MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Tissues
2.2. MALDI Mass Spectrometry Imaging
2.3. Bioinformatics and Statistical Analysis
3. Results
3.1. Sample Description and MALDI-MS Imaging Experiments
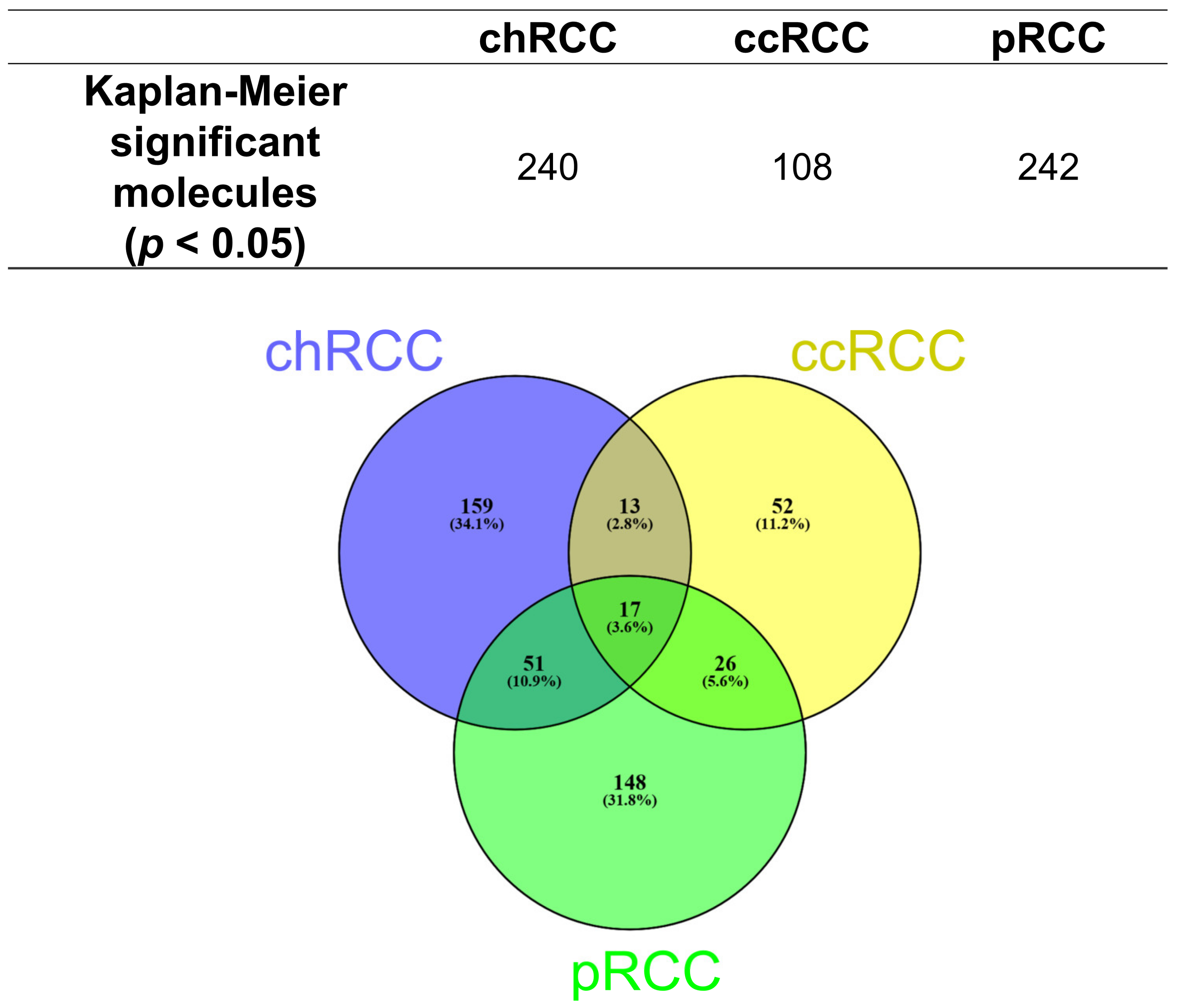
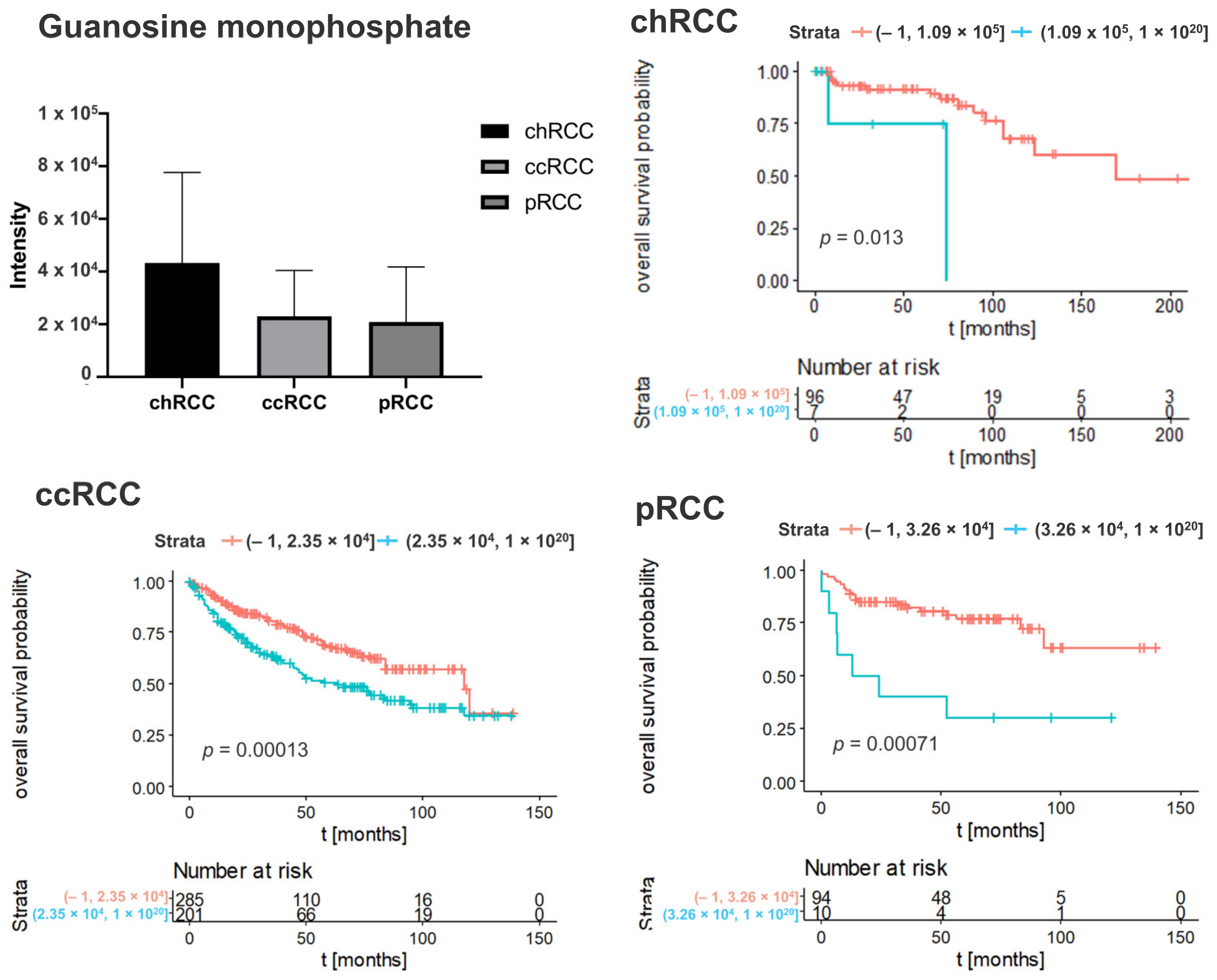
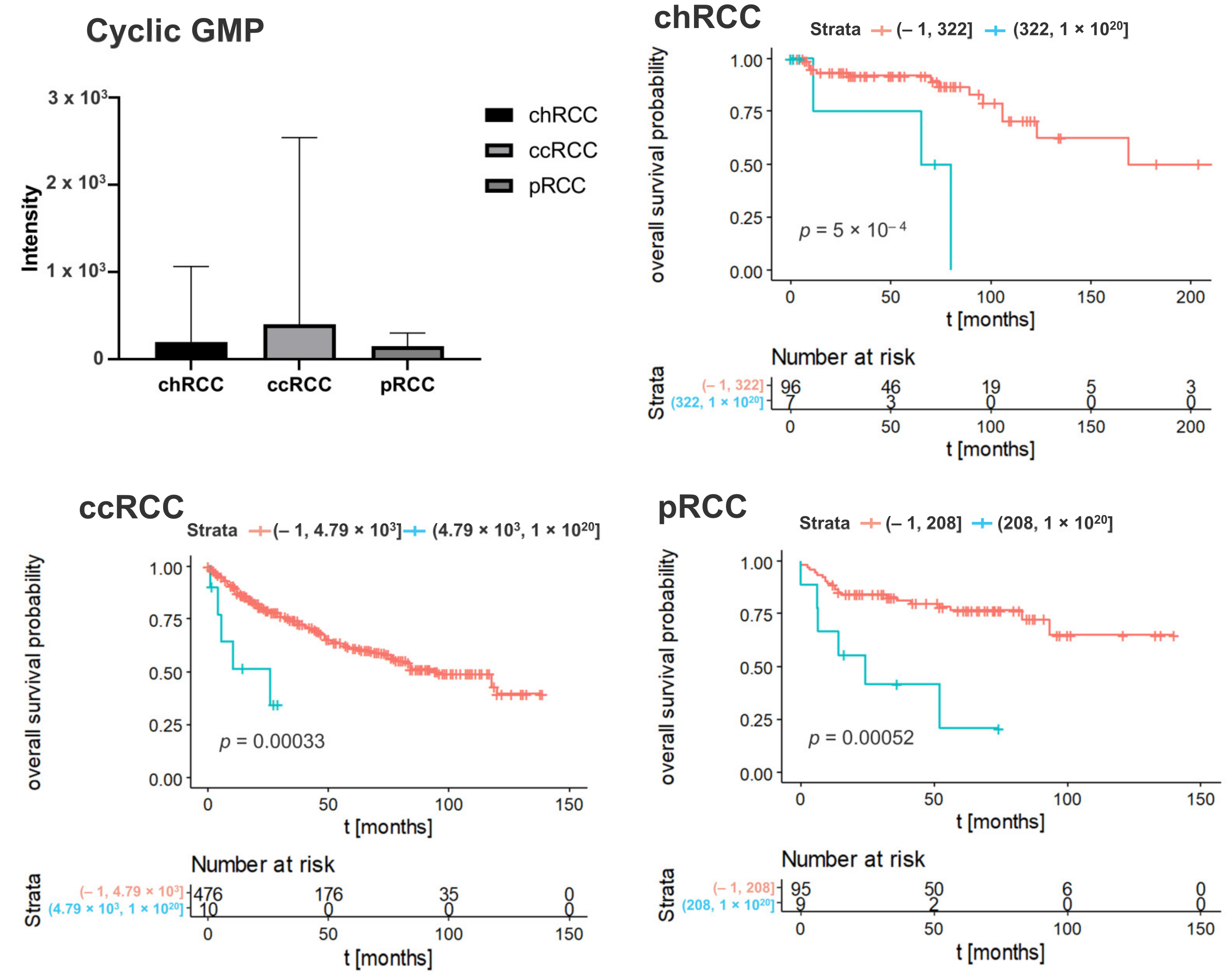
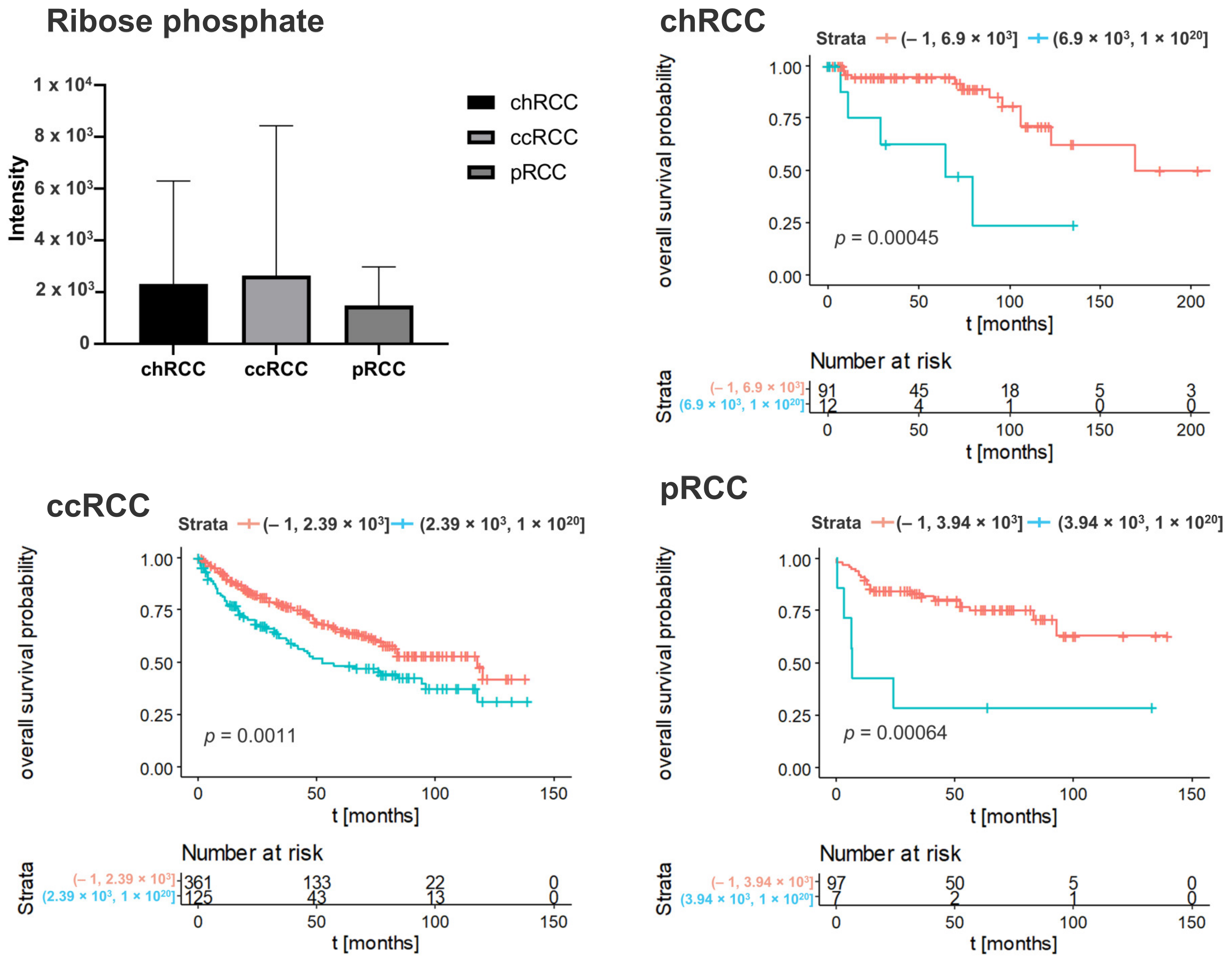
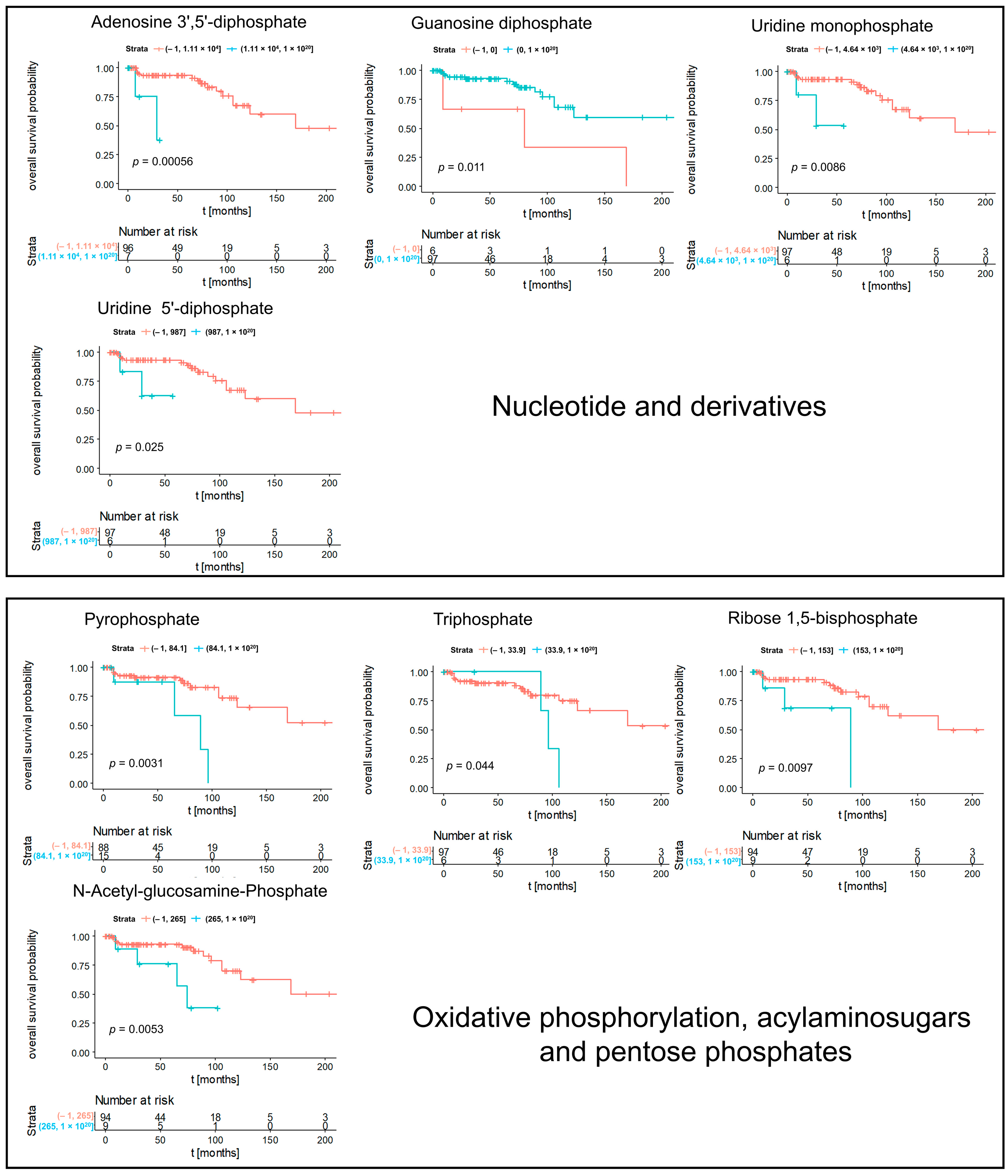
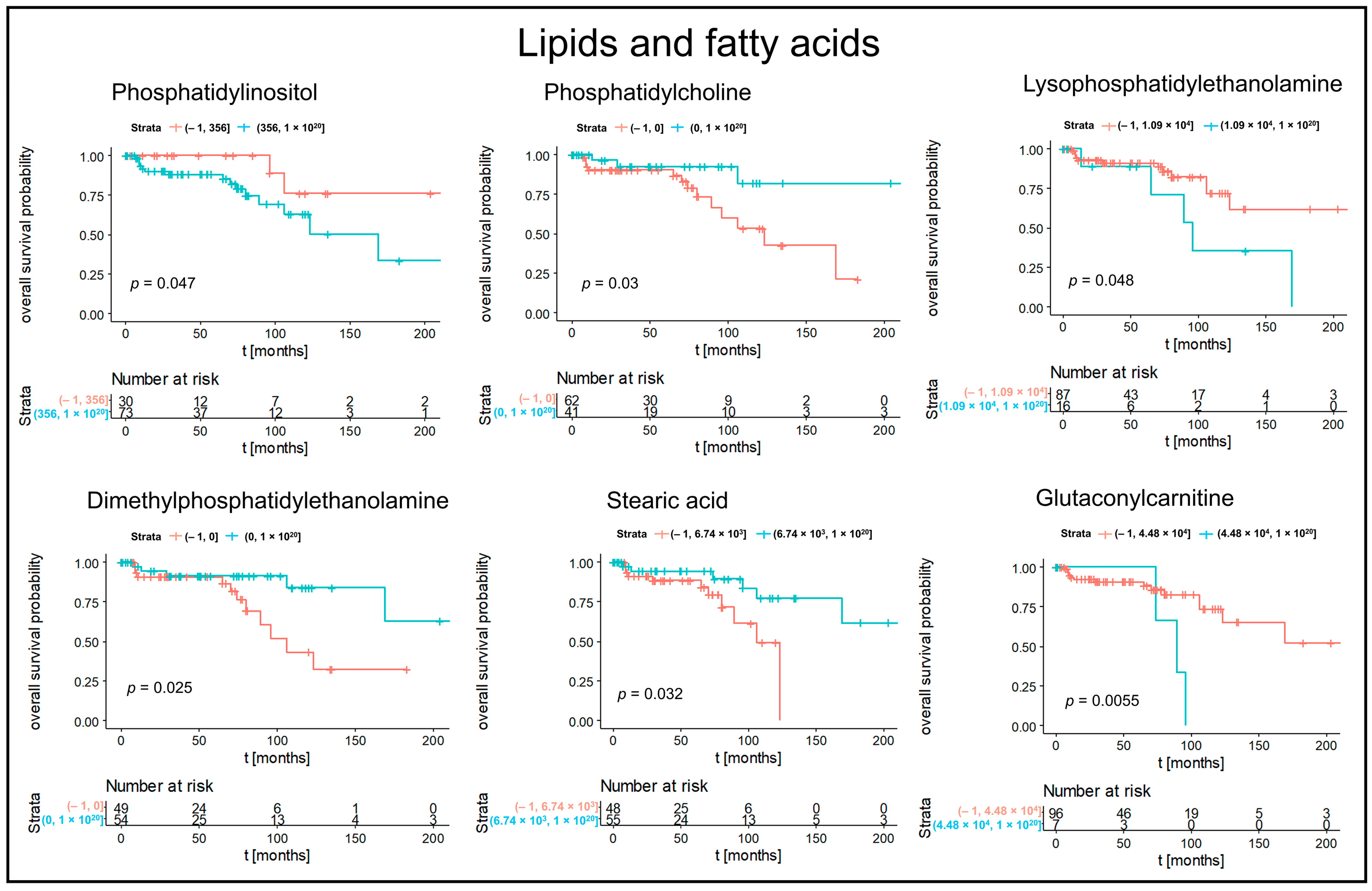
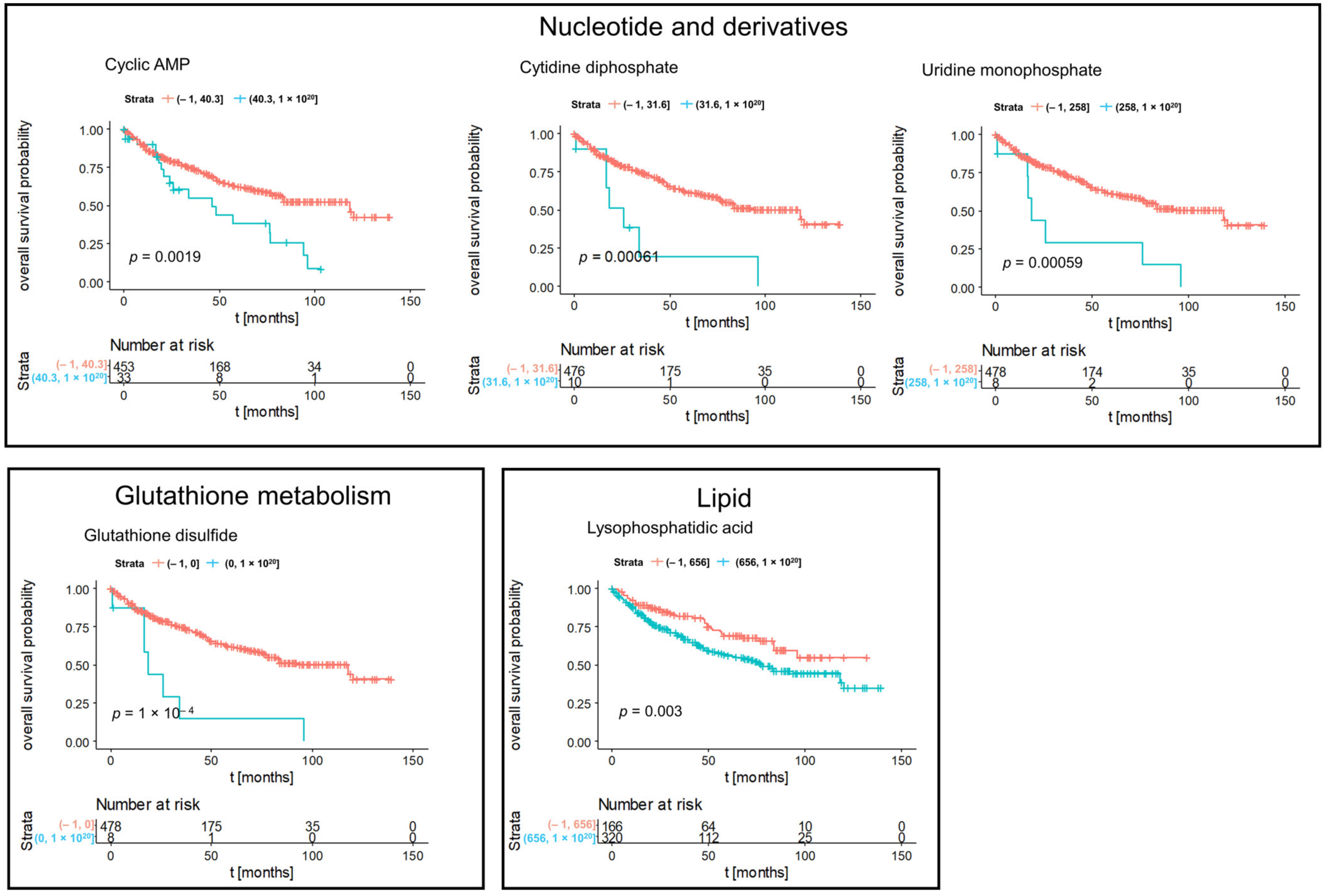
3.2. Kaplan–Meier Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMP | adenosin monophosphate |
| ATP | adenosintriphosphat |
| CLD7 | Claudin 7 |
| CSS | cancer-specific survival |
| ccRCC | clear cell renal cell carcinoma |
| chRCC | chromophobe renal cell carcinoma |
| cGAMP | cyclic guanosine monophosphate-adenosin monophosphate |
| cGMP | cyclic Guanosinmonophosphate |
| cGAS | cyclic guanosine monophosphate-adenosin monophosphate synthase |
| CTLA-4 | cytotoxic T-lymphocyte-associated protein 4 |
| FFPE | formalin-fixed paraffin-embedded |
| FT-ICR | Fourier-transform ion cyclotron resonance |
| GMP | guanosin monophosphate |
| GTP | guanosintriphosphate |
| IFN | interferon |
| IkB or IKK | IkappaB kinase |
| IRF3 | interferon regulatory factor 3 |
| ISUP | International Society of Urological Pathology |
| KM | Kaplan–Meier |
| MALDI | High mass resolution matrix-assisted laser desorption/ionization |
| MET | mesenchymal–epithelial transition factor |
| MSI | mass spectrometry imaging |
| NF-κB | nuclear factor kappa B |
| OS | overall survival |
| PD-1 | programmed death 1 |
| PD-L1 | programmed Death-Ligand 1 |
| pRCC | papillary renal cell carcinoma |
| PFS | progression free survival |
| RMC | renal medullary carcinoma |
| rO | renal oncocytoma |
| STING | stimulator of interferon genes |
| TBK1 | Tank-binding kinase 1 |
| TNFα | tumor necrosis factor α |
| TMA | tissue microarray |
| WHO | World Health Organization |
References
- Ljungberg, B.; Campbell, S.C.; Choi, H.Y.; Jacqmin, D.; Lee, J.E.; Weikert, S.; Kiemeney, L.A. The epidemiology of renal cell carcinoma. Eur. Urol. 2011, 60, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernandez-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Paner, G.P.; Alvarado-Cabrero, I.; Young, A.N.; Stricker, H.J.; Lyles, R.H.; Moch, H. Chromophobe renal cell carcinoma: Histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 145 cases. Am. J. Surg. Pathol. 2008, 32, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Ged, Y.; Chen, Y.B.; Knezevic, A.; Casuscelli, J.; Redzematovic, A.; DiNatale, R.G.; Carlo, M.I.; Lee, C.H.; Feldman, D.R.; Patil, S.; et al. Metastatic Chromophobe Renal Cell Carcinoma: Presence or Absence of Sarcomatoid Differentiation Determines Clinical Course and Treatment Outcomes. Clin. Genitourin. Cancer 2019, 17, e678–e688. [Google Scholar] [CrossRef] [PubMed]
- Paner, G.P.; Amin, M.B.; Alvarado-Cabrero, I.; Young, A.N.; Stricker, H.J.; Moch, H.; Lyles, R.H. A novel tumor grading scheme for chromophobe renal cell carcinoma: Prognostic utility and comparison with Fuhrman nuclear grade. Am. J. Surg. Pathol. 2010, 34, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Martignoni, G.; Hartmann, A.; Calio, A.; Segala, D.; Stohr, C.; Wach, S.; Erlmeier, F.; Weichert, W.; Autenrieth, M.; et al. Multi-institutional re-evaluation of prognostic factors in chromophobe renal cell carcinoma: Proposal of a novel two-tiered grading scheme. Virchows Arch. 2020, 476, 409–418. [Google Scholar] [CrossRef]
- Cheville, J.C.; Lohse, C.M.; Sukov, W.R.; Thompson, R.H.; Leibovich, B.C. Chromophobe renal cell carcinoma: The impact of tumor grade on outcome. Am. J. Surg. Pathol. 2012, 36, 851–856. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, Y.; Cui, J.; Wang, Y.; Xia, Y.; Song, J.; Cheng, S.; Zhou, C.; Zhang, D.; Zhang, B.; et al. The role of c-Met in prognosis and clinicopathology of renal cell carcinoma: Results from a single-centre study and systematic review. Urol. Oncol. 2017, 35, 532.e15–532.e23. [Google Scholar] [CrossRef]
- Erlmeier, F.; Weichert, W.; Autenrieth, M.; Ivanyi, P.; Hartmann, A.; Steffens, S. c-MET Oncogene in Renal Cell Carcinomas. Aktuelle Urol. 2016, 47, 475–479. [Google Scholar] [CrossRef]
- Carlsson, J.; Sundqvist, P.; Kosuta, V.; Falt, A.; Giunchi, F.; Fiorentino, M.; Davidsson, S. PD-L1 Expression is Associated with Poor Prognosis in Renal Cell Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 213–220. [Google Scholar] [CrossRef]
- Lavacchi, D.; Pellegrini, E.; Palmieri, V.E.; Doni, L.; Mela, M.M.; Di Maida, F.; Amedei, A.; Pillozzi, S.; Carini, M.; Antonuzzo, L. Immune Checkpoint Inhibitors in the Treatment of Renal Cancer: Current State and Future Perspective. Int. J. Mol. Sci. 2020, 21, 4691. [Google Scholar] [CrossRef]
- Shin, H.I.; Kim, B.H.; Chang, H.S.; Kim, C.I.; Jung, H.R.; Park, C.H. Expression of claudin-1 and -7 in clear cell renal cell carcinoma and its clinical significance. Korean J. Urol. 2011, 52, 317–322. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, S.Q.; Lin, J.Q.; Yu, W.; Eberlin, L.S. Mass Spectrometry Imaging Enables Discrimination of Renal Oncocytoma from Renal Cell Cancer Subtypes and Normal Kidney Tissues. Cancer Res. 2020, 80, 689–698. [Google Scholar] [CrossRef]
- Na, C.H.; Hong, J.H.; Kim, W.S.; Shanta, S.R.; Bang, J.Y.; Park, D.; Kim, H.K.; Kim, K.P. Identification of Protein Markers Specific for Papillary Renal Cell Carcinoma Using Imaging Mass Spectrometry. Mol. Cells 2015, 38, 624–629. [Google Scholar] [CrossRef] [Green Version]
- Kriegsmann, M.; Casadonte, R.; Maurer, N.; Stoehr, C.; Erlmeier, F.; Moch, H.; Junker, K.; Zgorzelski, C.; Weichert, W.; Schwamborn, K.; et al. Mass Spectrometry Imaging Differentiates Chromophobe Renal Cell Carcinoma and Renal Oncocytoma with High Accuracy. J. Cancer 2020, 11, 6081–6089. [Google Scholar] [CrossRef]
- Jorge, S.; Pereira, K.; Lopez-Fernandez, H.; LaFramboise, W.; Dhir, R.; Fernandez-Lodeiro, J.; Lodeiro, C.; Santos, H.M.; Capelo-Martinez, J.L. Ultrasonic-assisted extraction and digestion of proteins from solid biopsies followed by peptide sequential extraction hyphenated to MALDI-based profiling holds the promise of distinguishing renal oncocytoma from chromophobe renal cell carcinoma. Talanta 2020, 206, 120180. [Google Scholar] [CrossRef]
- Steurer, S.; Seddiqi, A.S.; Singer, J.M.; Bahar, A.S.; Eichelberg, C.; Rink, M.; Dahlem, R.; Huland, H.; Sauter, G.; Simon, R.; et al. MALDI imaging on tissue microarrays identifies molecular features associated with renal cell cancer phenotype. Anticancer Res. 2014, 34, 2255–2261. [Google Scholar]
- Stella, M.; Chinello, C.; Cazzaniga, A.; Smith, A.; Galli, M.; Piga, I.; Grasso, A.; Grasso, M.; Del Puppo, M.; Varallo, M.; et al. Histology-guided proteomic analysis to investigate the molecular profiles of clear cell Renal Cell Carcinoma grades. J. Proteom. 2019, 191, 38–47. [Google Scholar] [CrossRef]
- Erlmeier, F.; Hartmann, A.; Autenrieth, M.; Wiedemann, M.; Ivanyi, P.; Steffens, S.; Weichert, W. PD-1/PD-L1 expression in chromophobe renal cell carcinoma: An immunological exception? Med. Oncol. 2016, 33, 120. [Google Scholar] [CrossRef]
- Ly, A.; Buck, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.P.; Kuppen, P.J.; van de Velde, C.J.; Weirich, G.; et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat. Protoc. 2016, 11, 1428–1443. [Google Scholar] [CrossRef]
- Prade, V.M.; Kunzke, T.; Feuchtinger, A.; Rohm, M.; Luber, B.; Lordick, F.; Buck, A.; Walch, A. De novo discovery of metabolic heterogeneity with immunophenotype-guided imaging mass spectrometry. Mol. Metab. 2020, 36, 100953. [Google Scholar] [CrossRef]
- Kunzke, T.; Prade, V.M.; Buck, A.; Sun, N.; Feuchtinger, A.; Matzka, M.; Fernandez, I.E.; Wuyts, W.; Ackermann, M.; Jonigk, D.; et al. Patterns of Carbon-Bound Exogenous Compounds in Patients with Lung Cancer and Association with Disease Pathophysiology. Cancer Res. 2021, 81, 5862–5875. [Google Scholar] [CrossRef]
- Kunzke, T.; Balluff, B.; Feuchtinger, A.; Buck, A.; Langer, R.; Luber, B.; Lordick, F.; Zitzelsberger, H.; Aichler, M.; Walch, A. Native glycan fragments detected by MALDI-FT-ICR mass spectrometry imaging impact gastric cancer biology and patient outcome. Oncotarget 2017, 8, 68012–68025. [Google Scholar] [CrossRef] [Green Version]
- Chinello, C.; Gianazza, E.; Zoppis, I.; Mainini, V.; Galbusera, C.; Picozzi, S.; Rocco, F.; Galasso, G.; Bosari, S.; Ferrero, S.; et al. Serum biomarkers of renal cell carcinoma assessed using a protein profiling approach based on ClinProt technique. Urology 2010, 75, 842–847. [Google Scholar] [CrossRef]
- Junker, H.; Venz, S.; Zimmermann, U.; Thiele, A.; Scharf, C.; Walther, R. Stage-related alterations in renal cell carcinoma--comprehensive quantitative analysis by 2D-DIGE and protein network analysis. PLoS ONE 2011, 6, e21867. [Google Scholar] [CrossRef] [Green Version]
- Prade, V.M.; Sun, N.; Shen, J.; Feuchtinger, A.; Kunzke, T.; Buck, A.; Schraml, P.; Moch, H.; Schwamborn, K.; Autenrieth, M.; et al. The synergism of spatial metabolomics and morphometry improves machine learning-based renal tumour subtype classification. Clin. Transl. Med. 2022, 12, e666. [Google Scholar] [CrossRef]
- Wobker, S.E.; Williamson, S.R. Modern Pathologic Diagnosis of Renal Oncocytoma. J. Kidney Cancer VHL 2017, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Shi, H.; Wu, J.; Zhang, X.; Sun, L.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 2013, 51, 226–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diner, E.J.; Burdette, D.L.; Wilson, S.C.; Monroe, K.M.; Kellenberger, C.A.; Hyodo, M.; Hayakawa, Y.; Hammond, M.C.; Vance, R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013, 3, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef] [Green Version]
- Demaria, O.; De Gassart, A.; Coso, S.; Gestermann, N.; Di Domizio, J.; Flatz, L.; Gaide, O.; Michielin, O.; Hwu, P.; Petrova, T.V.; et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 15408–15413. [Google Scholar] [CrossRef] [Green Version]
- Corrales, L.; Glickman, L.H.; McWhirter, S.M.; Kanne, D.B.; Sivick, K.E.; Katibah, G.E.; Woo, S.R.; Lemmens, E.; Banda, T.; Leong, J.J.; et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015, 11, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Lemos, H.; Mohamed, E.; Huang, L.; Ou, R.; Pacholczyk, G.; Arbab, A.S.; Munn, D.; Mellor, A.L. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016, 76, 2076–2081. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.; Zheng, J.; Yao, X.; Weng, G.; Wu, L. Essential role of the cGMP/PKG signaling pathway in regulating the proliferation and survival of human renal carcinoma cells. Int. J. Mol. Med. 2014, 34, 1430–1438. [Google Scholar] [CrossRef] [Green Version]
- Msaouel, P.; Malouf, G.G.; Su, X.; Yao, H.; Tripathi, D.N.; Soeung, M.; Gao, J.; Rao, P.; Coarfa, C.; Creighton, C.J.; et al. Comprehensive Molecular Characterization Identifies Distinct Genomic and Immune Hallmarks of Renal Medullary Carcinoma. Cancer Cell 2020, 37, 720–734.e713. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Meierhofer, D. Glutathione Metabolism in Renal Cell Carcinoma Progression and Implications for Therapies. Int. J. Mol. Sci. 2019, 20, 3672. [Google Scholar] [CrossRef] [Green Version]
- Meister, A. On the discovery of glutathione. Trends Biochem. Sci. 1988, 13, 185–188. [Google Scholar] [CrossRef]
- Al Ahmad, A.; Paffrath, V.; Clima, R.; Busch, J.F.; Rabien, A.; Kilic, E.; Villegas, S.; Timmermann, B.; Attimonelli, M.; Jung, K.; et al. Papillary Renal Cell Carcinomas Rewire Glutathione Metabolism and Are Deficient in Both Anabolic Glucose Synthesis and Oxidative Phosphorylation. Cancers 2019, 11, 1298. [Google Scholar] [CrossRef] [Green Version]
- Priolo, C.; Khabibullin, D.; Reznik, E.; Filippakis, H.; Ogorek, B.; Kavanagh, T.R.; Nijmeh, J.; Herbert, Z.T.; Asara, J.M.; Kwiatkowski, D.J.; et al. Impairment of gamma-glutamyl transferase 1 activity in the metabolic pathogenesis of chromophobe renal cell carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, E6274–E6282. [Google Scholar] [CrossRef] [Green Version]
- Buck, A.; Ly, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.P.; Kuppen, P.J.; van de Velde, C.J.; Weirich, G.; et al. High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples. J. Pathol. 2015, 237, 123–132. [Google Scholar] [CrossRef] [Green Version]

| Patient Characteristics | n = 782 |
|---|---|
| Age median (range) (years) | 27–88 (64) |
| Gender | |
| Male | 269 (39.4%) |
| Female | 413 (60.6%) |
| ISUP Grade | |
| Grade 1 | 25 (4.1%) |
| Grade 2 | 241 (39.4%) |
| Grade 3 | 191 (31.3%) |
| Grade 4 | 154 (25.2%) |
| Pathological stage | |
| pT1 | 417 (53.6%) |
| pT2 | 90 (11.6%) |
| pT3 | 231 (29.7%) |
| pT4 | 10 (1.3%) |
| pN+ | 26 (15.0%) |
| pM+ | 4 (36.4%) |
| Subtype | |
| chRCC | 108 (13.8%) |
| ccRCC | 552 (70.6%) |
| ppRCC | 122 (15.6%) |
| Survival (Dead/Alive) | 214 (31.6%)/464 (68.4%) |
| Overall survival median (months) | 36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erlmeier, F.; Sun, N.; Shen, J.; Feuchtinger, A.; Buck, A.; Prade, V.M.; Kunzke, T.; Schraml, P.; Moch, H.; Autenrieth, M.; et al. MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas. Cancers 2022, 14, 1763. https://doi.org/10.3390/cancers14071763
Erlmeier F, Sun N, Shen J, Feuchtinger A, Buck A, Prade VM, Kunzke T, Schraml P, Moch H, Autenrieth M, et al. MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas. Cancers. 2022; 14(7):1763. https://doi.org/10.3390/cancers14071763
Chicago/Turabian StyleErlmeier, Franziska, Na Sun, Jian Shen, Annette Feuchtinger, Achim Buck, Verena M. Prade, Thomas Kunzke, Peter Schraml, Holger Moch, Michael Autenrieth, and et al. 2022. "MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas" Cancers 14, no. 7: 1763. https://doi.org/10.3390/cancers14071763
APA StyleErlmeier, F., Sun, N., Shen, J., Feuchtinger, A., Buck, A., Prade, V. M., Kunzke, T., Schraml, P., Moch, H., Autenrieth, M., Weichert, W., Hartmann, A., & Walch, A. (2022). MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas. Cancers, 14(7), 1763. https://doi.org/10.3390/cancers14071763








