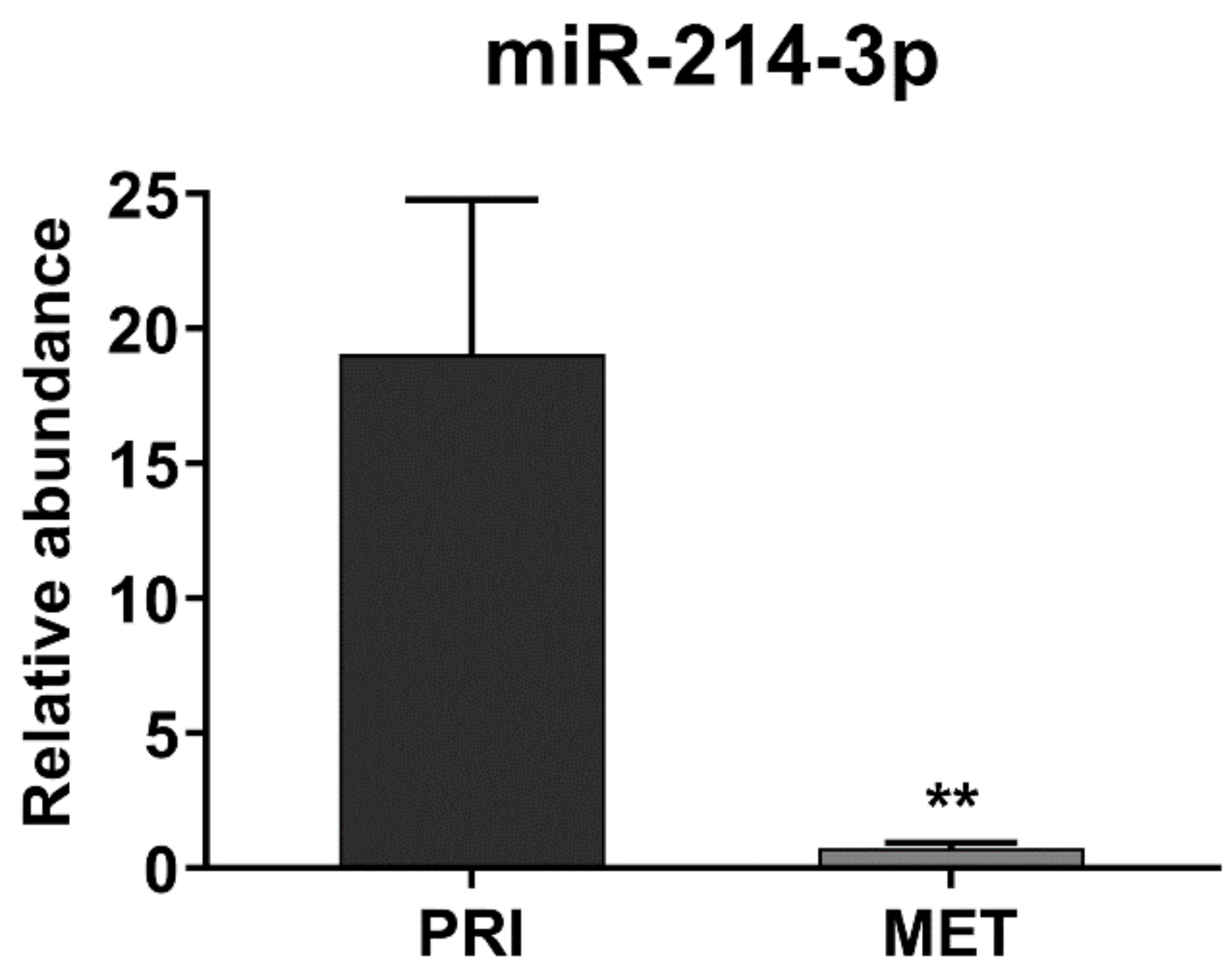
3.1. miR-214-3p Is Commonly Repressed by EWS-FLI1 and CD99 and Restoration of Its Expression Stalls Tumor Cell Growth and Cell Migration
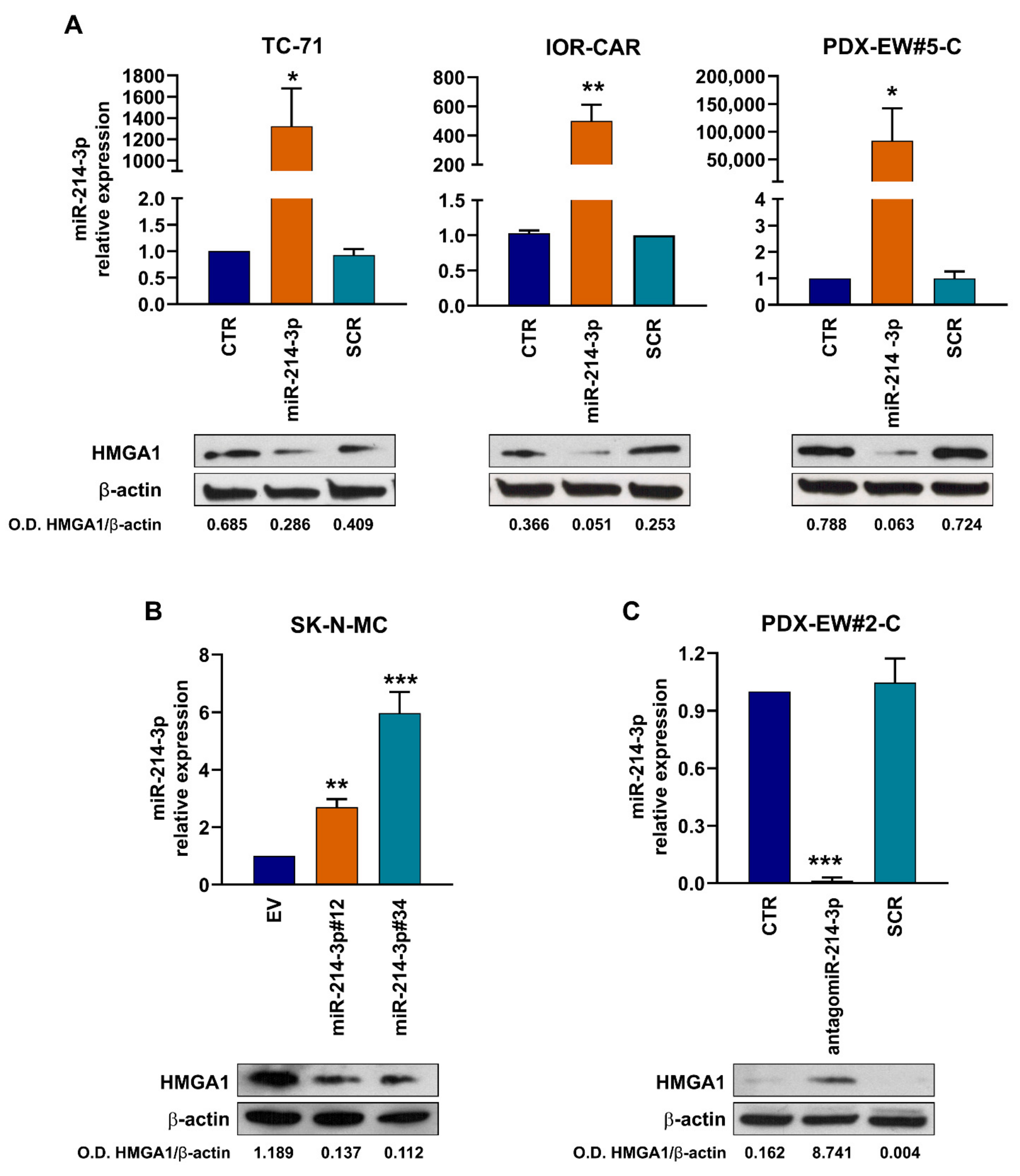
3.2. miR-214-3p Affects HMGA1 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pasello, M.; Manara, M.C.; Scotlandi, K. CD99 at the crossroads of physiology and pathology. J. Cell Commun. Signal. 2018, 12, 55–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetete-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef] [Green Version]
- Brohl, A.S.; Solomon, D.A.; Chang, W.; Wang, J.; Song, Y.; Sindiri, S.; Patidar, R.; Hurd, L.; Chen, L.; Shern, J.F.; et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014, 10, e1004475. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef]
- Kovar, H.; Bierbaumer, L.; Radic-Sarikas, B. The YAP/TAZ Pathway in Osteogenesis and Bone Sarcoma Pathogenesis. Cells 2020, 9, 972. [Google Scholar] [CrossRef] [Green Version]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suva, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef] [Green Version]
- Tomazou, E.M.; Sheffield, N.C.; Schmidl, C.; Schuster, M.; Schonegger, A.; Datlinger, P.; Kubicek, S.; Bock, C.; Kovar, H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015, 10, 1082–1095. [Google Scholar] [CrossRef] [Green Version]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schonegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef]
- Barrett, C.; Budhiraja, A.; Parashar, V.; Batish, M. The Landscape of Regulatory Noncoding RNAs in Ewing’s Sarcoma. Biomedicines 2021, 9, 933. [Google Scholar] [CrossRef] [PubMed]
- Dylla, L.; Moore, C.; Jedlicka, P. MicroRNAs in Ewing Sarcoma. Front. Oncol. 2013, 3, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Feo, A.; Sciandra, M.; Ferracin, M.; Felicetti, F.; Astolfi, A.; Pignochino, Y.; Picci, P.; Care, A.; Scotlandi, K. Exosomes from CD99-deprived Ewing sarcoma cells reverse tumor malignancy by inhibiting cell migration and promoting neural differentiation. Cell Death Dis. 2019, 10, 471. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.; Aryee, D.N.; Felicetti, F.; De Feo, A.; Mancarella, C.; Manara, M.C.; Picci, P.; Colombo, M.P.; Kovar, H.; Care, A.; et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-kappaB signaling. Oncogene 2016, 35, 3944–3954. [Google Scholar] [CrossRef]
- Riggi, N.; Suva, M.L.; Stamenkovic, I. Ewing’s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef]
- Manara, M.C.; Pasello, M.; Scotlandi, K. CD99: A Cell Surface Protein with an Oncojanus Role in Tumors. Genes 2018, 9, 159. [Google Scholar] [CrossRef] [Green Version]
- Franzetti, G.A.; Laud-Duval, K.; Bellanger, D.; Stern, M.H.; Sastre-Garau, X.; Delattre, O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene 2013, 32, 3915–3921. [Google Scholar] [CrossRef] [Green Version]
- Ban, J.; Jug, G.; Mestdagh, P.; Schwentner, R.; Kauer, M.; Aryee, D.N.; Schaefer, K.L.; Nakatani, F.; Scotlandi, K.; Reiter, M.; et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene 2011, 30, 2173–2180. [Google Scholar] [CrossRef] [Green Version]
- McKinsey, E.L.; Parrish, J.K.; Irwin, A.E.; Niemeyer, B.F.; Kern, H.B.; Birks, D.K.; Jedlicka, P. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene 2011, 30, 4910–4920. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [Green Version]
- Peng, R.Q.; Wan, H.Y.; Li, H.F.; Liu, M.; Li, X.; Tang, H. MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J. Biol. Chem. 2012, 287, 14301–14309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Liu, M.; Li, X.; Tang, H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013, 587, 488–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.; Lei, Z.; Jin-An, M.; Xue-Zhen, L.; Xing-Nan, Z.; Xiu-Wen, D. The inhibitory role of miR-214 in cervical cancer cells through directly targeting mitochondrial transcription factor A (TFAM). Eur. J. Gynaecol. Oncol. 2014, 35, 676–682. [Google Scholar] [PubMed]
- Penna, E.; Orso, F.; Cimino, D.; Tenaglia, E.; Lembo, A.; Quaglino, E.; Poliseno, L.; Haimovic, A.; Osella-Abate, S.; De Pitta, C.; et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011, 30, 1990–2007. [Google Scholar] [CrossRef] [Green Version]
- Penna, E.; Orso, F.; Taverna, D. miR-214 as a key hub that controls cancer networks: Small player, multiple functions. J. Investig. Dermatol. 2015, 135, 960–969. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.S.; Yang, X.H.; Wang, X.D.; Wang, Y.L.; Zhou, B.; Song, Z.S. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013, 13, 68. [Google Scholar] [CrossRef] [Green Version]
- Xia, H.; Ooi, L.L.; Hui, K.M. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS ONE 2012, 7, e44206. [Google Scholar] [CrossRef]
- Zhong, D.; Xu, G.Z.; Wu, J.Z.; Liu, H.; Tang, J.Y.; Wang, C.G. Circ-ITCH sponges miR-214 to promote the osteogenic differentiation in osteoporosis via upregulating YAP1. Cell Death Dis. 2021, 12, 340. [Google Scholar] [CrossRef]
- Li, D.; Liu, J.; Guo, B.; Liang, C.; Dang, L.; Lu, C.; He, X.; Cheung, H.Y.; Xu, L.; Lu, C.; et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 2016, 7, 10872. [Google Scholar] [CrossRef]
- Wang, X.; Guo, B.; Li, Q.; Peng, J.; Yang, Z.; Wang, A.; Li, D.; Hou, Z.; Lv, K.; Kan, G.; et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013, 19, 93–100. [Google Scholar] [CrossRef]
- Allen-Rhoades, W.; Kurenbekova, L.; Satterfield, L.; Parikh, N.; Fuja, D.; Shuck, R.L.; Rainusso, N.; Trucco, M.; Barkauskas, D.A.; Jo, E.; et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015, 4, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Poos, K.; Smida, J.; Nathrath, M.; Maugg, D.; Baumhoer, D.; Korsching, E. How microRNA and transcription factor co-regulatory networks affect osteosarcoma cell proliferation. PLoS Comput. Biol. 2013, 9, e1003210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, D.; Dang, L.; Liang, C.; Guo, B.; Lu, C.; He, X.; Cheung, H.Y.; He, B.; Liu, B.; et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci. Rep. 2017, 7, 40487. [Google Scholar] [CrossRef] [PubMed]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Quwaider, D.; Krzeminski, P.; Ticona, F.V.; Paino, T.; Delgado, M.; Aires, A.; Ocio, E.M.; Garcia-Sanz, R.; et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 2013, 98, 640–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignali, R.; Marracci, S. HMGA Genes and Proteins in Development and Evolution. Int. J. Mol. Sci. 2020, 21, 654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerzoni, C.; Fiori, V.; Terracciano, M.; Manara, M.C.; Moricoli, D.; Pasello, M.; Sciandra, M.; Nicoletti, G.; Gellini, M.; Dominici, S.; et al. CD99 triggering in Ewing sarcoma delivers a lethal signal through p53 pathway reactivation and cooperates with doxorubicin. Clin. Cancer Res. 2015, 21, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanni, P.; Landuzzi, L.; Manara, M.C.; Righi, A.; Nicoletti, G.; Cristalli, C.; Pasello, M.; Parra, A.; Carrabotta, M.; Ferracin, M.; et al. Bone sarcoma patient-derived xenografts are faithful and stable preclinical models for molecular and therapeutic investigations. Sci. Rep. 2019, 9, 12174. [Google Scholar] [CrossRef]
- Amaral, A.T.; Manara, M.C.; Berghuis, D.; Ordonez, J.L.; Biscuola, M.; Lopez-Garcia, M.A.; Osuna, D.; Lucarelli, E.; Alviano, F.; Lankester, A.; et al. Characterization of human mesenchymal stem cells from ewing sarcoma patients. Pathogenetic implications. PLoS ONE 2014, 9, e85814. [Google Scholar] [CrossRef] [Green Version]
- Sciandra, M.; Marino, M.T.; Manara, M.C.; Guerzoni, C.; Grano, M.; Oranger, A.; Lucarelli, E.; Lollini, P.L.; Dozza, B.; Pratelli, L.; et al. CD99 drives terminal differentiation of osteosarcoma cells by acting as a spatial regulator of ERK 1/2. J. Bone Miner. Res. 2014, 29, 1295–1309. [Google Scholar] [CrossRef]
- Rocchi, A.; Manara, M.C.; Sciandra, M.; Zambelli, D.; Nardi, F.; Nicoletti, G.; Garofalo, C.; Meschini, S.; Astolfi, A.; Colombo, M.P.; et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J. Clin. Investig. 2010, 120, 668–680. [Google Scholar] [CrossRef]
- Balestra, T.; Manara, M.C.; Laginestra, M.A.; Pasello, M.; De Feo, A.; Bassi, C.; Guerzoni, C.; Landuzzi, L.; Lollini, P.L.; Donati, D.M.; et al. Targeting CD99 Compromises the Oncogenic Effects of the Chimera EWS-FLI1 by Inducing Reexpression of Zyxin and Inhibition of GLI1 Activity. Mol. Cancer Ther. 2021, 21, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.; Garcia-Aragoncillo, E.; Azorin, D.; Agra, N.; Sastre, A.; Gonzalez-Mediero, I.; Garcia-Miguel, P.; Pestana, A.; Gallego, S.; Segura, D.; et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin. Cancer Res. 2007, 13, 2429–2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Tirode, F.; Laud-Duval, K.; Prieur, A.; Delorme, B.; Charbord, P.; Delattre, O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007, 11, 421–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surdez, D.; Landuzzi, L.; Scotlandi, K.; Manara, M.C. Ewing Sarcoma PDX Models. Methods Mol. Biol. 2021, 2226, 223–242. [Google Scholar] [CrossRef]
- Manara, M.C.; Terracciano, M.; Mancarella, C.; Sciandra, M.; Guerzoni, C.; Pasello, M.; Grilli, A.; Zini, N.; Picci, P.; Colombo, M.P.; et al. CD99 triggering induces methuosis of Ewing sarcoma cells through IGF-1R/RAS/Rac1 signaling. Oncotarget 2016, 7, 79925–79942. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br. J. Cancer 2016, 115, 741–751. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhao, X.; Guo, Z.; Ma, X.; Song, Y.; Guo, Y. Regulation of NEAT1/miR-214-3p on the growth, migration and invasion of endometrial carcinoma cells. Arch. Gynecol. Obstet. 2017, 295, 1469–1475. [Google Scholar] [CrossRef]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef] [Green Version]
- Richter, G.H.; Plehm, S.; Fasan, A.; Rossler, S.; Unland, R.; Bennani-Baiti, I.M.; Hotfilder, M.; Lowel, D.; von Luettichau, I.; Mossbrugger, I.; et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 5324–5329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.J.; Ye, H.; Zeng, C.W.; He, B.; Zhang, H.; Chen, Y.Q. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J. Hematol. Oncol. 2010, 3, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Cai, H.; Lin, L.; Tang, M.; Cai, H. Upregulated expression of microRNA-214 is linked to tumor progression and adverse prognosis in pediatric osteosarcoma. Pediatr. Blood Cancer 2014, 61, 206–210. [Google Scholar] [CrossRef]
- Xu, C.; He, T.; Li, Z.; Liu, H.; Ding, B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed. Pharmacother. 2017, 95, 1504–1513. [Google Scholar] [CrossRef]
- Gutierrez, N.C.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Delgado, M.; De Las Rivas, J.; Ticona, F.V.; Ferminan, E.; Martin-Jimenez, P.; Chillon, C.; Risueno, A.; et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010, 24, 629–637. [Google Scholar] [CrossRef]
- Elango, R.; Alsaleh, K.A.; Vishnubalaji, R.; Manikandan, M.; Ali, A.M.; Abd El-Aziz, N.; Altheyab, A.; Al-Rikabi, A.; Alfayez, M.; Aldahmash, A.; et al. MicroRNA Expression Profiling on Paired Primary and Lymph Node Metastatic Breast Cancer Revealed Distinct microRNA Profile Associated With LNM. Front. Oncol. 2020, 10, 756. [Google Scholar] [CrossRef]
- Sumter, T.F.; Xian, L.; Huso, T.; Koo, M.; Chang, Y.T.; Almasri, T.N.; Chia, L.; Inglis, C.; Reid, D.; Resar, L.M. The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr. Mol. Med. 2016, 16, 353–393. [Google Scholar] [CrossRef] [Green Version]
- Cleynen, I.; Van de Ven, W.J. The HMGA proteins: A myriad of functions (Review). Int. J. Oncol. 2008, 32, 289–305. [Google Scholar] [CrossRef] [Green Version]
- Fusco, A.; Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7, 899–910. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, L.; Zheng, Y.; Guo, L. HMGA1 in cancer: Cancer classification by location. J. Cell Mol. Med. 2019, 23, 2293–2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resar, L.M. The high mobility group A1 gene: Transforming inflammatory signals into cancer? Cancer Res. 2010, 70, 436–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pegoraro, S.; Ros, G.; Piazza, S.; Sommaggio, R.; Ciani, Y.; Rosato, A.; Sgarra, R.; Del Sal, G.; Manfioletti, G. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget 2013, 4, 1293–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgarra, R.; Pegoraro, S.; Ros, G.; Penzo, C.; Chiefari, E.; Foti, D.; Brunetti, A.; Manfioletti, G. High Mobility Group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 216–229. [Google Scholar] [CrossRef]
- De Martino, M.; Fusco, A.; Esposito, F. HMGA and Cancer: A Review on Patent Literatures. Recent Pat. Anticancer Drug Discov. 2019, 14, 258–267. [Google Scholar] [CrossRef]
- Colamaio, M.; Tosti, N.; Puca, F.; Mari, A.; Gattordo, R.; Kuzay, Y.; Federico, A.; Pepe, A.; Sarnataro, D.; Ragozzino, E.; et al. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin. Ther. Targets 2016, 20, 1169–1179. [Google Scholar] [CrossRef]
- Kim, D.K.; Seo, E.J.; Choi, E.J.; Lee, S.I.; Kwon, Y.W.; Jang, I.H.; Kim, S.C.; Kim, K.H.; Suh, D.S.; Seong-Jang, K.; et al. Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Exp. Mol. Med. 2016, 48, e255. [Google Scholar] [CrossRef] [Green Version]
- Loria, R.; Laquintana, V.; Bon, G.; Trisciuoglio, D.; Frapolli, R.; Covello, R.; Amoreo, C.A.; Ferraresi, V.; Zoccali, C.; Novello, M.; et al. HMGA1/E2F1 axis and NFkB pathways regulate LPS progression and trabectedin resistance. Oncogene 2018, 37, 5926–5938. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Feo, A.; Pazzaglia, L.; Ciuffarin, L.; Mangiagli, F.; Pasello, M.; Simonetti, E.; Pellegrini, E.; Ferrari, C.; Bianchi, G.; Spazzoli, B.; et al. miR-214-3p Is Commonly Downregulated by EWS-FLI1 and by CD99 and Its Restoration Limits Ewing Sarcoma Aggressiveness. Cancers 2022, 14, 1762. https://doi.org/10.3390/cancers14071762
De Feo A, Pazzaglia L, Ciuffarin L, Mangiagli F, Pasello M, Simonetti E, Pellegrini E, Ferrari C, Bianchi G, Spazzoli B, et al. miR-214-3p Is Commonly Downregulated by EWS-FLI1 and by CD99 and Its Restoration Limits Ewing Sarcoma Aggressiveness. Cancers. 2022; 14(7):1762. https://doi.org/10.3390/cancers14071762
Chicago/Turabian StyleDe Feo, Alessandra, Laura Pazzaglia, Lisa Ciuffarin, Fabio Mangiagli, Michela Pasello, Elisa Simonetti, Evelin Pellegrini, Cristina Ferrari, Giuseppe Bianchi, Benedetta Spazzoli, and et al. 2022. "miR-214-3p Is Commonly Downregulated by EWS-FLI1 and by CD99 and Its Restoration Limits Ewing Sarcoma Aggressiveness" Cancers 14, no. 7: 1762. https://doi.org/10.3390/cancers14071762
APA StyleDe Feo, A., Pazzaglia, L., Ciuffarin, L., Mangiagli, F., Pasello, M., Simonetti, E., Pellegrini, E., Ferrari, C., Bianchi, G., Spazzoli, B., & Scotlandi, K. (2022). miR-214-3p Is Commonly Downregulated by EWS-FLI1 and by CD99 and Its Restoration Limits Ewing Sarcoma Aggressiveness. Cancers, 14(7), 1762. https://doi.org/10.3390/cancers14071762