Regulation of Let-7a-5p and miR-199a-5p Expression by Akt1 Modulates Prostate Cancer Epithelial-to-Mesenchymal Transition via the Transforming Growth Factor-β Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, shRNA Gene Silencing, Antibodies, and Other Reagents
2.2. NanoString-Based miRNA and mRNA Profiling
2.3. In Vivo miRNA Isolation and Quantitative RT-PCR
2.4. miRNA Transfection
2.5. Western Blotting
2.6. mirPath v.3 and dbEMT Analysis
2.7. Migration Assay
2.8. Matrigel® Invasion Assay
2.9. Ingenuity® Pathway Analysis
2.10. cBioportal Genomic Data Analysis of the MSKCC Study
2.11. Statistical Analysis
3. Results
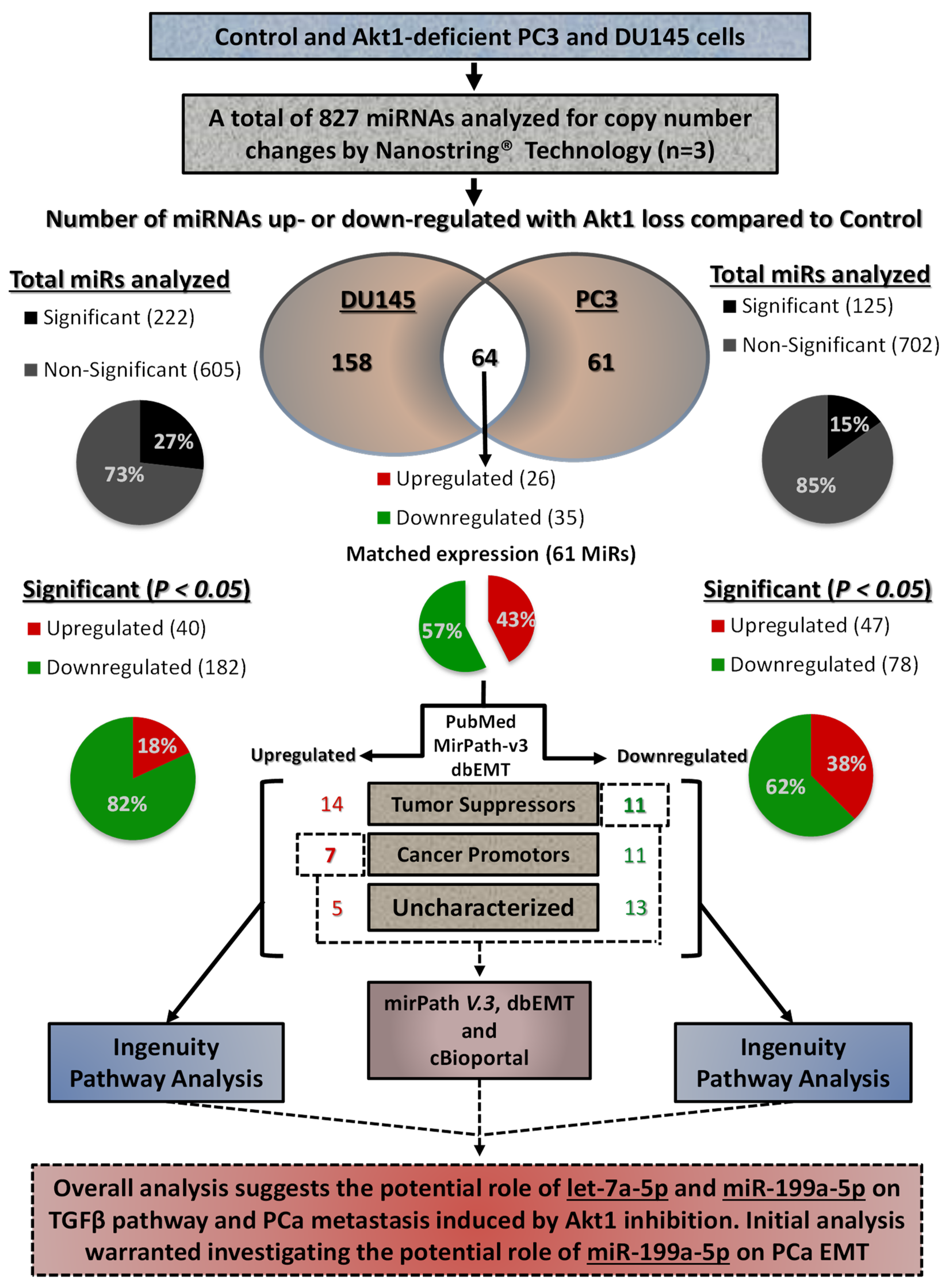
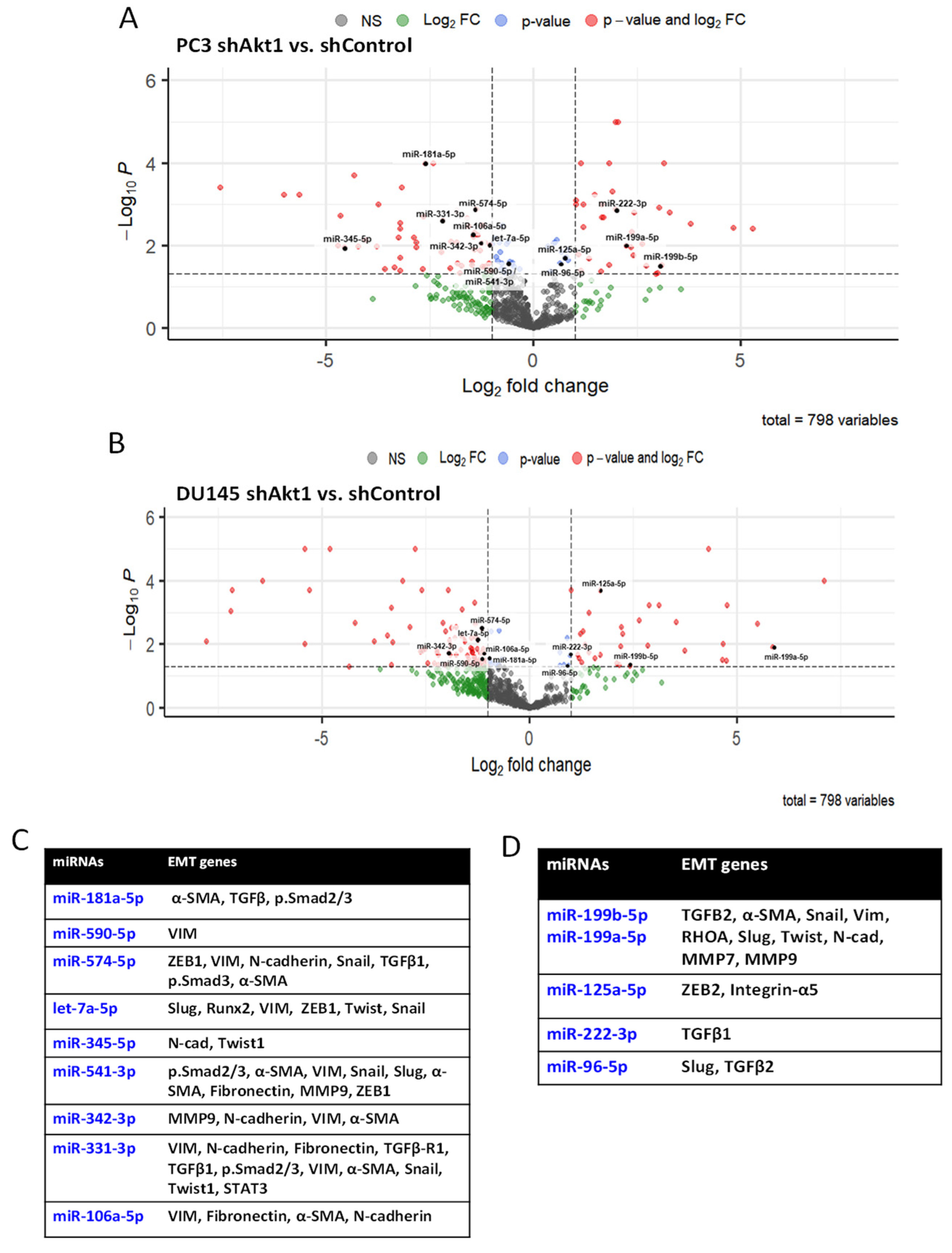
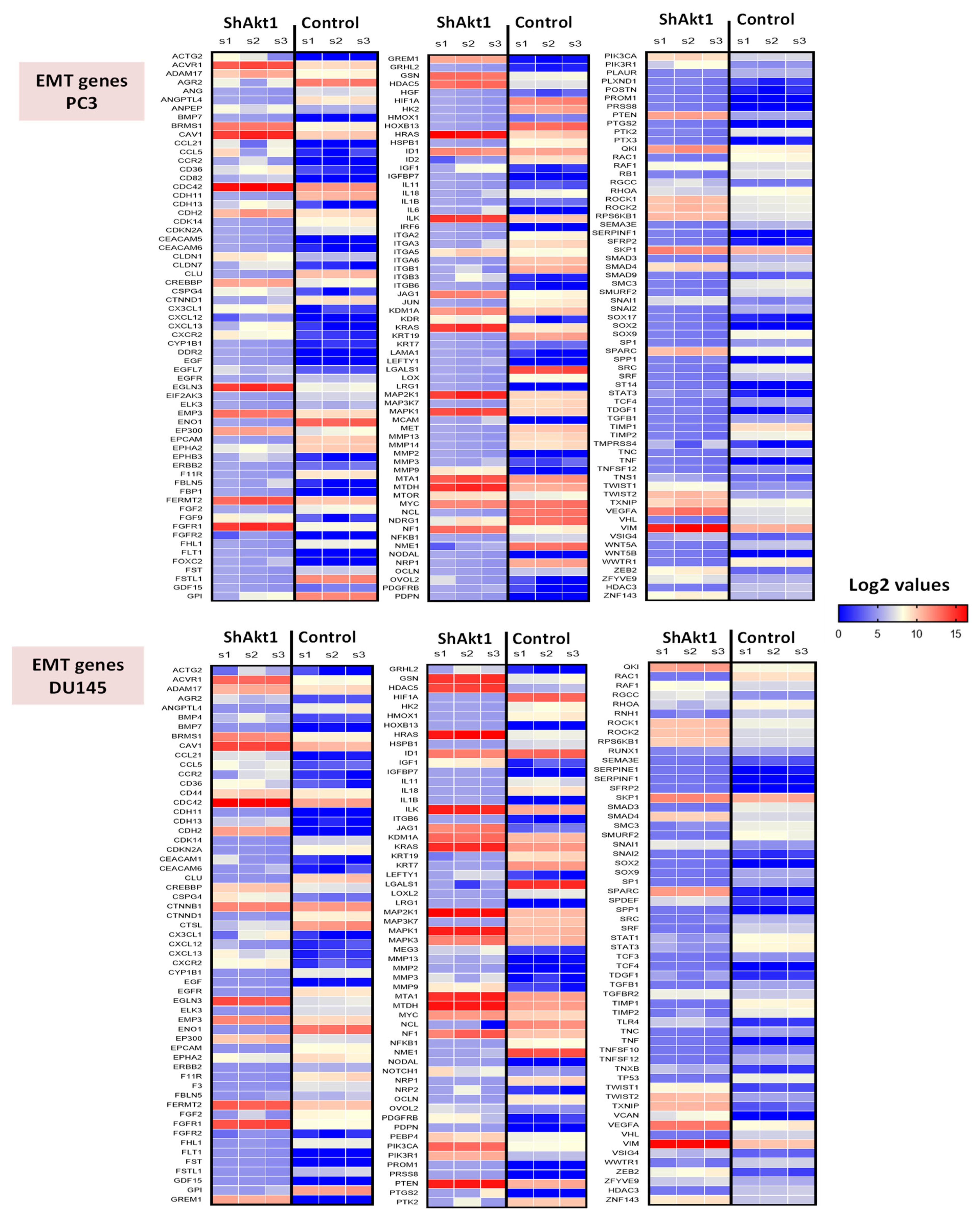
3.1. Akt1 Inhibition Changes the microRNA Expression Profile in PC3 and DU145 Cells
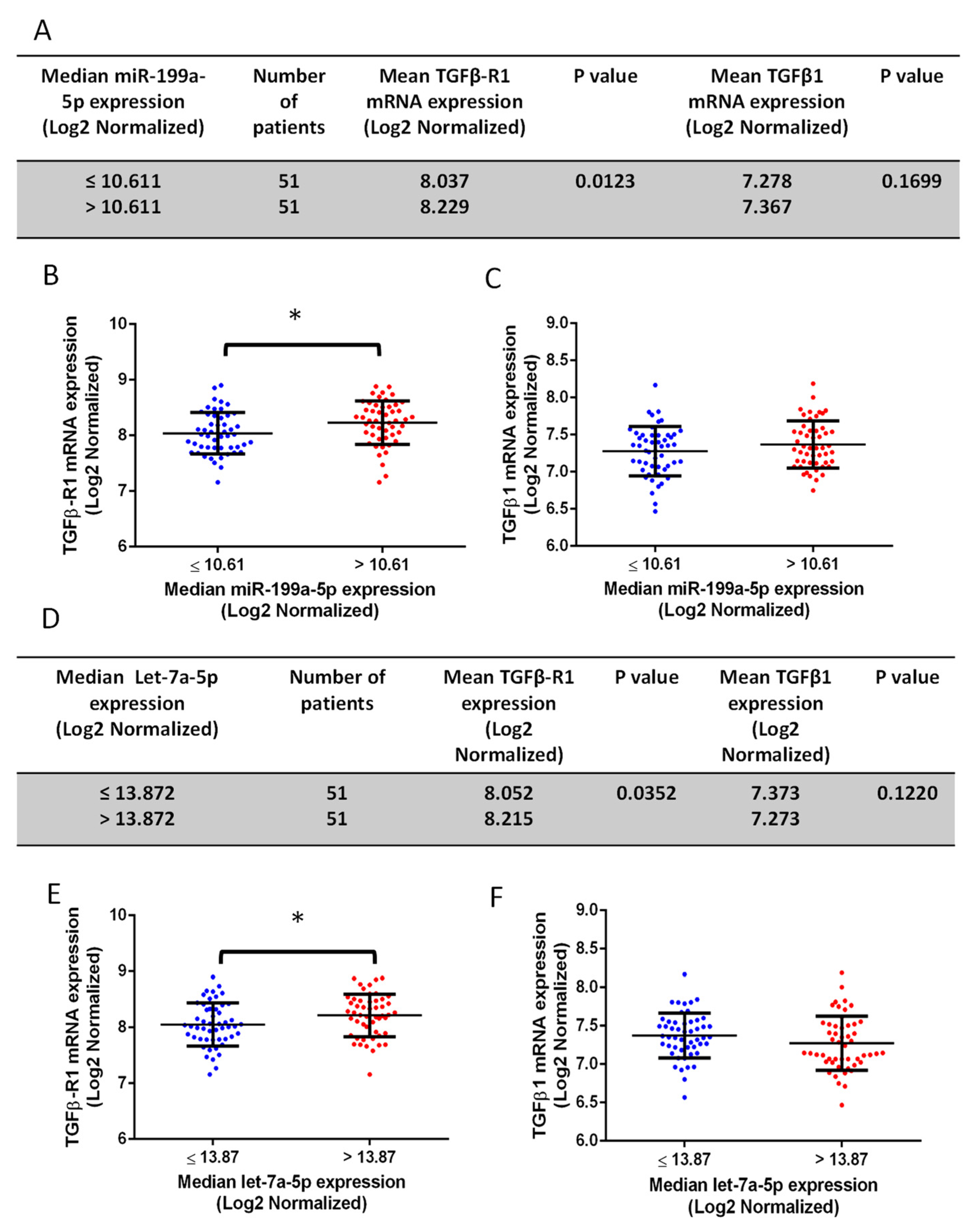
3.2. Human PCa Tissues with a High Gleason Score Express Higher miR-199a-5p and Lower Let-7a-5p Compared to the Low Gleason Score Tissues
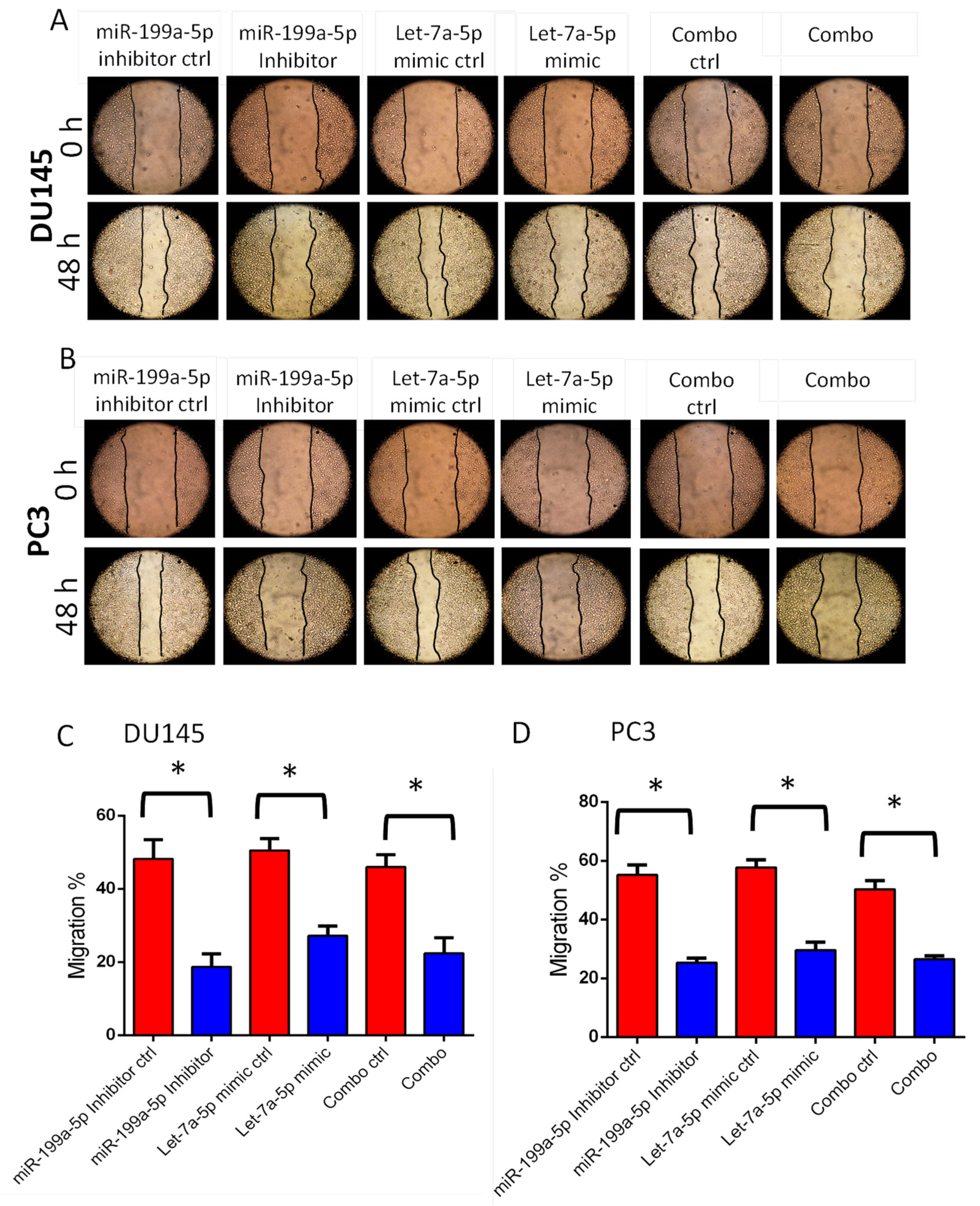
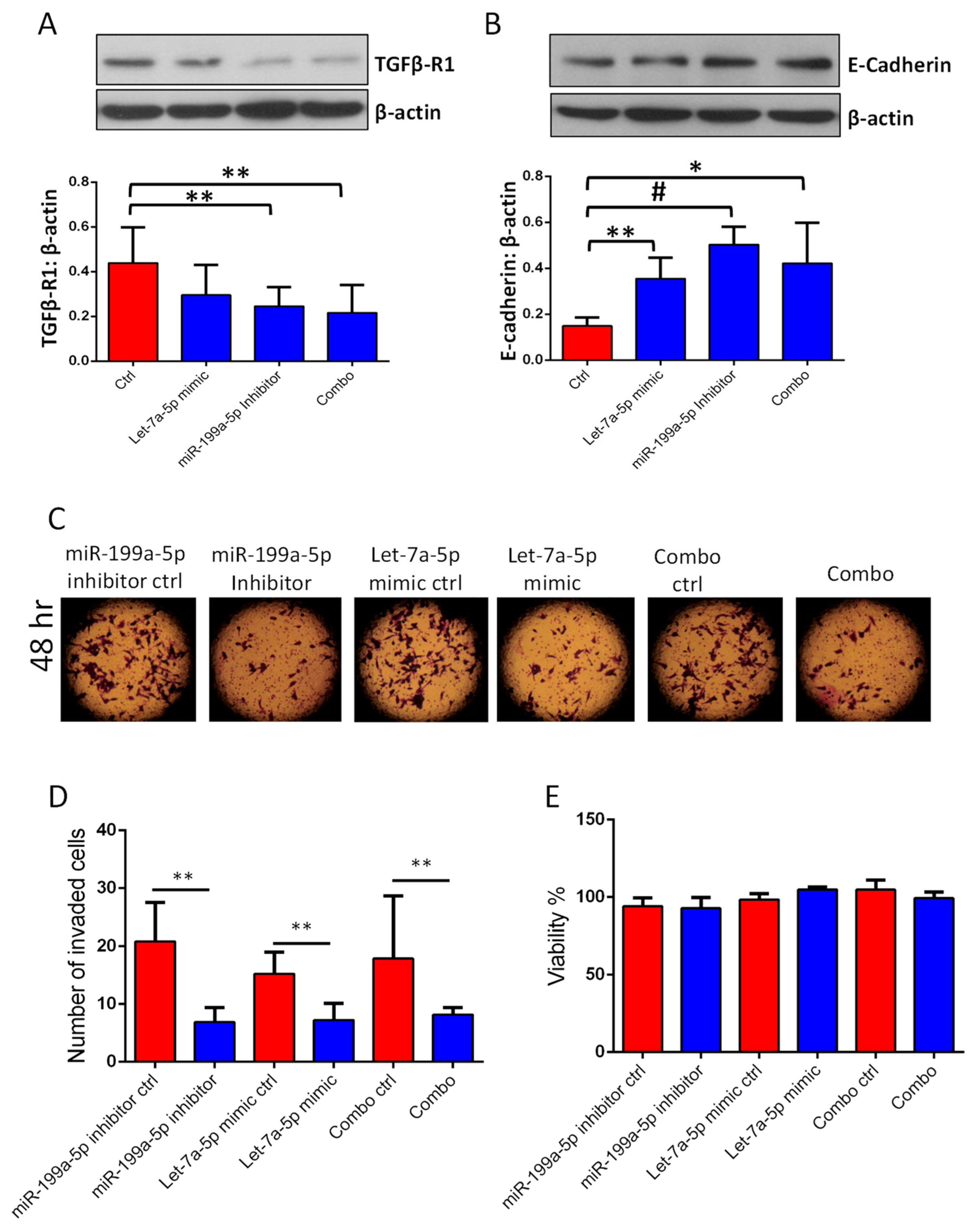
3.3. Transfections with miR-199a-5p Inhibitor and/or let-7a-5p Mimic Impairs Migration and Invasion of PC3 and DU145 Cell Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Guan, X. Cancer metastases: Challenges and opportunities. Acta Pharm. Sin. B 2015, 5, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Swanton, C. Metastasis as an evolutionary process. Science 2016, 352, 169–175. [Google Scholar] [CrossRef]
- Narain, T.A.; Sooriakumaran, P. Beyond Prostate Specific Antigen: New Prostate Cancer Screening Options. World J. Mens Health 2022, 40, 66–73. [Google Scholar] [CrossRef]
- Ha Chung, B.; Horie, S.; Chiong, E. The incidence, mortality, and risk factors of prostate cancer in Asian men. Prostate Int. 2019, 7, 1–8. [Google Scholar] [CrossRef]
- Benafif, S.; Eeles, R. Genetic predisposition to prostate cancer. Br. Med. Bull. 2016, 120, 75–89. [Google Scholar] [CrossRef]
- Jiang, Y.; Meyers, T.J.; Emeka, A.A.; Cooley, L.F.; Cooper, P.R.; Lancki, N.; Helenowski, I.; Kachuri, L.; Lin, D.W.; Stanford, J.L.; et al. Genetic Factors Associated with Prostate Cancer Conversion from Active Surveillance to Treatment. HGG Adv. 2022, 3, 100070. [Google Scholar] [CrossRef]
- Leslie, S.W.; Soon-Sutton, T.L.; Sajjad, H.; Siref, L.E. Prostate Cancer. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Mello-Grand, M.; Gregnanin, I.; Sacchetto, L.; Ostano, P.; Zitella, A.; Bottoni, G.; Oderda, M.; Marra, G.; Munegato, S.; Pardini, B.; et al. Circulating microRNAs combined with PSA for accurate and non-invasive prostate cancer detection. Carcinogenesis 2019, 40, 246–253. [Google Scholar] [CrossRef]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the epithelial-mesenchymal transition by TGF-beta. Future Oncol 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, J.J. The Dual Role of TGFbeta in Human Cancer: From Tumor Suppression to Cancer Metastasis. ISRN Mol. Biol. 2012, 2012, 381428. [Google Scholar] [CrossRef] [PubMed]
- Al-Azayzih, A.; Gao, F.; Goc, A.; Somanath, P.R. TGFbeta1 induces apoptosis in invasive prostate cancer and bladder cancer cells via Akt-independent, p38 MAPK and JNK/SAPK-mediated activation of caspases. Biochem. Biophys. Res. Commun. 2012, 427, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Colak, S.; Ten Dijke, P. Targeting TGF-beta Signaling in Cancer. Trends Cancer 2017, 3, 56–71. [Google Scholar] [CrossRef]
- Trivedi, T.; Pagnotti, G.M.; Guise, T.A.; Mohammad, K.S. The Role of TGF-beta in Bone Metastases. Biomolecules 2021, 11, 1643. [Google Scholar] [CrossRef]
- Al-Azayzih, A.; Gao, F.; Somanath, P.R. P21 activated kinase-1 mediates transforming growth factor beta1-induced prostate cancer cell epithelial to mesenchymal transition. Biochim. Biophys. Acta 2015, 1853, 1229–1239. [Google Scholar] [CrossRef]
- Si, L.; Yang, Z.; Ding, L.; Zhang, D. Regulatory effects of lncRNAs and miRNAs on the crosstalk between autophagy and EMT in cancer: A new era for cancer treatment. J. Cancer Res. Clin. Oncol. 2022, 148, 547–564. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef]
- Graziani, V.; Rodriguez-Hernandez, I.; Maiques, O.; Sanz-Moreno, V. The amoeboid state as part of the epithelial-to-mesenchymal transition programme. Trends Cell Biol. 2022, 32, 228–242. [Google Scholar] [CrossRef]
- Wang, J.; Yang, L.; Yang, J.; Kuropatwinski, K.; Wang, W.; Liu, X.Q.; Hauser, J.; Brattain, M.G. Transforming growth factor beta induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Res. 2008, 68, 3152–3160. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Sabbineni, H.; Artham, S.; Somanath, P.R. Modulation of long-term endothelial-barrier integrity is conditional to the cross-talk between Akt and Src signaling. J. Cell. Physiol. 2017, 232, 2599–2609. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Alwhaibi, A.; Sabbineni, H.; Verma, A.; Eldahshan, W.; Somanath, P.R. Suppression of Akt1-beta-catenin pathway in advanced prostate cancer promotes TGFbeta1-mediated epithelial to mesenchymal transition and metastasis. Cancer Lett. 2017, 402, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Sabbineni, H.; Verma, A.; Somanath, P.R. Isoform-specific effects of transforming growth factor beta on endothelial-to-mesenchymal transition. J. Cell. Physiol. 2018, 233, 8418–8428. [Google Scholar] [CrossRef]
- Sabbineni, H.; Verma, A.; Artham, S.; Anderson, D.; Amaka, O.; Liu, F.; Narayanan, S.P.; Somanath, P.R. Pharmacological inhibition of beta-catenin prevents EndMT in vitro and vascular remodeling in vivo resulting from endothelial Akt1 suppression. Biochem. Pharmacol. 2019, 164, 205–215. [Google Scholar] [CrossRef]
- Gao, F.; Alwhaibi, A.; Artham, S.; Verma, A.; Somanath, P.R. Endothelial Akt1 loss promotes prostate cancer metastasis via beta-catenin-regulated tight-junction protein turnover. Br. J. Cancer 2018, 118, 1464–1475. [Google Scholar] [CrossRef]
- Alwhaibi, A.; Kolhe, R.; Gao, F.; Cobran, E.K.; Somanath, P.R. Genome atlas analysis based profiling of Akt pathway genes in the early and advanced human prostate cancer. Oncoscience 2019, 6, 317–336. [Google Scholar] [CrossRef]
- Alwhaibi, A.; Verma, A.; Artham, S.; Adil, M.S.; Somanath, P.R. Nodal pathway activation due to Akt1 suppression is a molecular switch for prostate cancer cell epithelial-to-mesenchymal transition and metastasis. Biochem. Pharmacol. 2019, 168, 1–13. [Google Scholar] [CrossRef]
- Alwhaibi, A.; Gao, F.; Artham, S.; Hsia, B.M.; Mondal, A.; Kolhe, R.; Somanath, P.R. Modulation in the microRNA repertoire is responsible for the stage-specific effects of Akt suppression on murine neuroendocrine prostate cancer. Heliyon 2018, 4, e00796. [Google Scholar] [CrossRef]
- Alwhaibi, A.; Verma, A.; Adil, M.S.; Somanath, P.R. The unconventional role of Akt1 in the advanced cancers and in diabetes-promoted carcinogenesis. Pharmacol. Res. 2019, 145, 104270. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.S.; Khulood, D.; Somanath, P.R. Targeting Akt-associated microRNAs for cancer therapeutics. Biochem. Pharmacol. 2021, 189, 114384. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Kasprzak, W.K.; Bhandari, Y.; Fan, L.; Cavanaugh, Q.; Jiang, M.; Dai, L.; Yang, A.; Shao, T.J.; Shapiro, B.A.; et al. Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires. Cell Rep. 2019, 26, 447–459.e4. [Google Scholar] [CrossRef]
- Suzuki, H.I. MicroRNA Control of TGF-beta Signaling. Int. J. Mol. Sci. 2018, 19, 1901. [Google Scholar] [CrossRef]
- Janakiraman, H.; House, R.P.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H.; Palanisamy, V. The Long (lncRNA) and Short (miRNA) of It: TGFbeta-Mediated Control of RNA-Binding Proteins and Noncoding RNAs. Mol. Cancer Res. 2018, 16, 567–579. [Google Scholar] [CrossRef]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs—The micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar] [CrossRef]
- Sekhon, K.; Bucay, N.; Majid, S.; Dahiya, R.; Saini, S. MicroRNAs and epithelial-mesenchymal transition in prostate cancer. Oncotarget 2016, 7, 67597–67611. [Google Scholar] [CrossRef]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.; Visakorpi, T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007, 67, 6130–6135. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Polytarchou, C.; Hatziapostolou, M.; Kottakis, F.; Maroulakou, I.G.; Struhl, K.; Tsichlis, P.N. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci. Signal. 2009, 2, ra62. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Bard, J.E.; Nowak, N.J.; Buck, M.J.; Kandel, E.S. FOXO1 regulates expression of a microRNA cluster on X chromosome. Aging (Albany NY) 2013, 5, 347–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, F.; Al-Azayzih, A.; Somanath, P.R. Discrete functions of GSK3alpha and GSK3beta isoforms in prostate tumor growth and micrometastasis. Oncotarget 2015, 6, 5947–5962. [Google Scholar] [CrossRef]
- Chen, M.L.; Xu, P.Z.; Peng, X.D.; Chen, W.S.; Guzman, G.; Yang, X.; Di Cristofano, A.; Pandolfi, P.P.; Hay, N. The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/- mice. Genes Dev. 2006, 20, 1569–1574. [Google Scholar] [CrossRef]
- Goc, A.; Liu, J.; Byzova, T.V.; Somanath, P.R. Akt1 mediates prostate cancer cell microinvasion and chemotaxis to metastatic stimuli via integrin beta(3) affinity modulation. Br. J. Cancer 2012, 107, 713–723. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Aghdam, S.G.; Ebrazeh, M.; Hemmatzadeh, M.; Seyfizadeh, N.; Shabgah, A.G.; Azizi, G.; Ebrahimi, N.; Babaie, F.; Mohammadi, H. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J. Cell. Physiol. 2019, 234, 9927–9942. [Google Scholar] [CrossRef]
- Lombe, C.P.; Meyer, M.; Pretorius, A. Bioinformatics Prediction and Analysis of MicroRNAs and Their Targets as Biomarkers for Prostate Cancer: A Preliminary Study. Mol. Biotechnol. 2022, 64, 401–412. [Google Scholar] [CrossRef]
- Bai, T.; Liu, Y.; Li, B. LncRNA LOXL1-AS1/miR-let-7a-5p/EGFR-related pathway regulates the doxorubicin resistance of prostate cancer DU-145 cells. IUBMB Life 2019, 71, 1537–1551. [Google Scholar] [CrossRef]
- Liu, T.P.; Huang, C.C.; Yeh, K.T.; Ke, T.W.; Wei, P.L.; Yang, J.R.; Cheng, Y.W. Down-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: Implications for chemotherapy. Surg. Oncol. 2016, 25, 429–434. [Google Scholar] [CrossRef]
- Ghanbari, R.; Mosakhani, N.; Sarhadi, V.K.; Armengol, G.; Nouraee, N.; Mohammadkhani, A.; Khorrami, S.; Arefian, E.; Paryan, M.; Malekzadeh, R.; et al. Simultaneous Underexpression of let-7a-5p and let-7f-5p microRNAs in Plasma and Stool Samples from Early Stage Colorectal Carcinoma. Biomark. Cancer 2015, 7, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Huang, R.; Su, Z.; Zhang, M.; Xu, M.; Gong, J.; Chen, N.; Zeng, H.; Chen, X.; Zhou, Q. Downregulation of miR-199a-5p promotes prostate adeno-carcinoma progression through loss of its inhibition of HIF-1alpha. Oncotarget 2017, 8, 83523–83538. [Google Scholar] [CrossRef] [PubMed]
- Goc, A.; Al-Husein, B.; Kochuparambil, S.T.; Liu, J.; Heston, W.W.; Somanath, P.R. PI3 kinase integrates Akt and MAP kinase signaling pathways in the regulation of prostate cancer. Int. J. Oncol. 2010, 38, 267–277. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alwhaibi, A.; Parvathagiri, V.; Verma, A.; Artham, S.; Adil, M.S.; Somanath, P.R. Regulation of Let-7a-5p and miR-199a-5p Expression by Akt1 Modulates Prostate Cancer Epithelial-to-Mesenchymal Transition via the Transforming Growth Factor-β Pathway. Cancers 2022, 14, 1625. https://doi.org/10.3390/cancers14071625
Alwhaibi A, Parvathagiri V, Verma A, Artham S, Adil MS, Somanath PR. Regulation of Let-7a-5p and miR-199a-5p Expression by Akt1 Modulates Prostate Cancer Epithelial-to-Mesenchymal Transition via the Transforming Growth Factor-β Pathway. Cancers. 2022; 14(7):1625. https://doi.org/10.3390/cancers14071625
Chicago/Turabian StyleAlwhaibi, Abdulrahman, Varun Parvathagiri, Arti Verma, Sandeep Artham, Mir S. Adil, and Payaningal R. Somanath. 2022. "Regulation of Let-7a-5p and miR-199a-5p Expression by Akt1 Modulates Prostate Cancer Epithelial-to-Mesenchymal Transition via the Transforming Growth Factor-β Pathway" Cancers 14, no. 7: 1625. https://doi.org/10.3390/cancers14071625
APA StyleAlwhaibi, A., Parvathagiri, V., Verma, A., Artham, S., Adil, M. S., & Somanath, P. R. (2022). Regulation of Let-7a-5p and miR-199a-5p Expression by Akt1 Modulates Prostate Cancer Epithelial-to-Mesenchymal Transition via the Transforming Growth Factor-β Pathway. Cancers, 14(7), 1625. https://doi.org/10.3390/cancers14071625






