PARP-1 Expression and BRCA1 Mutations in Breast Cancer Patients’ CTCs
Abstract
:Simple Summary
Abstract
1. Introduction
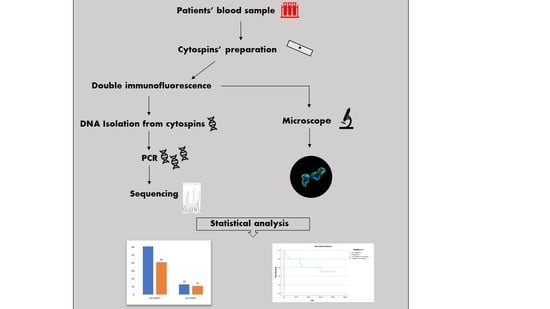
2. Materials and Methods
2.1. Patients’ Samples and Cytospins’ Preparation
2.2. Double Immunofluorescence
2.3. DNA Isolation from Cytospins
2.4. Amplification and Preparation of PCR Products
2.5. Genotyping—Sanger Sequencing
2.6. Statistical Analysis
3. Results
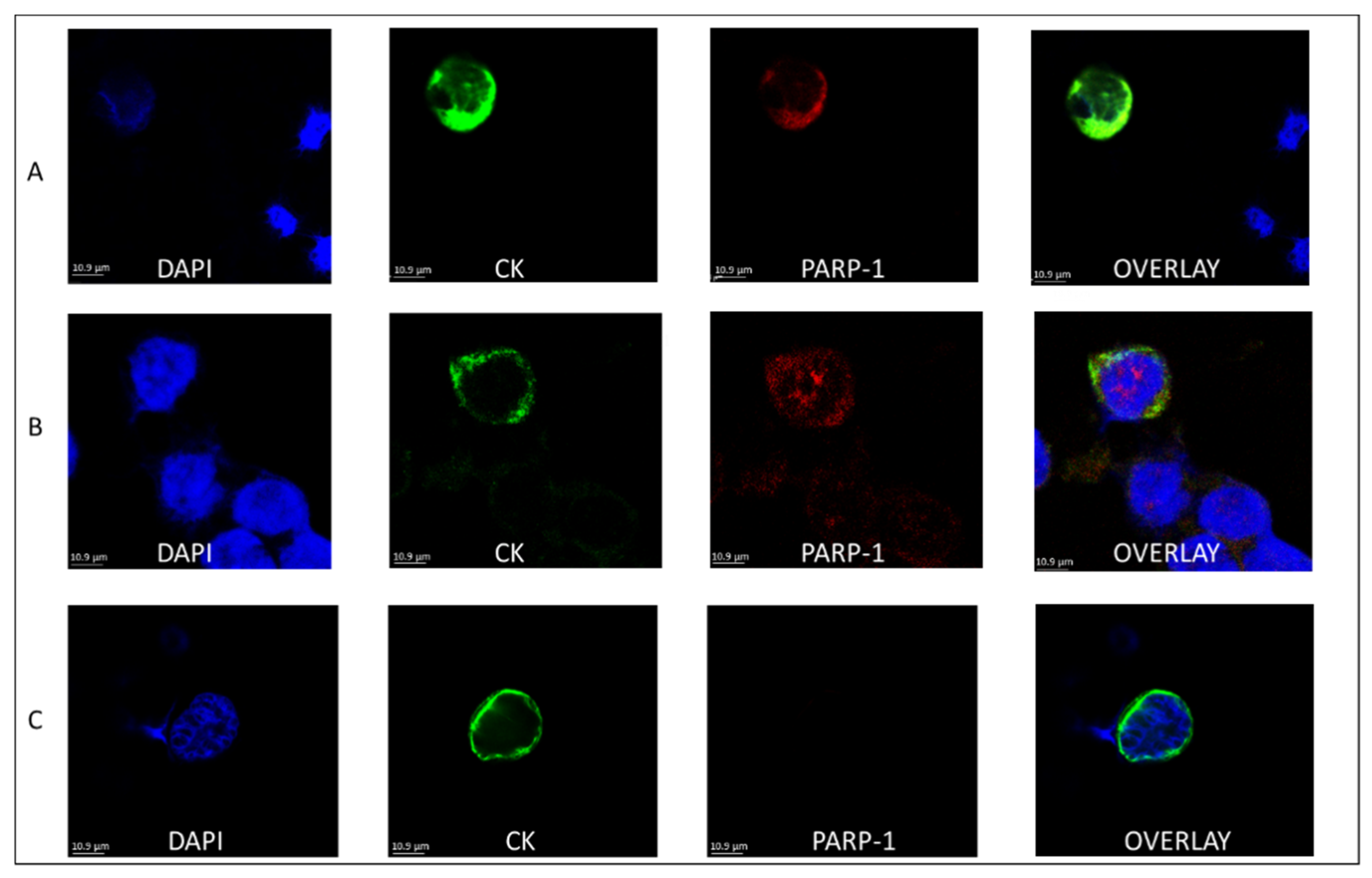
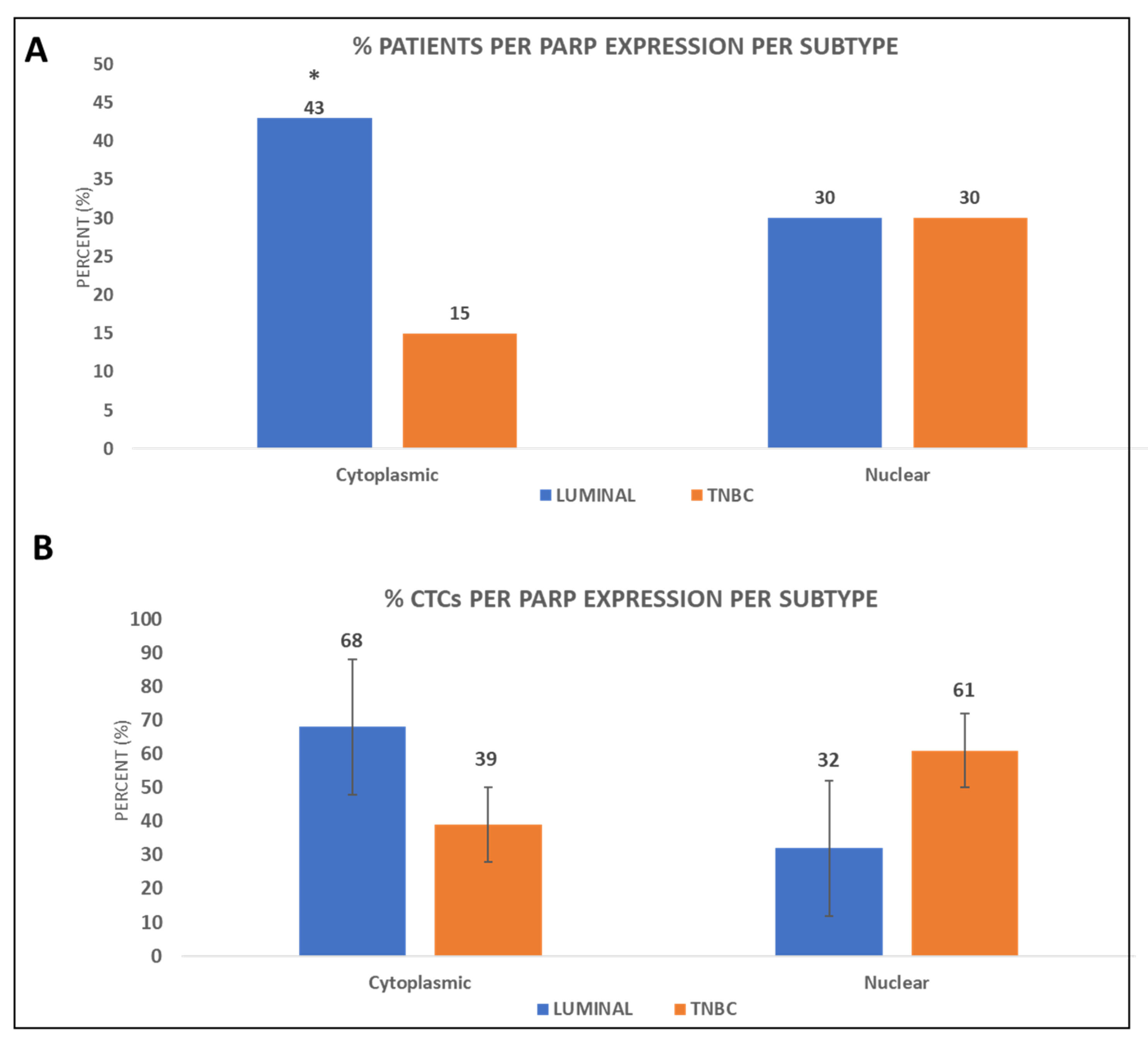
3.1. PARP-1 Expression in the CTCs of BC Patients
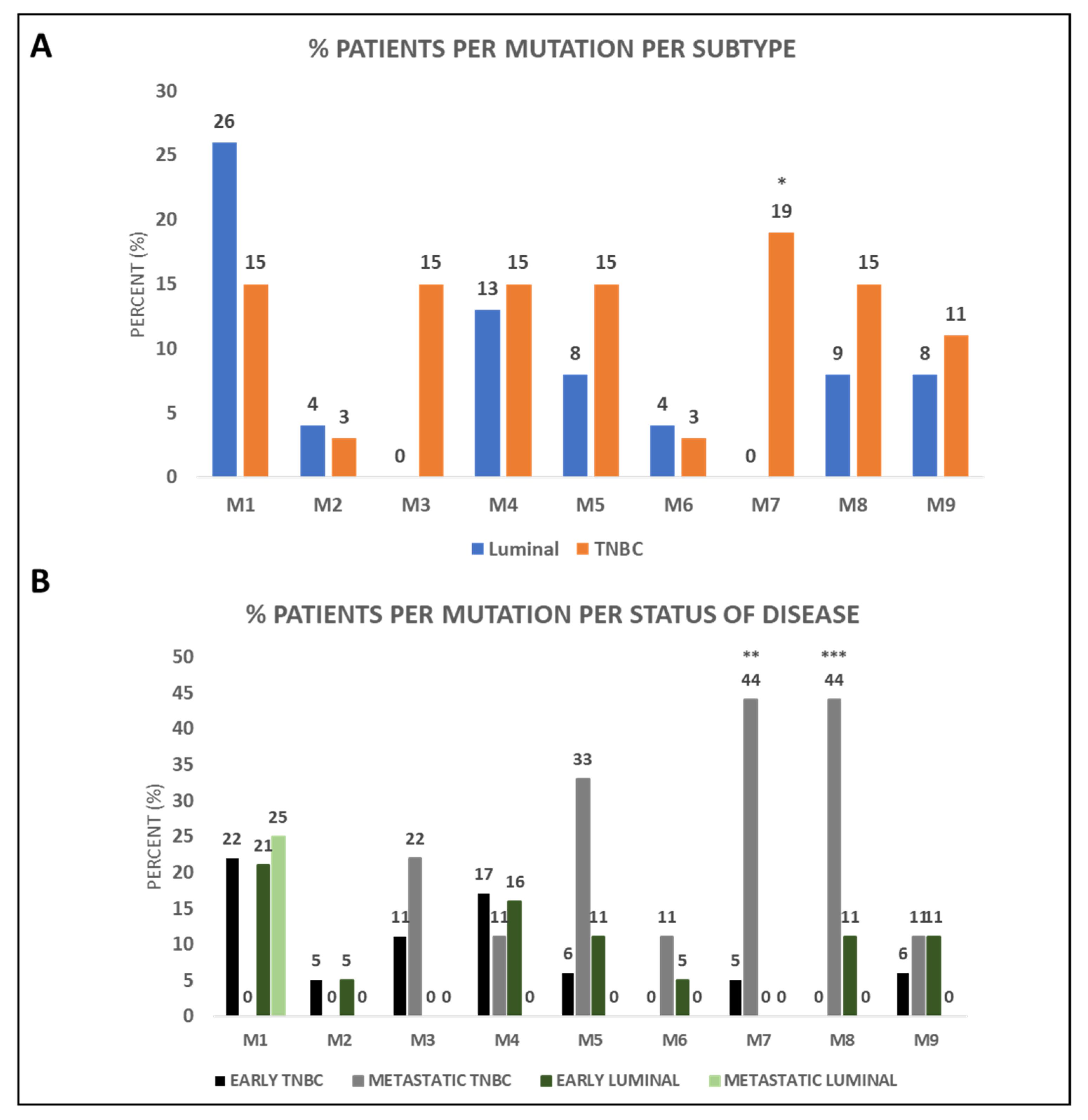
3.2. BRCA1 Mutations Identified in CTCs of BC Patients
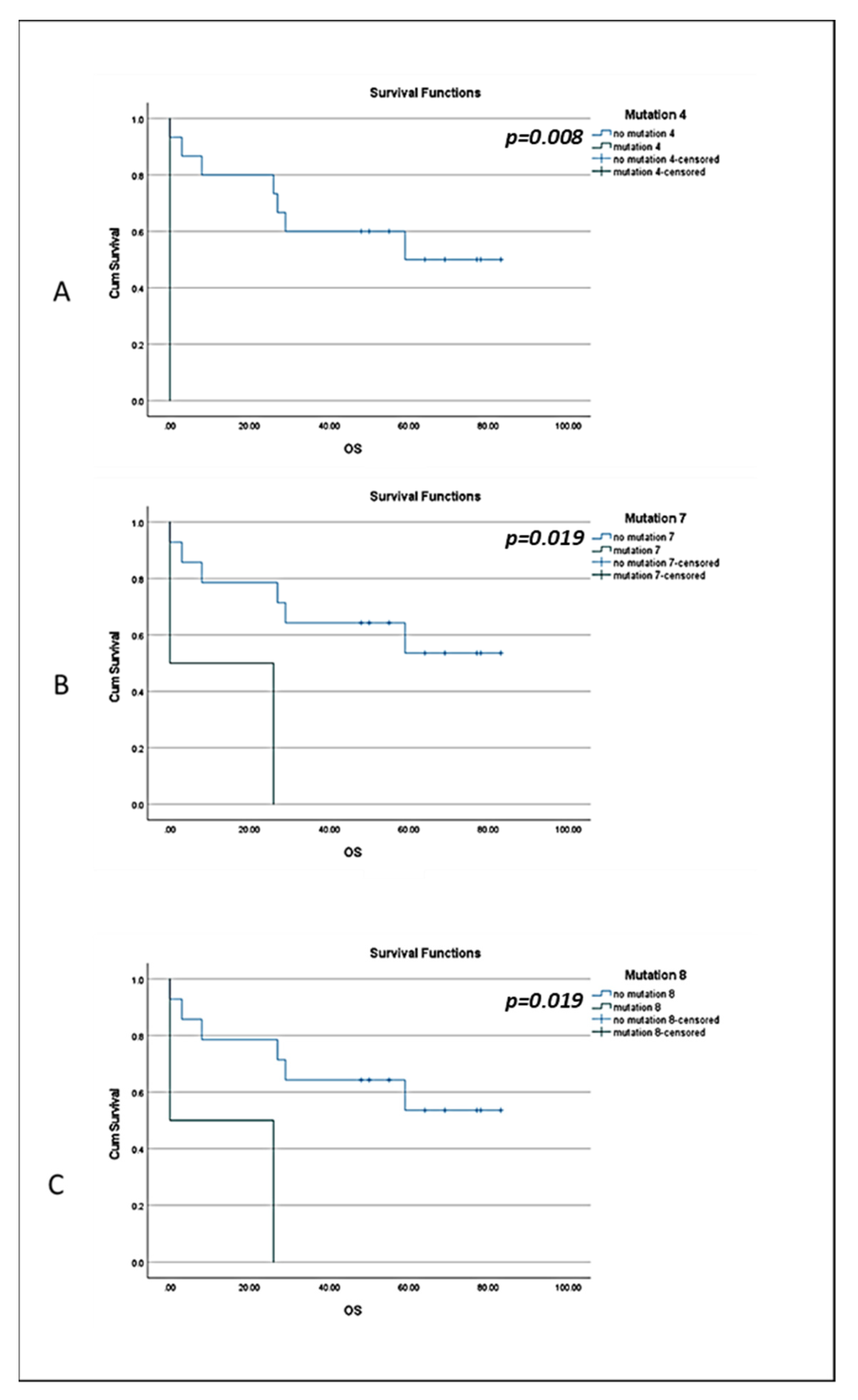
3.3. BRCA1 Mutations in CTCs of BC Patients and Clinical Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Sekine, M.; Nishino, K.; Enomoto, T. Differences in Ovarian and Other Cancers Risks by Population and BRCA Mutation Location. Genes 2021, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Godet, I.; Gilkes, D.M. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr. Cancer Sci. Ther. 2017, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, Y.; Pegram, M. Research advances and new challenges in overcoming triple-negative breast cancer. Cancer Drug Resist 2021, 4, 517–542. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, B. Untangling Triple-Negative Breast Cancer Molecular Peculiarity and Chemo-Resistance: Trailing Towards Marker-Based Targeted Therapies. Cureus 2021, 13, e16636. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, D.; Sendi, M.A.; Kelly, C.M. Overview of recent advances in metastatic triple negative breast cancer. World J. Clin. Oncol. 2021, 12, 164–182. [Google Scholar] [CrossRef] [PubMed]
- Sukumar, J.; Gast, K.; Quiroga, D.; Lustberg, M.; Williams, N. Triple-negative breast cancer: Promising prognostic biomarkers currently in development. Expert Rev. Anticancer Ther. 2021, 21, 135–148. [Google Scholar] [CrossRef]
- Krishnan, R.; Patel, P.S.; Hakem, R. BRCA1 and Metastasis: Outcome of Defective DNA Repair. Cancers 2021, 14, 108. [Google Scholar] [CrossRef]
- Santana Dos Santos, E.; Lallemand, F.; Burke, L.; Stoppa-Lyonnet, D.; Brown, M.; Caputo, S.M.; Rouleau, E. Non-Coding Variants in BRCA1 and BRCA2 Genes: Potential Impact on Breast and Ovarian Cancer Predisposition. Cancers 2018, 10, 453. [Google Scholar] [CrossRef] [Green Version]
- Xavier, M.A.; Rezende, F.; Titze-de-Almeida, R.; Cornelissen, B. BRCAness as a Biomarker of Susceptibility to PARP Inhibitors in Glioblastoma Multiforme. Biomolecules 2021, 11, 1188. [Google Scholar] [CrossRef]
- Layman, R.M.; Arun, B. PARP Inhibitors in Triple-Negative Breast Cancer Including Those with BRCA Mutations. Cancer J. 2021, 27, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.D.; Parveen, A.; Yadav, D.K. Role of PARP in TNBC: Mechanism of Inhibition, Clinical Applications, and Resistance. Biomedicines 2021, 9, 1512. [Google Scholar] [CrossRef] [PubMed]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Beek, L.; McClay, E.; Patel, S.; Schimpl, M.; Spagnolo, L.; Maia de Oliveira, T. PARP Power: A Structural Perspective on PARP1, PARP2, and PARP3 in DNA Damage Repair and Nucleosome Remodelling. Int. J. Mol. Sci. 2021, 22, 5112. [Google Scholar] [CrossRef]
- Kim, D.S.; Camacho, C.V.; Kraus, W.L. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp. Mol. Med. 2021, 53, 42–51. [Google Scholar] [CrossRef]
- Donizy, P.; Pietrzyk, G.; Halon, A.; Kozyra, C.; Gansukh, T.; Lage, H.; Surowiak, P.; Matkowski, R. Nuclear-cytoplasmic PARP-1 expression as an unfavorable prognostic marker in lymph nodenegative early breast cancer: 15-year follow-up. Oncol. Rep. 2014, 31, 1777–1787. [Google Scholar] [CrossRef] [Green Version]
- Von Minckwitz, G.; Muller, B.M.; Loibl, S.; Budczies, J.; Hanusch, C.; Darb-Esfahani, S.; Hilfrich, J.; Weiss, E.; Huober, J.; Blohmer, J.U.; et al. Cytoplasmic poly(adenosine diphosphate-ribose) polymerase expression is predictive and prognostic in patients with breast cancer treated with neoadjuvant chemotherapy. J. Clin. Oncol. 2011, 29, 2150–2157. [Google Scholar] [CrossRef]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013, 4, 2240. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, H.; Xu, Z.; Tang, H.; Geng, A.; Cai, B.; Su, T.; Shi, J.; Jiang, C.; Tian, X.; et al. A PARP1-BRG1-SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites. Nucleic Acids Res. 2019, 47, 8563–8580. [Google Scholar]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Palleschi, M.; Tedaldi, G.; Sirico, M.; Virga, A.; Ulivi, P.; De Giorgi, U. Moving beyond PARP Inhibition: Current State and Future Perspectives in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 7884. [Google Scholar] [CrossRef]
- Cortesi, L.; Rugo, H.S.; Jackisch, C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target. Oncol. 2021, 16, 255–282. [Google Scholar] [CrossRef]
- Azim, H.A.; Kassem, L.; Azim, H., Jr. Integrating PARP inhibitors into the management of breast cancer: Where are we? Chin. Clin. Oncol. 2021, 10, 50. [Google Scholar] [CrossRef]
- Xia, M.; Guo, Z.; Hu, Z. The Role of PARP Inhibitors in the Treatment of Prostate Cancer: Recent Advances in Clinical Trials. Biomolecules 2021, 11, 722. [Google Scholar] [CrossRef]
- Guney Eskiler, G.; Cecener, G.; Egeli, U.; Tunca, B. Triple negative breast cancer: New therapeutic approaches and BRCA status. APMIS 2018, 126, 371–379. [Google Scholar] [CrossRef]
- Sun, X.; Wang, X.; Zhang, J.; Zhao, Z.; Feng, X.; Liu, L.; Ma, Z. Efficacy and safety of PARP inhibitors in patients with BRCA-mutated advanced breast cancer: A meta-analysis and systematic review. Breast 2021, 60, 26–34. [Google Scholar] [CrossRef]
- Pop, L.; Suciu, I.; Ionescu, O.; Bacalbasa, N.; Ionescu, P. The role of novel poly (ADP-ribose) inhibitors in the treatment of locally advanced and metastatic Her-2/neu negative breast cancer with inherited germline BRCA1/2 mutations. A review of the literature. J. Med. Life 2021, 14, 17–20. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.E.; Tung, N.; Conte, P.; Im, S.A.; Senkus, E.; Xu, B.; Masuda, N.; Delaloge, S.; Li, W.; Armstrong, A.; et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann. Oncol. 2019, 30, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Goncalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Hurvitz, S.A.; Mina, L.A.; Rugo, H.S.; Lee, K.H.; Goncalves, A.; Diab, S.; Woodward, N.; Goodwin, A.; Yerushalmi, R.; et al. Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer: Final overall survival results from the EMBRACA trial. Ann. Oncol. 2020, 31, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Stathopoulou, A.; Vlachonikolis, I.; Mavroudis, D.; Perraki, M.; Kouroussis, C.; Apostolaki, S.; Malamos, N.; Kakolyris, S.; Kotsakis, A.; Xenidis, N.; et al. Molecular detection of cytokeratin-19-positive cells in the peripheral blood of patients with operable breast cancer: Evaluation of their prognostic significance. J. Clin. Oncol. 2002, 20, 3404–3412. [Google Scholar] [CrossRef]
- Mego, M.; Karaba, M.; Sedlackova, T.; Benca, J.; Repiska, G.; Krasnicanova, L.; Macuch, J.; Sieberova, G.; Jurisova, S.; Pindak, D.; et al. Circulating tumor cells and breast cancer-specific mutations in primary breast cancer. Mol. Clin. Oncol. 2020, 12, 565–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocco, S.; Piezzo, M.; Calabrese, A.; Cianniello, D.; Caputo, R.; Lauro, V.D.; Fusco, G.; Gioia, G.D.; Licenziato, M.; De Laurentiis, M. Biomarkers in Triple-Negative Breast Cancer: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2020, 21, 4579. [Google Scholar] [CrossRef] [PubMed]
- Chantzara, E.; Xenidis, N.; Kallergi, G.; Georgoulias, V.; Kotsakis, A. Circulating tumor cells as prognostic biomarkers in breast cancer: Current status and future prospects. Expert Rev. Mol. Diagn. 2021, 21, 1037–1048. [Google Scholar] [CrossRef]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef] [Green Version]
- Agelaki, S.; Dragolia, M.; Markonanolaki, H.; Alkahtani, S.; Stournaras, C.; Georgoulias, V.; Kallergi, G. Phenotypic characterization of circulating tumor cells in triple negative breast cancer patients. Oncotarget 2017, 8, 5309–5322. [Google Scholar] [CrossRef] [Green Version]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.; Gratama, J.W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef]
- Kallergi, G.; Agelaki, S.; Kalykaki, A.; Stournaras, C.; Mavroudis, D.; Georgoulias, V. Phosphorylated EGFR and PI3K/Akt signaling kinases are expressed in circulating tumor cells of breast cancer patients. Breast Cancer Res. 2008, 10, R80. [Google Scholar] [CrossRef] [Green Version]
- Kallergi, G.; Konstantinidis, G.; Markomanolaki, H.; Papadaki, M.A.; Mavroudis, D.; Stournaras, C.; Georgoulias, V.; Agelaki, S. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol. Cancer Ther. 2013, 12, 1886–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallergi, G.; Mavroudis, D.; Georgoulias, V.; Stournaras, C. Phosphorylation of FAK, PI-3K, and impaired actin organization in CK-positive micrometastatic breast cancer cells. Mol. Med. 2007, 13, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Chimonidou, M.; Kallergi, G.; Georgoulias, V.; Welch, D.R.; Lianidou, E.S. Breast cancer metastasis suppressor-1 promoter methylation in primary breast tumors and corresponding circulating tumor cells. Mol. Cancer Res. 2013, 11, 1248–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroupis, C.; Christopoulos, K.; Devetzoglou, M.; Tsiagas, I.; Lianidou, E.S. Asymmetric real-time PCR detection of BRCA1 5382insC mutation by melting curve analysis in the LightCycler. Clin. Chim. Acta 2008, 390, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.S.; Abreu, R.B.; Velkova, A.; Marsillac, S.; Rodarte, R.S.; Suarez-Kurtz, G.; Iversen, E.S.; Monteiro, A.N.; Carvalho, M.A. Probing structure-function relationships in missense variants in the carboxy-terminal region of BRCA1. PLoS ONE 2014, 9, e97766. [Google Scholar] [CrossRef]
- Chevalier, L.M.; Billaud, A.; Fronteau, S.; Dauve, J.; Patsouris, A.; Verriele, V.; Morel, A. Somatic mRNA Analysis of BRCA1 Splice Variants Provides a Direct Theranostic Impact on PARP Inhibitors. Mol. Diagn. Ther. 2020, 24, 233–243. [Google Scholar] [CrossRef]
- Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Rojo, F.; Garcia-Parra, J.; Zazo, S.; Tusquets, I.; Ferrer-Lozano, J.; Menendez, S.; Eroles, P.; Chamizo, C.; Servitja, S.; Ramirez-Merino, N.; et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann. Oncol. 2012, 23, 1156–1164. [Google Scholar] [CrossRef]
- Stanley, J.; Klepczyk, L.; Keene, K.; Wei, S.; Li, Y.; Forero, A.; Grizzle, W.; Wielgos, M.; Brazelton, J.; LoBuglio, A.F.; et al. PARP1 and phospho-p65 protein expression is increased in human HER2-positive breast cancers. Breast Cancer Res. Treat. 2015, 150, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Rajiah, I.R.; Skepper, J. Differential localisation of PARP-1 N-terminal fragment in PARP-1(+/+) and PARP-1(−/−) murine cells. Mol. Cells 2014, 37, 526–531. [Google Scholar] [CrossRef] [Green Version]
- Domagala, P.; Huzarski, T.; Lubinski, J.; Gugala, K.; Domagala, W. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: Possible implications for PARP-1 inhibitor therapy. Breast Cancer Res. Treat. 2011, 127, 861–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, N.; Yim, K.; Abdul-Ghafar, J.; Seo, K.J.; Chong, Y. High Poly(ADP-Ribose) Polymerase Expression Does Relate to Poor Survival in Solid Cancers: A Systematic Review and Meta-Analysis. Cancers 2021, 13, 5594. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [PubMed] [Green Version]
- Chen, H.; Wu, J.; Zhang, Z.; Tang, Y.; Li, X.; Liu, S.; Cao, S.; Li, X. Association Between BRCA Status and Triple-Negative Breast Cancer: A Meta-Analysis. Front. Pharmacol. 2018, 9, 909. [Google Scholar] [CrossRef] [PubMed]

| Number | Mutation | rs Code | Number of Patients | Exon/Intron | Biological Effect |
|---|---|---|---|---|---|
| M1 | G > A | c.5277 + 65C > T | 10 | Intron | Unknown 1 |
| M2 | A > G | c.5277 + 67T > C | 2 | Intron | Unknown 1 |
| M3 | A > T | rs2051507989 | 5 | Exon | Synonymous |
| M4 | A > T | rs80358069 | 7 | Intron | Pathogenic |
| M5 | G > C | rs1567764460 | 6 | Exon | Missense Variant |
| M6 | T > C | rs1057524456 | 2 | Intron | No Information |
| M7 | G > T | rs80358173 | 5 | Intron | Pathogenic |
| M8 | G > C | rs377595653 | 6 | Exon | Pathogenic |
| M9 | A > C | rs80357270 | 5 | Exon | No functional impact |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sklias, T.; Vardas, V.; Pantazaka, E.; Christopoulou, A.; Georgoulias, V.; Kotsakis, A.; Vasilopoulos, Y.; Kallergi, G. PARP-1 Expression and BRCA1 Mutations in Breast Cancer Patients’ CTCs. Cancers 2022, 14, 1731. https://doi.org/10.3390/cancers14071731
Sklias T, Vardas V, Pantazaka E, Christopoulou A, Georgoulias V, Kotsakis A, Vasilopoulos Y, Kallergi G. PARP-1 Expression and BRCA1 Mutations in Breast Cancer Patients’ CTCs. Cancers. 2022; 14(7):1731. https://doi.org/10.3390/cancers14071731
Chicago/Turabian StyleSklias, Thodoris, Vasileios Vardas, Evangelia Pantazaka, Athina Christopoulou, Vassilis Georgoulias, Athanasios Kotsakis, Yiannis Vasilopoulos, and Galatea Kallergi. 2022. "PARP-1 Expression and BRCA1 Mutations in Breast Cancer Patients’ CTCs" Cancers 14, no. 7: 1731. https://doi.org/10.3390/cancers14071731
APA StyleSklias, T., Vardas, V., Pantazaka, E., Christopoulou, A., Georgoulias, V., Kotsakis, A., Vasilopoulos, Y., & Kallergi, G. (2022). PARP-1 Expression and BRCA1 Mutations in Breast Cancer Patients’ CTCs. Cancers, 14(7), 1731. https://doi.org/10.3390/cancers14071731