A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker
Abstract
:Simple Summary
Abstract
1. Introduction
2. Related Works
2.1. Selection of Antibodies
2.2. Deep Learning in Application to Gynecologic Oncology
2.3. Weakly Supervised Learning
3. Materials and Methods
3.1. Study Population and Experimental Setup
3.2. Weakly Supervised Learning Framework
Boosting Learning, Focusing Sampling, Boosted Data Augmentation and Transfer Learning
3.3. A Hybrid Deep Learning Model
3.3.1. Tissue Core Detection Model
3.3.2. Weakly Supervised Tumorlike Tissue Selection Model
3.3.3. Treatment Effectiveness Prediction Model
3.3.4. Model Selection Method with Early Stop Mechanism
4. Results
4.1. Quantitative Evaluation
4.2. Statistical Analysis
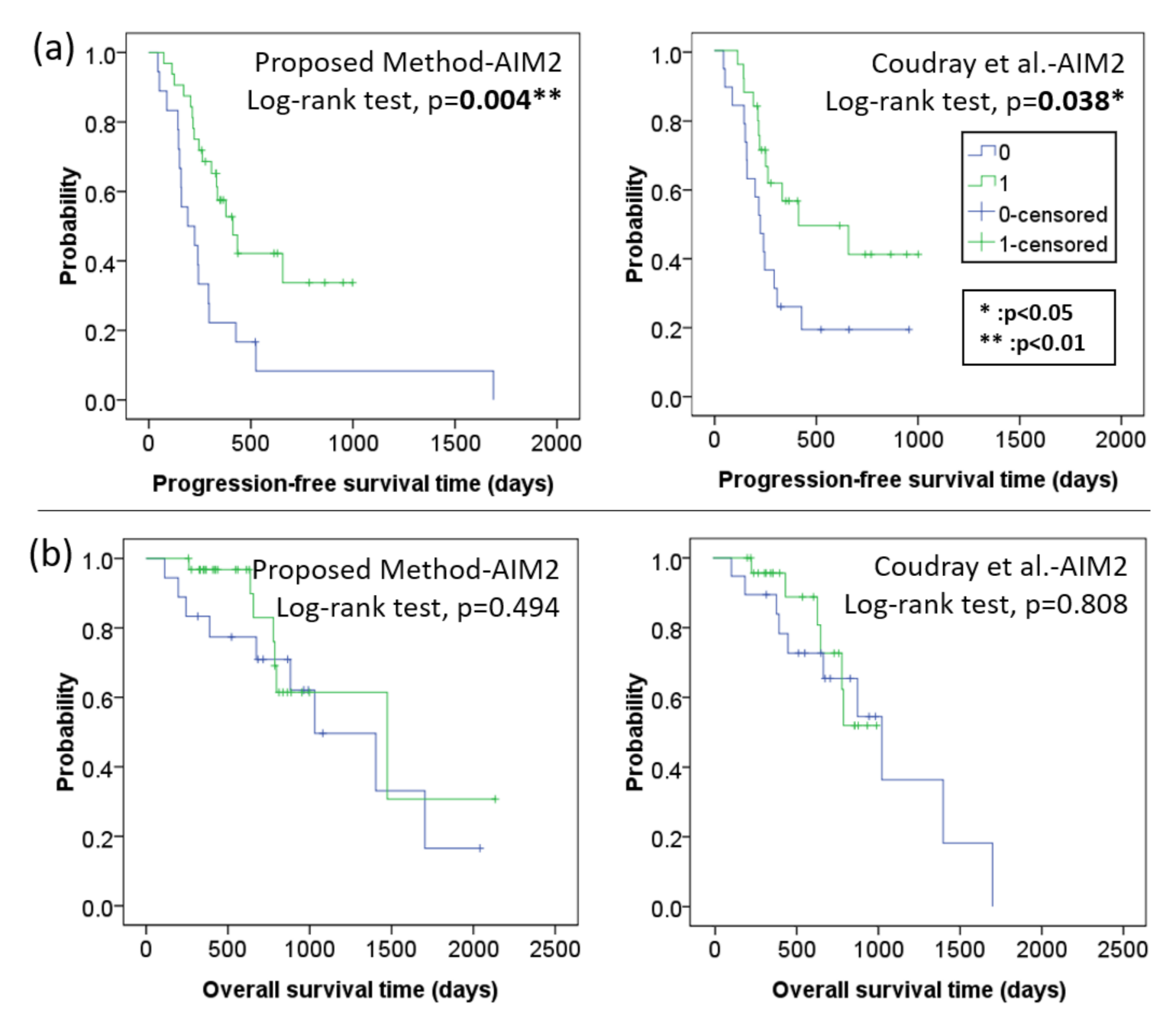
4.2.1. Kaplan–Meier Progression Free Survival and Overall Survival Analysis
4.2.2. Cox Proportional Hazards Model Analysis
5. Conclusions and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [PubMed]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2018, 81, 17–38. [Google Scholar] [PubMed] [Green Version]
- Ledermann, J.; Raja, F.; Fotopoulou, C.; Gonzalez-Martin, A.; Colombo, N.; Sessa, C.; Group, E.G.W. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013, 24, vi24–vi32. [Google Scholar] [PubMed]
- Pentheroudakis, G.; Pavlidis, N. Serous papillary peritoneal carcinoma: Unknown primary tumour, ovarian cancer counterpart or a distinct entity? A systematic review. Crit. Rev. Oncol./Hematol. 2010, 75, 27–42. [Google Scholar]
- Lim, H.J.; Ledger, W. Targeted therapy in ovarian cancer. Women’s Health 2016, 12, 363–378. [Google Scholar]
- Rüegg, C. Leukocytes, inflammation, and angiogenesis in cancer: Fatal attractions. J. Leukoc. Biol. 2006, 80, 682–684. [Google Scholar]
- Kim, Y.W.; West, X.Z.; Byzova, T.V. Inflammation and oxidative stress in angiogenesis and vascular disease. J. Mol. Med. 2013, 91, 323–328. [Google Scholar]
- Park, J.S.; Kim, I.K.; Han, S.; Park, I.; Kim, C.; Bae, J.; Oh, S.J.; Lee, S.; Kim, J.H.; Woo, D.C.; et al. Normalization of tumor vessels by Tie2 activation and Ang2 inhibition enhances drug delivery and produces a favorable tumor microenvironment. Cancer Cell 2016, 30, 953–967. [Google Scholar]
- Burger, R.A.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Monk, B.J.; Huang, H.; Mannel, R.S.; Homesley, H.D.; Fowler, J.; Greer, B.E.; et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar]
- Claussen, C.; Rody, A.; Hanker, L. Treatment of Recurrent Epithelial Ovarian Cancer. Geburtshilfe Frauenheilkd. 2020, 80, 1195–1204. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar]
- Garcia, J.; Hurwitz, H.I.; Sandler, A.B.; Miles, D.; Coleman, R.L.; Deurloo, R.; Chinot, O.L. Bevacizumab (Avastin®) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar]
- Karki, R.; Kanneganti, T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 2019, 19, 197–214. [Google Scholar]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar]
- Man, S.M.; Karki, R.; Kanneganti, T.D. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 2016, 46, 269–280. [Google Scholar]
- Lee, C.; Do, H.T.T.; Her, J.; Kim, Y.; Seo, D.; Rhee, I. Inflammasome as a promising therapeutic target for cancer. Life Sci. 2019, 231, 116593. [Google Scholar]
- Ponomareva, L.; Liu, H.; Duan, X.; Dickerson, E.; Shen, H.; Panchanathan, R.; Choubey, D. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol. Cancer Res. 2013, 11, 1193–1202. [Google Scholar]
- Chang, C.M.; Wang, M.L.; Lu, K.H.; Yang, Y.P.; Juang, C.M.; Wang, P.H.; Hsu, R.J.; Yu, M.H.; Chang, C.C. Integrating the dysregulated inflammasome-based molecular functionome in the malignant transformation of endometriosis-associated ovarian carcinoma. Oncotarget 2018, 9, 3704–3726. [Google Scholar]
- Bjørge, L.; Hakulinen, J.; Vintermyr, O.; Jarva, H.; Jensen, T.S.; Iversen, O.E.; Meri, S. Ascitic complement system in ovarian cancer. Br. J. Cancer 2005, 92, 895–905. [Google Scholar]
- Su, K.M.; Lin, T.W.; Liu, L.C.; Yang, Y.P.; Wang, M.L.; Tsai, P.H.; Wang, P.H.; Yu, M.H.; Chang, C.M.; Chang, C.C. The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis. Int. J. Mol. Sci. 2020, 21, 2824. [Google Scholar]
- Pio, R.; Corrales, L.; Lambris, J.D. The role of complement in tumor growth. In Tumor Microenvironment and Cellular Stress; Springer: Berlin/Heidelberg, Germany, 2014; pp. 229–262. [Google Scholar]
- Langer, H.F.; Chung, K.J.; Orlova, V.V.; Choi, E.Y.; Kaul, S.; Kruhlak, M.J.; Alatsatianos, M.; DeAngelis, R.A.; Roche, P.A.; Magotti, P.; et al. Complement-mediated inhibition of neovascularization reveals a point of convergence between innate immunity and angiogenesis. Blood J. Am. Soc. Hematol. 2010, 116, 4395–4403. [Google Scholar]
- Nozaki, M.; Raisler, B.J.; Sakurai, E.; Sarma, J.V.; Barnum, S.R.; Lambris, J.D.; Chen, Y.; Zhang, K.; Ambati, B.K.; Baffi, J.Z.; et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc. Natl. Acad. Sci. USA 2006, 103, 2328–2333. [Google Scholar]
- Wang, S.; Liu, Z.; Rong, Y.; Zhou, B.; Bai, Y.; Wei, W.; Wang, M.; Guo, Y.; Tian, J. Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother. Oncol. 2019, 132, 171–177. [Google Scholar]
- Sato, M.; Horie, K.; Hara, A.; Miyamoto, Y.; Kurihara, K.; Tomio, K.; Yokota, H. Application of deep learning to the classification of images from colposcopy. Oncol. Lett. 2018, 15, 3518–3523. [Google Scholar]
- Matsuo, K.; Purushotham, S.; Jiang, B.; Mandelbaum, R.S.; Takiuchi, T.; Liu, Y.; Roman, L.D. Survival outcome prediction in cervical cancer: Cox models vs deep-learning model. Am. J. Obstet. Gynecol. 2019, 220, 381.e1–381.e14. [Google Scholar] [CrossRef]
- Ke, J.; Shen, Y.; Lu, Y.; Deng, J.; Wright, J.D.; Zhang, Y.; Huang, Q.; Wang, D.; Jing, N.; Liang, X.; et al. Quantitative analysis of abnormalities in gynecologic cytopathology with deep learning. Lab. Investig. 2021, 101, 513–524. [Google Scholar]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 2018, 38, BSR20180289. [Google Scholar]
- Ghoniem, R.M.; Algarni, A.D.; Refky, B.; Ewees, A.A. Multi-Modal Evolutionary Deep Learning Model for Ovarian Cancer Diagnosis. Symmetry 2021, 13, 643. [Google Scholar]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [PubMed]
- Li, Y.F.; Guo, L.Z.; Zhou, Z.H. Towards safe weakly supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 334–346. [Google Scholar]
- Kim, J.; Kim, H.J.; Kim, C.; Lee, J.H.; Kim, K.W.; Park, Y.M.; Kim, H.W.; Ki, S.Y.; Kim, Y.M.; Kim, W.H. Weakly-supervised deep learning for ultrasound diagnosis of breast cancer. Sci. Rep. 2021, 11, 24382. [Google Scholar] [PubMed]
- Liu, M.Z.; Swintelski, C.; Sun, S.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R. Weakly supervised deep learning approach to breast MRI assessment. Acad. Radiol. 2022, 29, S166–S172. [Google Scholar]
- Lu, M.Y.; Williamson, D.F.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar]
- Li, Z.; Zhang, J.; Tan, T.; Teng, X.; Sun, X.; Zhao, H.; Liu, L.; Xiao, Y.; Lee, B.; Li, Y.; et al. Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images—the ACDC@LungHP Challenge 2019. IEEE J. Biomed. Health Inform. 2021, 25, 429–440. [Google Scholar]
- Wang, C.W.; Liou, Y.A.; Lin, Y.J.; Chang, C.C.; Chu, P.H.; Lee, Y.C.; Wang, C.H.; Chao, T.K. Artificial intelligence-assisted fast screening cervical high grade squamous intraepithelial lesion and squamous cell carcinoma diagnosis and treatment planning. Sci. Rep. 2021, 11, 16244. [Google Scholar]
- Lin, Y.J.; Chao, T.K.; Khalil, M.A.; Lee, Y.C.; Hong, D.Z.; Wu, J.J.; Wang, C.W. Deep Learning Fast Screening Approach on Cytological Whole Slides for Thyroid Cancer Diagnosis. Cancers 2021, 13, 3891. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: High quality object detection and instance segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 1483–1498. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Dudley, W.N.; Wickham, R.; Coombs, N. An introduction to survival statistics: Kaplan-Meier analysis. J. Adv. Pract. Oncol. 2016, 7, 91. [Google Scholar]
- Monk, B.J.; Dalton, H.; Farley, J.H.; Chase, D.M.; Benjamin, I. Antiangiogenic agents as a maintenance strategy for advanced epithelial ovarian cancer. Crit. Rev. Oncol. 2013, 86, 161–175. [Google Scholar]
- Hamanishi, J.; Mandai, M.; Konishi, I. Immune checkpoint inhibition in ovarian cancer. Int. Immunol. 2016, 28, 339–348. [Google Scholar]
- Oza, A.M.; Cook, A.D.; Pfisterer, J.; Embleton, A.; Ledermann, J.A.; Pujade-Lauraine, E.; Kristensen, G.; Carey, M.S.; Beale, P.; Cervantes, A.; et al. Standard chemotherapy with or without bevacizumab for women with newly diagnosed ovarian cancer (ICON7): Overall survival results of a phase 3 randomised trial. Lancet Oncol. 2015, 16, 928–936. [Google Scholar]
- Jayson, G.C.; Kerbel, R.; Ellis, L.M.; Harris, A.L. Antiangiogenic therapy in oncology: Current status and future directions. Lancet 2016, 388, 518–529. [Google Scholar]
- Szajnik, M.; Czystowska-Kuźmicz, M.; Elishaev, E.; Whiteside, T.L. Biological markers of prognosis, response to therapy and outcome in ovarian carcinoma. Expert Rev. Mol. Diagn. 2016, 16, 811–826. [Google Scholar]
- Lambrechts, D.; Lenz, H.J.; de Haas, S.; Carmeliet, P.; Scherer, S.J. Markers of response for the antiangiogenic agent bevacizumab. J. Clin. Oncol. 2013, 31, 1219–1230. [Google Scholar]
- Backen, A.; Renehan, A.G.; Clamp, A.R.; Berzuini, C.; Zhou, C.; Oza, A.; Bannoo, S.; Scherer, S.J.; Banks, R.E.; Dive, C.; et al. The combination of circulating Ang1 and Tie2 levels predicts progression-free survival advantage in bevacizumab-treated patients with ovarian cancer. Clin. Cancer Res. 2014, 20, 4549–4558. [Google Scholar]
- Steffensen, K.D.; Madsen, C.V.; Andersen, R.F.; Waldstrøm, M.; Adimi, P.; Jakobsen, A. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur. J. Cancer 2014, 50, 2611–2618. [Google Scholar]
- Comunanza, V.; Bussolino, F. Therapy for cancer: Strategy of combining anti-angiogenic and target therapies. Front. Cell Dev. Biol. 2017, 5, 101. [Google Scholar]
- Carmeliet, P.; Jain, R.K. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat. Rev. Drug Discov. 2011, 10, 417–427. [Google Scholar]
- Clarke, J.M.; Hurwitz, H.I. Understanding and targeting resistance to anti-angiogenic therapies. J. Gastrointest. Oncol. 2013, 4, 253–263. [Google Scholar]
- Crea, F.; Fornaro, L.; Bocci, G.; Sun, L.; Farrar, W.L.; Falcone, A.; Danesi, R. EZH2 inhibition: Targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012, 31, 753–761. [Google Scholar] [PubMed]
- Hsu, P.C.; Chao, T.K.; Chou, Y.C.; Yu, M.H.; Wang, Y.C.; Lin, Y.H.; Lee, Y.L.; Liu, L.C.; Chang, C.C. AIM2 Inflammasome in Tumor Cells as a Biomarker for Predicting the Treatment Response to Antiangiogenic Therapy in Epithelial Ovarian Cancer Patients. J. Clin. Med. 2021, 10, 4529. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar]
- Beck, A.H.; Sangoi, A.R.; Leung, S.; Marinelli, R.J.; Nielsen, T.O.; Van De Vijver, M.J.; West, R.B.; Van De Rijn, M.; Koller, D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Transl. Med. 2011, 3, 108ra113. [Google Scholar]
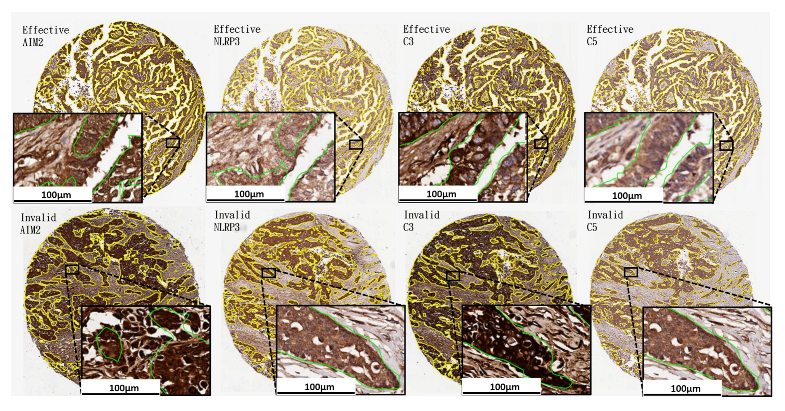
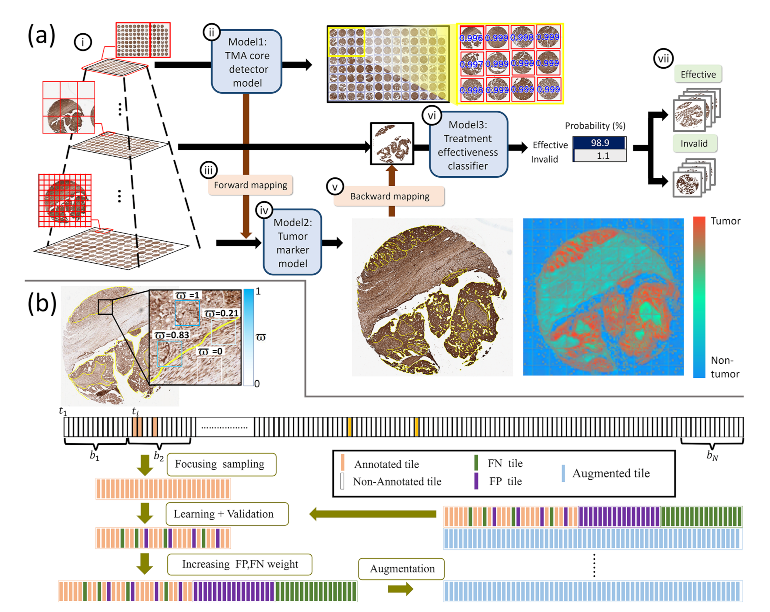
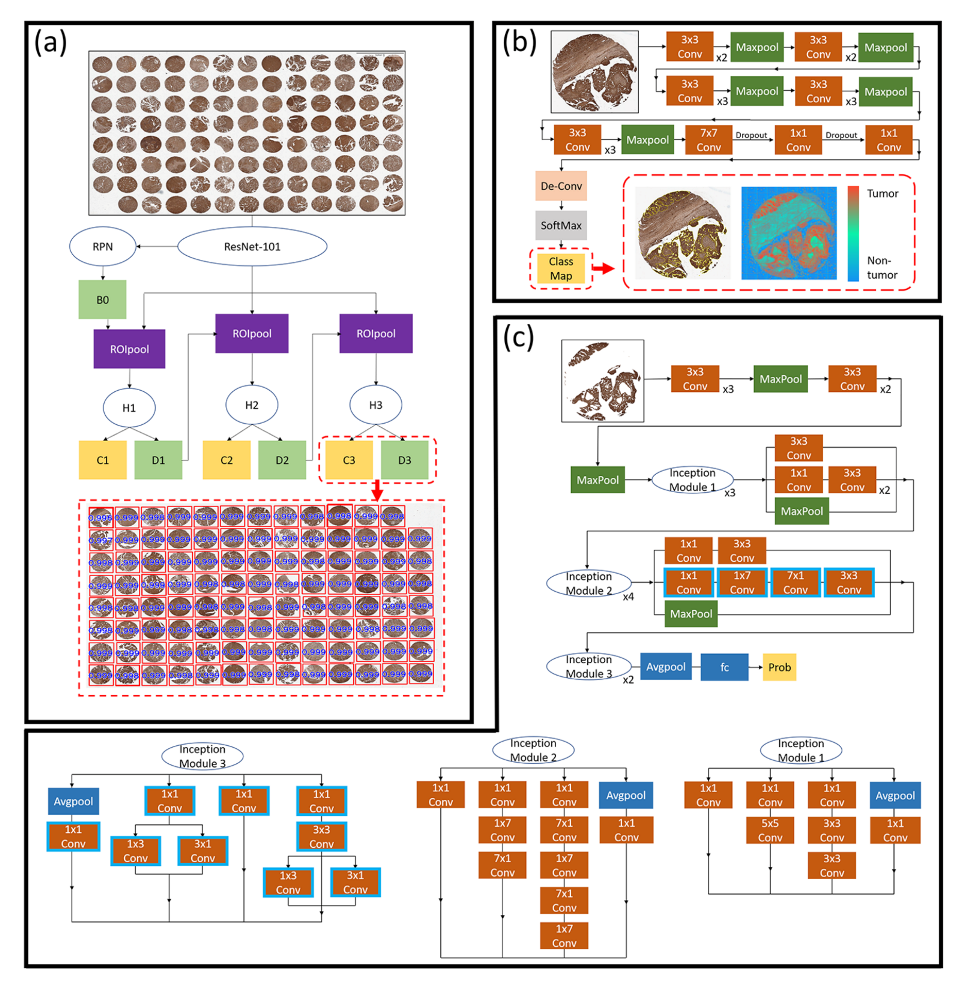
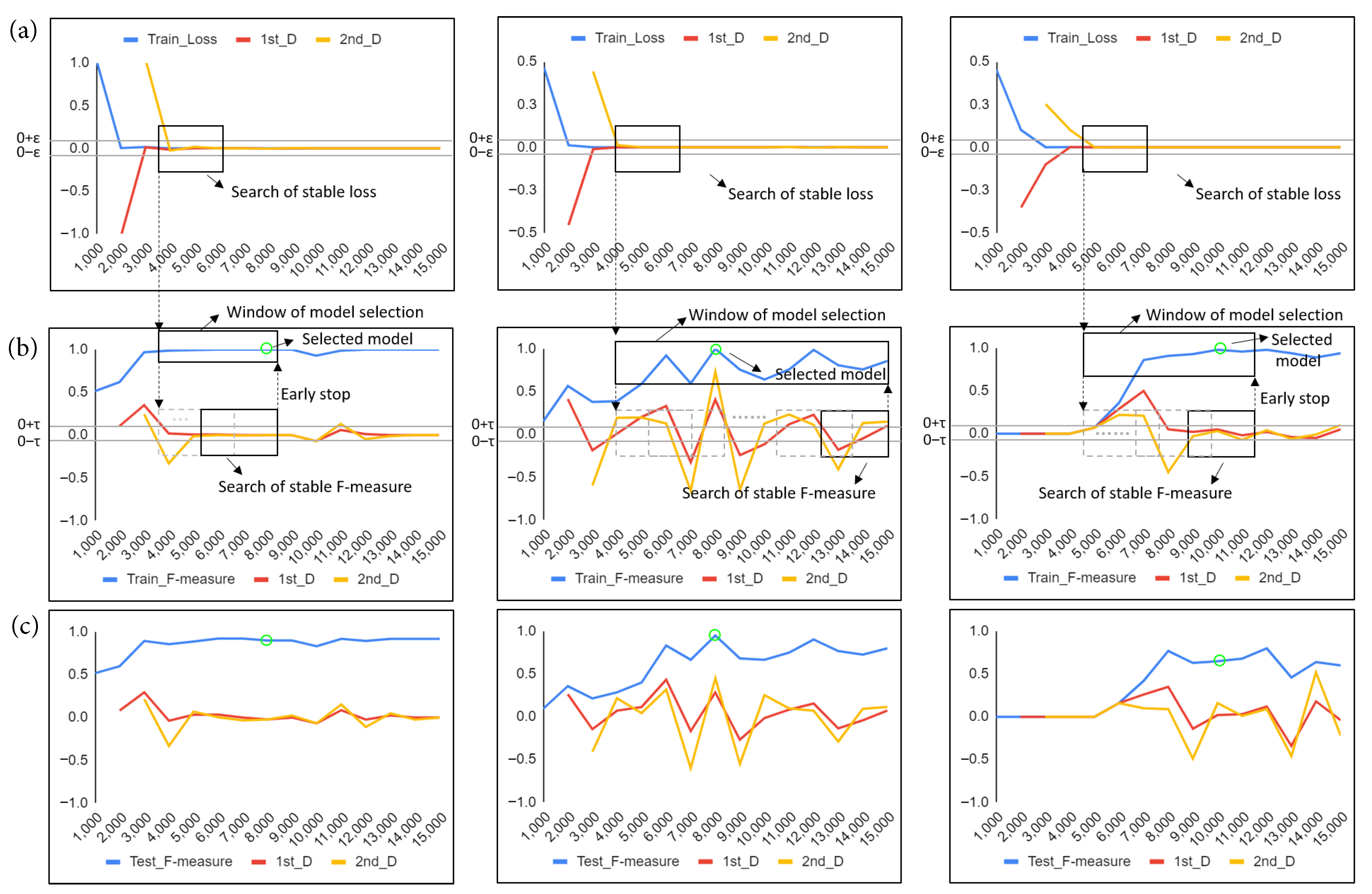

| Characteristics | N |
|---|---|
| Tissue Core | 720 |
| Patient age (mean, range) | (59.1, 23–79) |
| Diagnosis (%) | |
| Papillary serous carcinoma | 444 (61.6) |
| Peritoneal serous papillary carcinoma | 89 (12.3) |
| Clear cell carcinoma | 69 (9.6) |
| Unclassified carcinoma | 69 (9.6) |
| Endometrioid carcinoma | 39 (5.5) |
| MC | 10 (1.4) |
| FIGO stage (%) | |
| I | 69 (9.6) |
| II | 39 (5.4) |
| III | 454 (63) |
| IV | 158 (22) |
| Surgery (%) | |
| Optimal debulking | 306 (42.5) |
| CRS+HIPEC | 217 (30.1) |
| Suboptimal debulking | 197 (27.4) |
| Treatment effectiveness (%) | |
| Effective | 412 (57.2) |
| Invalid | 308 (42.8) |
| Treatment Outcome | AIM2 | C3 | C5 | NLRP3 | |
|---|---|---|---|---|---|
| Training (66%) | Effective | 68 | 68 | 68 | 68 |
| Invalid | 50 | 50 | 50 | 50 | |
| Testing (34%) | Effective | 35 | 35 | 35 | 35 |
| Invalid | 27 | 27 | 27 | 27 | |
| Total | 180 | 180 | 180 | 180 | |
| Method in Combination with Potential Biomarker | Accuracy | Precision | Recall | F-Measure | AUC |
|---|---|---|---|---|---|
| Proposed Weakly Supervised DL Method—AIM2 | 0.92 | 0.89 | 0.97 | 0.93 | 0.97 |
| Coudray et al. [44]—AIM2 | 0.90 | 0.97 | 0.86 | 0.91 | 0.97 |
| Proposed Weakly Supervised DL Method—C3 | 0.69 | 0.69 | 0.83 | 0.75 | 0.75 |
| Coudray et al. [44]—C3 | 0.90 | 0.91 | 0.91 | 0.91 | 0.94 |
| Proposed Weakly Supervised DL Method—C5 | 0.63 | 0.67 | 0.69 | 0.68 | 0.65 |
| Coudray et al. [44]—C5 | 0.69 | 0.68 | 0.86 | 0.76 | 0.78 |
| Proposed Weakly Supervised DL Method—NLRP3 | 0.52 | 0.56 | 0.71 | 0.63 | 0.50 |
| Coudray et al. [44]—NLRP3 | 0.71 | 0.76 | 0.71 | 0.74 | 0.73 |
| Method in Combination with Potential Biomarker | Accuracy | Precision | Recall | F-Measure | AUC |
|---|---|---|---|---|---|
| ProposedWeakly Supervised DL Method—AIM2 | 0.86 ± 0.07 | 0.9± 0.07 | 0.85 ± 0.06 | 0.87 ± 0.06 | 0.91 ± 0.05 |
| Coudray et al. [44]—AIM2 | 0.73 ± 0.17 | 0.76 ± 0.19 | 0.71 ± 0.39 | 0.68 ± 0.33 | 0.9 ± 0.07 |
| Proposed Weakly Supervised DL Method—C3 | 0.75 ± 0.1 | 0.77 ± 0.1 | 0.79 ± 0.1 | 0.78 ± 0.09 | 0.78 ± 0.12 |
| Coudray et al. [44]—C3 | 0.73 ± 0.08 | 0.77 ± 0.06 | 0.74 ± 0.21 | 0.74 ± 0.1 | 0.84 ± 0.09 |
| Proposed Weakly Supervised DL Method—C5 | 0.65 ± 0.03 | 0.66 ± 0.03 | 0.8 ± 0.04 | 0.72 ± 0.02 | 0.66 ± 0.07 |
| Coudray et al. [44]—C5 | 0.56 ± 0.13 | 0.69 ± 0.22 | 0.51 ± 0.3 | 0.54 ± 0.22 | 0.52 ± 0.23 |
| Proposed Weakly Supervised DL Method—NLRP3 | 0.56 ± 0.08 | 0.59 ± 0.05 | 0.77 ± 0.08 | 0.67 ± 0.06 | 0.55 ± 0.08 |
| Coudray et al. [44]—NLRP3 | 0.63 ± 0.18 | 0.68 ± 0.19 | 0.66 ± 0.32 | 0.63 ± 0.28 | 0.73 ± 0.24 |
| Adjusted HR (95% C.I.) | p Value | |
|---|---|---|
| Age | 1.03 (0.98–1.07) | 0.212 |
| BMI | 1.01 (0.92–1.11) | 0.848 |
| Number of bevacizumab used times | 0.97 (0.89–1.05) | 0.472 |
| FIGO (III + IV vs. I + II) | 4.23 (0.82–21.86) | 0.085 |
| Histology (others vs. serous) | 0.82 (0.26–2.62) | 0.737 |
| Surgery | ||
| CRS + HIPEC | 1.00 (reference) | reference |
| optimal | 0.92 (0.35–2.42) | 0.869 |
| suboptimal | 1.18 (0.42–3.30) | 0.753 |
| Theraphy | ||
| Concurrent therapy | 1.00 (reference) | reference |
| Second-line therapy | 1.71 (0.66–4.41) | 0.265 |
| Maintenance therapy | 0.23 (0.04–1.40) | 0.110 |
| DL model prediction | ||
| Proposed DL Method-AIM2 (effective v.s. invalid) | 0.18 (0.06–0.55) | 0.003 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-W.; Lee, Y.-C.; Chang, C.-C.; Lin, Y.-J.; Liou, Y.-A.; Hsu, P.-C.; Chang, C.-C.; Sai, A.-K.-O.; Wang, C.-H.; Chao, T.-K. A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker. Cancers 2022, 14, 1651. https://doi.org/10.3390/cancers14071651
Wang C-W, Lee Y-C, Chang C-C, Lin Y-J, Liou Y-A, Hsu P-C, Chang C-C, Sai A-K-O, Wang C-H, Chao T-K. A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker. Cancers. 2022; 14(7):1651. https://doi.org/10.3390/cancers14071651
Chicago/Turabian StyleWang, Ching-Wei, Yu-Ching Lee, Cheng-Chang Chang, Yi-Jia Lin, Yi-An Liou, Po-Chao Hsu, Chun-Chieh Chang, Aung-Kyaw-Oo Sai, Chih-Hung Wang, and Tai-Kuang Chao. 2022. "A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker" Cancers 14, no. 7: 1651. https://doi.org/10.3390/cancers14071651
APA StyleWang, C.-W., Lee, Y.-C., Chang, C.-C., Lin, Y.-J., Liou, Y.-A., Hsu, P.-C., Chang, C.-C., Sai, A.-K.-O., Wang, C.-H., & Chao, T.-K. (2022). A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker. Cancers, 14(7), 1651. https://doi.org/10.3390/cancers14071651








