Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. Sample Preparation and Measurement Conditions for NMR
2.3. NMR Related Statistical Analysis
2.4. MetPA Analysis of Metabolite Profiles for Metabolic Pathway Detection
2.5. Quantitative Real Time PCR
2.6. Gene Set Enrichment Analysis (GSEA)
2.7. Transcriptomics Related Statistical Analysis
3. Results
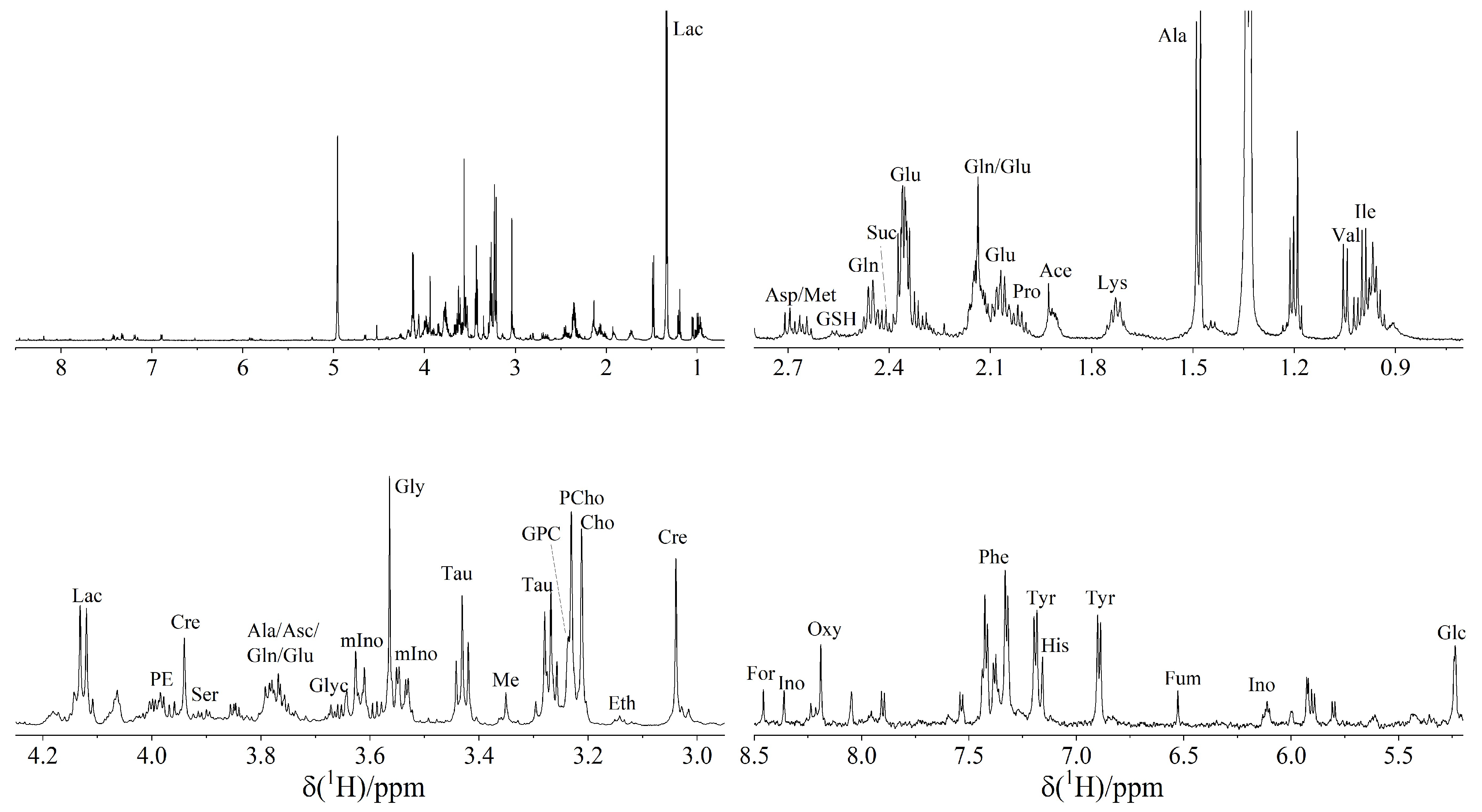
3.1. 1H NMR Spectroscopy Reveals 37 Metabolites in TETs
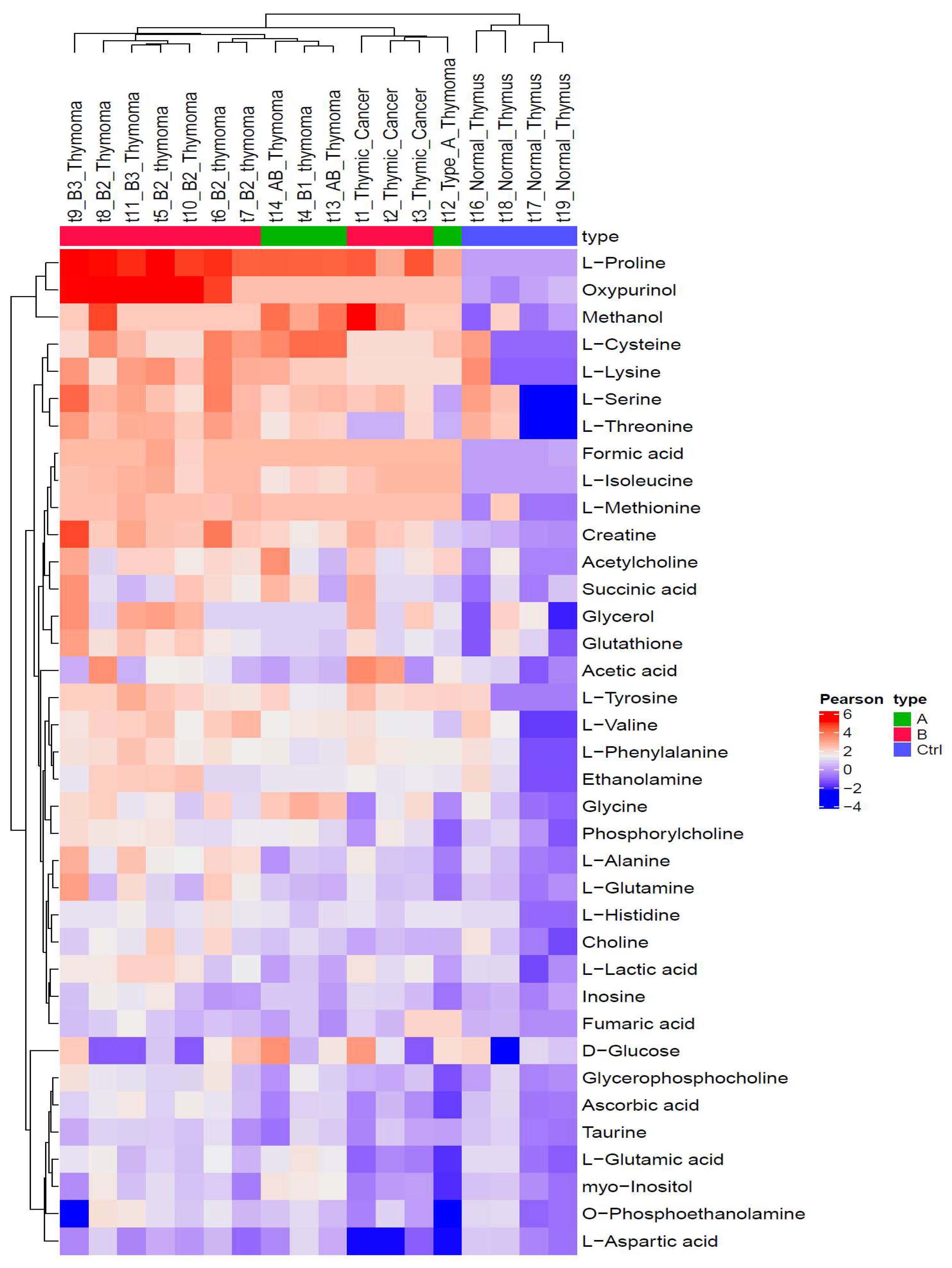
3.2. Metabolic Profiles Are Closely Associated with WHO Histotypes
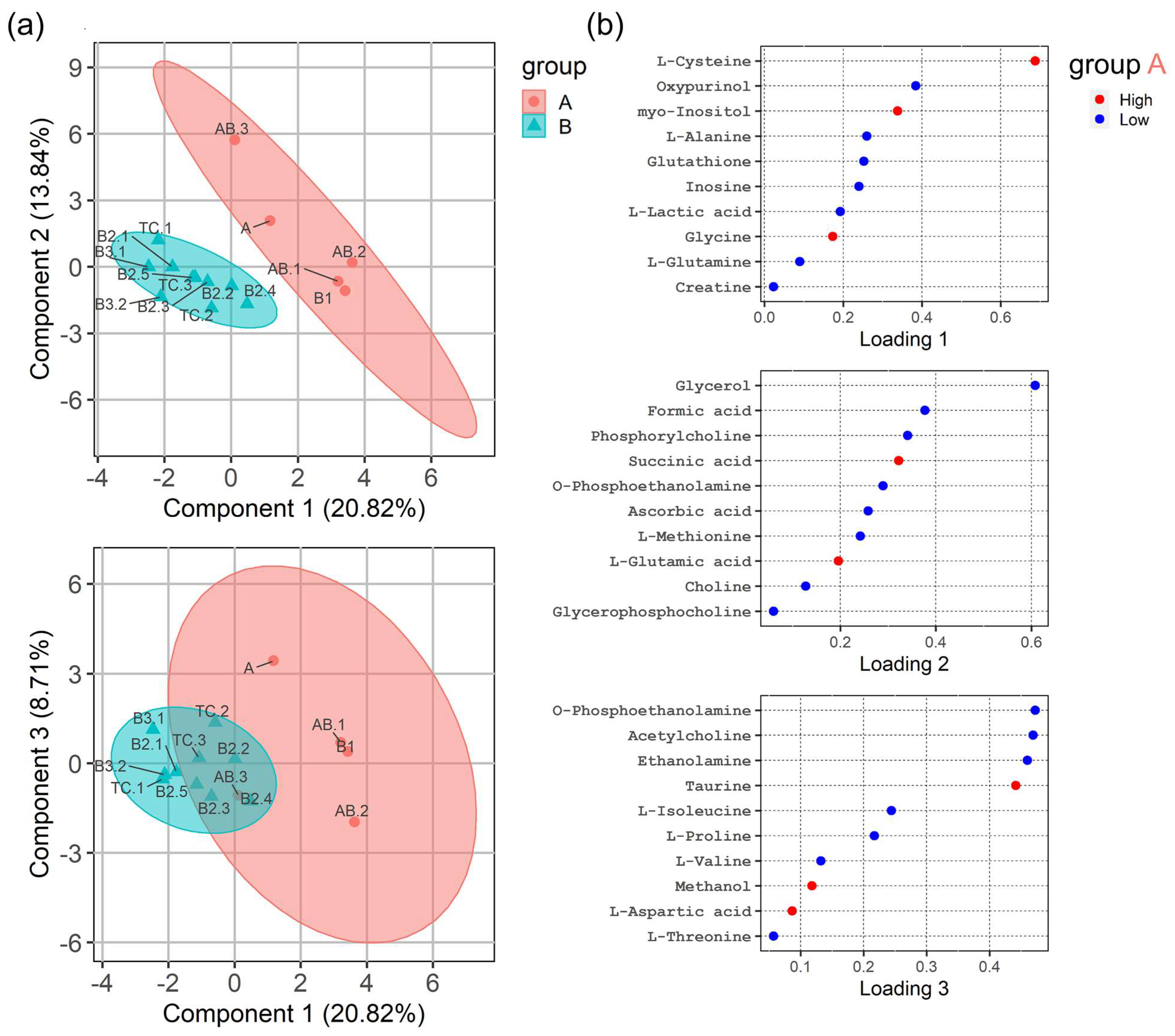
3.3. HRMAS 1H-NMR Analysis Discriminates ‘Indolent’ from ’Aggressive’ Groups of TETs
3.4. HRMAS 1H-NMR Analysis Reveals Differentially Activated Metabolic Pathways in TETs
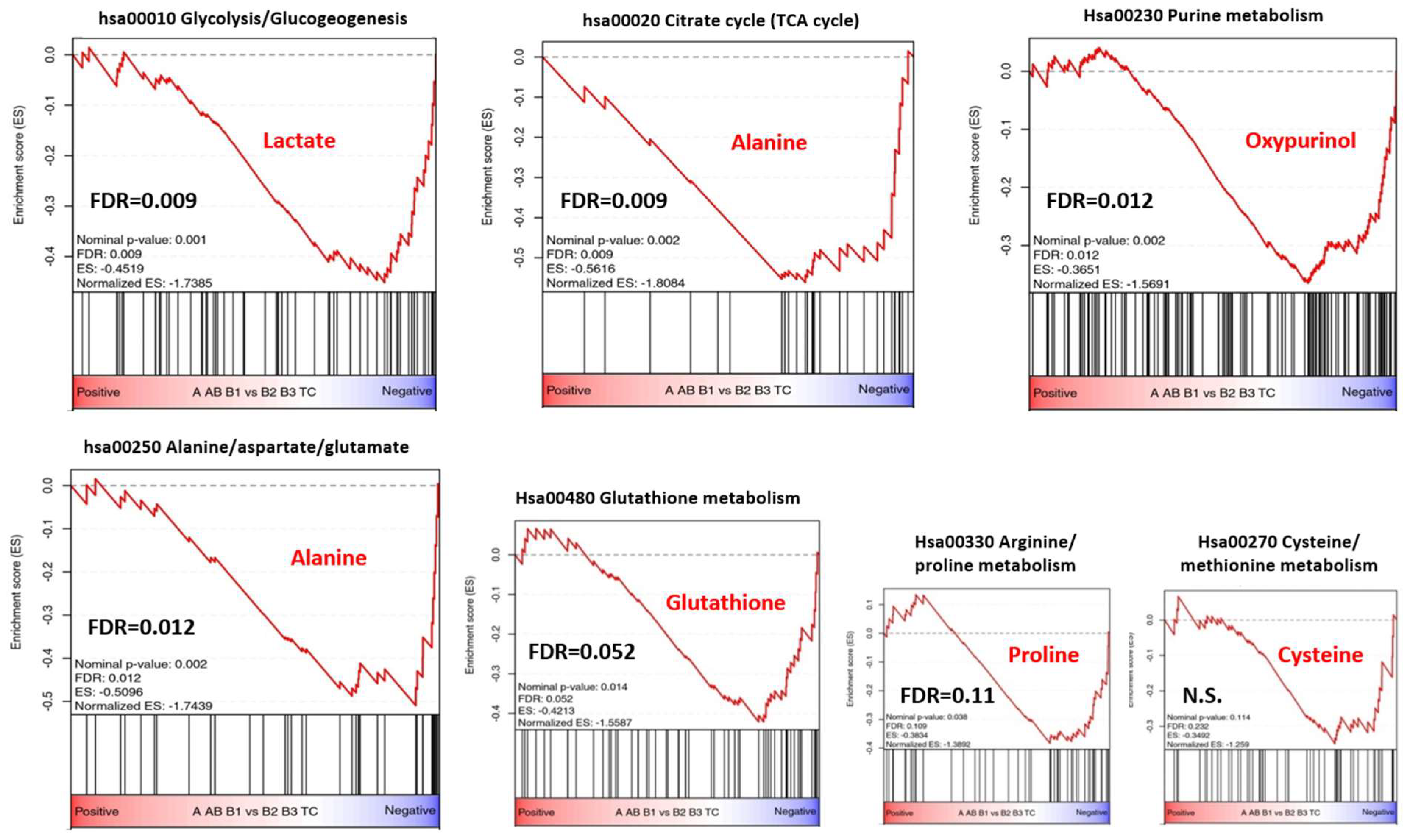
3.5. Metabolic Gene Set Enrichment Analysis of the TCGA Transcriptomic Dataset
4. Discussion
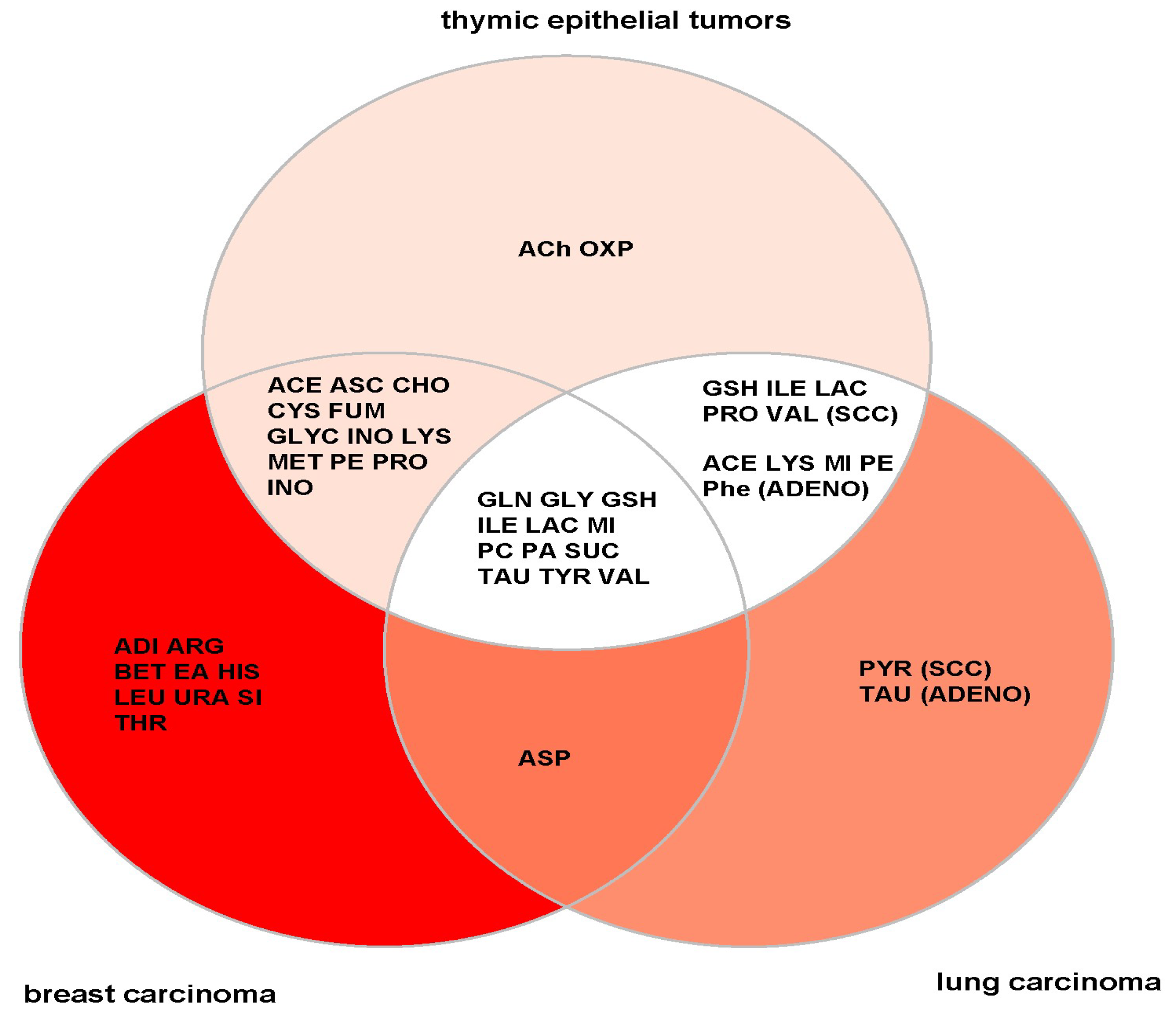
4.1. General versus TET-Specific Metabolites
4.2. Metabolite Profiles Meet KEGG Pathway and Gene Expression
4.3. Therapeutic Perspectives
4.4. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marx, A.; Chan, J.K.C.; Chalabreysse, L.; Dacic, S.; Detterbeck, F.; French, C.A.; Hornick, J.L.; Inagaki, H.; Jain, D.; Lazar, A.J.; et al. The 2021 WHO Classification of Tumors of the Thymus and Mediastinum: What Is New in Thymic Epithelial, Germ Cell, and Mesenchymal Tumors? J. Thorac. Oncol. 2021, 17, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I.; Meltzer, P.S.; Kim, I.-K.; Lucchi, M.; Park, K.-S.; Fontanini, G.; Gao, J.; Zucali, A.P.; Calabrese, F.; Favaretto, A.; et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nat. Genet. 2014, 46, 844–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belharazem, D.; Grass, A.; Paul, C.; Vitacolonna, M.; Schalke, B.; Rieker, R.J.; Körner, D.; Jungebluth, P.; Simon-Keller, K.; Hohenberger, P.; et al. Increased cFLIP expression in thymic epithelial tumors blocks autophagy via NF-κB signalling. Oncotarget 2017, 8, 89580–89594. [Google Scholar] [CrossRef] [Green Version]
- Radovich, M.; Pickering, C.R.; Felau, I.; Ha, G.; Zhang, H.; Jo, H.; Hoadley, K.A.; Anur, P.; Zhang, J.; McLellan, M.; et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell 2018, 33, 244–258.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Weis, C.-A.; Thelen, J.; Sticht, C.; Schalke, B.; Ströbel, P.; Marx, A. Thymoma Associated Myasthenia Gravis (TAMG): Differential Expression of Functional Pathways in Relation to MG Status in Different Thymoma Histotypes. Front. Immunol. 2020, 11, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Lin, X.; Cai, M.; Zheng, X.; Lian, L.; Fan, D.; Wu, X.; Lan, P.; Wang, J. Overexpression of Hexokinase 1 as a poor prognosticator in human colorectal cancer. Tumor Biol. 2016, 37, 3887–3895. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bingöl, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Miao, Y.; Liu, M.; Zeng, Y.; Gao, Z.; Peng, D.; Hu, B.; Li, X.; Zheng, Y.; Xue, Y.; et al. Pan-Cancer Analysis Reveals the Functional Importance of Protein Lysine Modification in Cancer Development. Front. Genet. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L.; Pineda-Lucena, A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Dong, B.; Ning, J.; Shao, X.; Zhao, L.; Jiang, Q.; Ji, H.; Cai, A.; Xue, W.; Gao, H. NMR-based metabolomics analysis identifies discriminatory metabolic disturbances in tissue and biofluid samples for progressive prostate cancer. Clin. Chim. Acta 2020, 501, 241–251. [Google Scholar] [CrossRef]
- Bruzzone, C.; Loizaga-Iriarte, A.; Sánchez-Mosquera, P.; Gil-Redondo, R.; Astobiza, I.; Diercks, T.; Cortazar, A.R.; Ugalde-Olano, A.; Schäfer, H.; Blanco, F.J.; et al. 1H NMR-Based Urine Metabolomics Reveals Signs of Enhanced Carbon and Nitrogen Recycling in Prostate Cancer. J. Proteome Res. 2020, 19, 2419–2428. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current metabolomics: Technological advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Thurtle, D.R.; Jenkins, V.; Pharoah, P.D.; Gnanapragasam, V.J. Understanding of prognosis in non-metastatic prostate cancer: A randomised comparative study of clinician estimates measured against the PREDICT prostate prognostic model. Br. J. Cancer 2019, 121, 715–718. [Google Scholar] [CrossRef] [Green Version]
- Detterbeck, F.C.; Nicholson, A.G.; Kondo, K.; van Schil, P.; Moran, C. The Masaoka-Koga stage classification for thymic malignancies: Clarification and definition of terms. J. Thorac. Oncol. 2011, 6, S1710–S1716. [Google Scholar] [CrossRef] [Green Version]
- Girard, N.; Ruffini, E.; Marx, A.; Faivre-Finn, C.; Peters, S. Thymic epithelial tumors: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. 5), v40–v55. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.H.; Palma, D.A.; Camidge, D.R.; Jones, B.L.; Robin, T.P.; Sher, D.J.; Koshy, M.; Kavanagh, B.D.; Gaspar, L.E.; Rusthoven, C.G. The Impact of Postoperative Radiotherapy for Thymoma and Thymic Carcinoma. J. Thorac. Oncol. 2017, 17, 734–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. Thymomas and Thymic Carcinomas. In National Comprehensive Cancer Network: Version 1.2021–December 4, 2020; NCCN Guidelines; NCCN: Plymouth Meeting, PA, USA, 2021. [Google Scholar]
- Conforti, F.; Pala, L.; Giaccone, G.; de Pas, T. Thymic epithelial tumors: From biology to treatment. Cancer Treat. Rev. 2020, 86, 102014. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Girard, N.; Marx, A. State of the Art of Genetic Alterations in Thymic Epithelial Tumors. J. Thorac. Oncol. 2014, 9, S131–S136. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.; Rajan, A.; Berman, A.; Tomita, Y.; Brzezniak, C.; Lee, M.J.; Lee, S.; Ling, A.; Spittler, A.J.; Carter, C.A.; et al. Sunitinib in patients with chemotherapy-refractory thymoma and thymic carcinoma: An open-label phase 2 trial. Lancet Oncol. 2015, 16, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Sato, J.; Satouchi, M.; Itoh, S.; Okuma, Y.; Niho, S.; Mizugaki, H.; Murakami, H.; Fujisaka, Y.; Kozuki, T.; Nakamura, K.; et al. Lenvatinib in patients with advanced or metastatic thymic carcinoma (REMORA): A multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 843–850. [Google Scholar] [CrossRef]
- Zucali, P.A.; De Pas, T.; Palmieri, G.; Favaretto, A.; Chella, A.; Tiseo, M.; Caruso, M.; Simonelli, M.; Perrino, M.; De Vincenzo, F.; et al. Phase II Study of Everolimus in Patients With Thymoma and Thymic Carcinoma Previously Treated with Cisplatin-Based Chemotherapy. J. Clin. Oncol. 2018, 36, 342–349. [Google Scholar] [CrossRef]
- Cho, J.; Kim, H.S.; Ku, B.M.; Choi, Y.L.; Cristescu, R.; Han, J.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; et al. Pembrolizumab for Patients With Refractory or Relapsed Thymic Epithelial Tumor: An Open-Label Phase II Trial. J. Clin. Oncol. 2019, 37, 2162–2170. [Google Scholar] [CrossRef] [PubMed]
- Giaccone, C.; Kim, G.; Thompson, J.; McGuire, C.; Kallakury, B.; Chahine, J.J.; Manning, M.; Mogg, R.; Blumenschein, W.M.; Tan, M.T.; et al. Pembrolizumab in patients with thymic carcinoma: A single-arm, single-centre, phase 2 study. Lancet Oncol. 2018, 19, 347–355. [Google Scholar] [CrossRef]
- Rajan, A.; Heery, R.C.; Thomas, A.; Mammen, A.L.; Perry, S.; O’Sullivan Coyne, G.; Guha, U.; Berman, A.; Szabo, E.; Madan, R.A.; et al. Efficacy and tolerability of anti-programmed death-ligand 1 (PD-L1) antibody (Avelumab) treatment in advanced thymoma. J. Immunother. Cancer 2019, 7, 269. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Mullenix, C.; Sheelat, M.; Zhao, C. The role of immunotherapy for management of advanced thymic epithelial tumors: A narrative review. Mediastinum 2021, 5, 23. [Google Scholar] [CrossRef]
- Emwas, A.H. The strengths and weaknesses of NMR spectrocopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [PubMed]
- Griffin, J.; Shockcor, J. Metabolic profiles of cancer cells. Nat. Rev. Cancer 2004, 4, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Gogiashvili, M.; Nowacki, J.; Hergenröder, R.; Hengstler, J.G.; Lambert, J.; Edlund, K. HR-MAS NMR Based Quantitative Metabolomics in Breast Cancer. Metabolites 2019, 9, 19. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.L.; Lean, C.L.; Bogdanova, A. Enhanced resolution of proton NMR spectra of malignant lymph nodes using magic-angle spinning. Magn. Reson. Med. 1996, 36, 653–658. [Google Scholar] [CrossRef]
- Duarte, I.F.; Stanley, E.G.; Holmes, E. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal. Chem. 2005, 77, 5570–5578. [Google Scholar] [CrossRef] [PubMed]
- Sitter, B.; Lundgren, S.; Bathen, T.F. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006, 19, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.W.; He, W.; Halpern, E.F.; Wu, C.L.; Cheng, L.L. Evaluation of Tissue Metabolites with High Resolution Magic Angle Spinning MR Spectroscopy Human Prostate Samples after Three-Year Storage at −80 °C. Biomark. Insights 2007, 2, 117727190700200006. [Google Scholar] [CrossRef]
- AlWahsh, M.; Othman, A.; Hamadneh, L.; Telfah, A.; Lambert, J.; Hikmat, S.; Alassi, A.; Mohamed, F.E.Z.; Hergenröder, R.; Al-Qirim, T.; et al. Second Exposure to Acetaminophen Overdose Is Associated With Liver Fibrosis in Mice. EXCLI J. 2018, 18, 51–62. [Google Scholar]
- Beckonert, O.; Coen, M.; Keun, H.C.; Wang, Y.; Ebbels, T.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Prot. 2010, 5, 1019–1032. [Google Scholar] [CrossRef]
- Akoka, S.; Barantin, L.; Trierweiler, M. Concentration Measurement by Proton NMR Using the ERETIC Method. Anal. Chem. 1999, 71, 2554–2557. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Prot. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.-A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [Green Version]
- Krzanowski, W.J.; Hand, D.J. ROC Curves for Continuous Data; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, C.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Sergushichev, A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 2016, 60012, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Geistlinger, L.; Csaba, G.; Zimmer, R. Bioconductor’s EnrichmentBrowser: Seamless navigation through combined results of set- & network-based enrichment analysis. BMC Bioinform. 2016, 17, 45. [Google Scholar] [CrossRef] [Green Version]
- Kaji, S.; Irino, T.; Kusuhara, M.; Makuuchi, R.; Yamakawa, Y.; Tokunaga, M.; Tanizawa, Y.; Bando, E.; Kawamura, T.; Kami, K.; et al. Metabolomic profiling of gastric cancer tissues identified potential biomarkers for predicting peritoneal recurrence. Gastric Cancer 2020, 23, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Spakova, I.; Rabajdova, M.; Mickova, H.; Graier, W.F.; Mareková, M. Effect of hypoxia factors gene silencing on ROS production and metabolic status of A375 malignant melanoma cells. Sci. Rep. 2021, 11, 10325. [Google Scholar] [CrossRef]
- Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Baek, H.-M.; Kim, S.; Kim, M.J.; Youk, J.H.; Moon, H.J.; Kim, E.K.; Han, K.H.; Kim, D.H.; Kim, S.I.; et al. HR-MAS MR spectroscopy of breast cancer tissue obtained with core needle biopsy: Correlation with prognostic factors. PLoS ONE 2012, 7, e51712. [Google Scholar] [CrossRef] [PubMed]
- Park, V.Y.; Yoon, D.; Koo, J.S.; Kim, E.-K.; Kim, S.I.; Choi, J.S.; Park, S.; Park, H.S.; Kim, S.; Kim, M.J. Intratumoral Agreement of High-Resolution Magic Angle Spinning Magnetic Resonance Spectroscopic Profiles in the Metabolic Characterization of Breast Cancer. Medicine 2016, 95, e3398. [Google Scholar] [CrossRef]
- Gogiashvili, M.; Horsch, S.; Marchan, R.; Gianmoena, K.; Cadenas, C.; Tanner, B.; Naumann, S.; Ersova, D.; Lippek, F.; Rahnenführer, J.; et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 2018, 31, e3862. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J. Metabolic implications of non-Electrogenic ATP/ADP exchange in Cancer cells: A mechanistic basis for the Warburg effect. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148410. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.-E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Bizzarri, M.; Dinicola, S.; Bevilacqua, A.; Cucina, A. Broad Spectrum Anticancer Activity of Myo-Inositol and Inositol Hexakisphosphate. Int. J. Endocrinol. 2016, 2016, 5616807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daher, B.; Vučetić, M.; Pouysségur, J. Cysteine Depletion, a Key Action to Challenge Cancer Cells to Ferroptotic Cell Death. Front. Oncol. 2020, 10, 723. [Google Scholar] [CrossRef] [PubMed]
- Bamji-Stocke, S.; van Berkel, V.; Miller, D.M.; Frieboes, H.B. A review of metabolism-associated biomarkers in lung cancer diagnosis and treatment. Metabolomics 2018, 14, 81. [Google Scholar] [CrossRef]
- Marx, A.; Yamada, Y.; Simon-Keller, K.; Schalke, B.; Willcox, N.; Ströbel, P.; Weis, C.-A. Thymus and autoimmunity. Semin. Immunopathol. 2021, 43, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.; Lips, K.S. The non-neuronal cholinergic system in health and disease. Pharmacology 2013, 92, 286–302. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Bai, P.; Xia, Y.; Lai, Q.W.S.; Guo, M.S.S.; Dai, K.; Zheng, Z.; Ling, C.S.J.; Dong, T.T.X.; Pi, R.; et al. Solar light induces expression of acetylcholinesterase in skin keratinocytes: Signalling mediated by activator protein 1 transcription factor. Neurochem. Int. 2020, 141, 104861. [Google Scholar] [CrossRef]
- Wilson, F.P.; Berns, J.S. Onco-nephrology: Tumor lysis syndrome. Clin. J. Am. Soc. Nephrol. 2012, 7, 1730–1739. [Google Scholar] [CrossRef] [Green Version]
- Fridley, B.; Batzler, A.; Li, L.; Li, F.; Matimba, A.; Jenkins, G.; Ji, Y.; Wang, L.; Weinshilboum, R.M. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet. Genom. 2011, 21, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [Green Version]
- Baresova, V.; Skopova, V.; Sikora, J.; Patterson, D.; Sovova, J.; Zikanova, M.; Kmoch, S. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum. Mol. Genet. 2011, 21, 1534–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taha-Mehlitz, S.; Bianco, G.; Coto-Llerena, M.; Kancherla, V.; Bantug, G.R.; Gallon, J.; Ercan, C.; Panebianco, F.; Eppenberger-Castori, S.; von Strauss, M.; et al. Adenylosuccinate lyase is oncogenic in colorectal cancer by causing mitochondrial dysfunction and independent activation of NRF2 and mTOR-MYC-axis. Theranostics 2021, 11, 4011–4021. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Gillespie, D.G.; Reichert, T.E.; Jackson, E.K.; Whiteside, T.L. Purine Metabolites in Tumor-Derived Exosomes May Facilitate Immune Escape of Head and Neck Squamous Cell Carcinoma. Cancers 2020, 12, 1602. [Google Scholar] [CrossRef] [PubMed]
- Dumanskiy, Y.V.; Stoliarova, O.Y.; Syniachenko, O.V.; Aliev, R.F.; Iermolaeva, M.V.; Sokrut, O.P. Comparative evaluation of purine dysmetabolism in gastric and pulmonary adenocarcinomas. Exp. Oncol. 2020, 42, 220–223. [Google Scholar] [CrossRef]
- de Montpréville, V.T.; Quilhot, P.; Chalabreysse, L.; de Muret, A.; Hofman, V.; Lantuéjoul, S.; Parrens, M.; Payan, M.J.; Rouquette, I.; Secq, V.; et al. Glut-1 intensity and pattern of expression in thymic epithelial tumors are predictive of WHO subtypes. Pathol. Res. Pract. 2015, 211, 996–1002. [Google Scholar] [CrossRef]
- Kaira, K.; Endo, M.; Abe, M.; Nakagawa, K.; Ohde, Y.; Okumura, T.; Takahashi, T.; Murakami, H.; Tsuya, A.; Nakamura, Y.; et al. Biologic correlation of 2-18F-fluoro-2-deoxy-D-glucose uptake on positron emission tomography in thymic epithelial tumors. J. Clin. Oncol. 2010, 28, 3746–3753. [Google Scholar] [CrossRef]
- Kaira, K.; Abe, M.; Nakagawa, K.; Ohde, Y.; Okumura, T.; Takahashi, T.; Murakami, H.; Shukuya, T.; Kenmotsu, H.; Naito, T.; et al. 18F-FDG uptake on PET in primary mediastinal non-thymic neoplasm: A clinicopathological study. Eur. J. Radiol. 2012, 81, 2423–2429. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Marx, A.; Belharazem, D.; Lee, D.-H.; Popovic, Z.V.; Reißfelder, C.; Schalke, B.; Schölch, S.; Ströbel, P.; Weis, C.A.; Yamada, Y. Molecular pathology of thymomas: Implications for diagnosis and therapy. Virchows Arch. 2021, 478, 101–110. [Google Scholar] [CrossRef]
- D’Aniello, C.; Patriarca, E.J.; Phang, J.M.; Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front. Oncol. 2020, 10, 776. [Google Scholar] [CrossRef]
- Phang, J.M. Proline Metabolism in Cell Regulation and Cancer Biology: Recent Advances and Hypotheses. Antioxid. Redox Signal. 2019, 30, 635–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-W.; Hua, K.-T.; Li, K.-C.; Kao, H.-F.; Hong, R.-L.; Ko, J.-Y.; Hsiao, M.; Kuo, M.L.; Tan, C.T. Histone Methyltransferase G9a Drives Chemotherapy Resistance by Regulating the Glutamate-Cysteine Ligase Catalytic Subunit in Head and Neck Squamous Cell Carcinoma. Mol. Cancer Ther. 2017, 16, 1421–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, E.M.; Bogner, A.N.; Vandekeere, A.; Tam, G.S.; Patel, S.M.; Becker, D.F.; Fendt, S.M.; Tanner, J.J. In crystallo screening for proline analog inhibitors of the proline cycle enzyme PYCR1. J. Biol. Chem. 2020, 295, 18316–18327. [Google Scholar] [CrossRef]
- Narayanan, D.; Ma, S.; Özcelik, D. Targeting the Redox Landscape in Cancer Therapy. Cancers 2020, 12, 1706. [Google Scholar] [CrossRef]
- van Loenhout, J.; Peeters, M.; Bogaerts, A.; Smits, E.; Deben, C. Oxidative Stress-Inducing Anticancer Therapies: Taking a Closer Look at Their Immunomodulating Effects. Antioxidants 2020, 9, 1188. [Google Scholar] [CrossRef]
- Zhang, J.; Duan, D.; Song, Z.-L.; Liu, T.; Hou, Y.; Fang, J. Small molecules regulating reactive oxygen species homeostasis for cancer therapy. Med. Res. Rev. 2021, 41, 342–394. [Google Scholar] [CrossRef]
- Sukjoi, W.; Ngamkham, J.; Attwood, P.V.; Jitrapakdee, S. Targeting Cancer Metabolism and Current Anti-Cancer Drugs. Adv. Exp. Med. Biol. 2021, 1286, 15–48. [Google Scholar] [CrossRef]
- Akins, N.S.; Nielson, T.C.; Le, H.V. Inhibition of Glycolysis and Glutaminolysis: An Emerging Drug Discovery Approach to Combat Cancer. Curr. Top. Med. Chem. 2018, 18, 494–504. [Google Scholar] [CrossRef]
- Jin, H.; Wang, S.; Zaal, E.A.; Wang, C.; Wu, H.; Bosma, A.; Jochems, F.; Isima, N.; Jin, G.; Lieftink, C.; et al. A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. eLife 2020, 9, e56749. [Google Scholar] [CrossRef]
- Calvert, A.E.; Chalastanis, A.; Wu, Y.; Hurley, L.A.; Kouri, F.M.; Bi, Y.; Kachman, M.; May, J.L.; Bartom, E.; Hua, Y.; et al. Cancer-Associated IDH1 Promotes Growth and Resistance to Targeted Therapies in the Absence of Mutation. Cell Rep. 2017, 19, 1858–1873. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.S.; Gupta, V.K.; Garrido, V.T.; Hadad, R.; Durden, B.C.; Kesh, K.; Giri, B.; Ferrantella, A.; Dudeja, V.; Saluja, A.; et al. Targeting tumor-intrinsic hexosamine biosynthesis sensitizes pancreatic cancer to anti-PD1 therapy. J. Clin. Investig. 2020, 130, 451–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Chen, S.; Liu, W.; Ma, Y.; Li, J.; Fisher, P.B.; Fang, X.; Wang, X.Y. Immunometabolism: A new target for improving cancer immunotherapy. Adv. Cancer Res. 2019, 143, 195–253. [Google Scholar] [CrossRef]
- Soreq, H. Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci. 2015, 38, 448–458. [Google Scholar] [CrossRef] [PubMed]

| Diagnosis | N | Age (Years) | Sex (M/F) | MG Status | Stage (I–IV) | Lymphocyte Content (%) |
|---|---|---|---|---|---|---|
| Type A | 1 | 77 | M | − | n.k. | 10 |
| Type AB | 3 (AB.1–AB.3) * | 46 77 77 | M M M | − + − | II II II | 30 60 30 |
| Type B1 | 1 | 72 | M | − | III | 90 |
| Type B2 | 5 (B2.1–B2.5) * | 75 73 51 72 73 | F F M F F | + + − − n.k. | III III IVa IVa III | 60 60 80 70 80 |
| Type B3 | 2 (B3.1 and B3.2) * | 80 58 | F M | − − | I II | 10 20 |
| TC | 3 (TC.1–TC.3) * | 75 69 78 | M M M | − − − | III II II | 10 10 10 |
| NT | 4 (NT.1–NT.4) * | 27 ** 42 39 20 | M M *** F ** F * | − − + + | 90 90 90 90 |
| Key Metabolite | MetPA-Derived KEGG Pathway | Assumed Function | p-Value | Impact |
|---|---|---|---|---|
| Cysteine | Cysteine/methionine metabolism | Transulfuration Pathway | <0.001 | 0.22 |
| Glycine | Glycine, serine and threonine metabolism | Homocysteine cycle | 0.012 | 0.46 |
| Glutathione | Glutathione metabolism | Redox state | 0.013 | 0.37 |
| Alanine | Alanine, aspartate and glutamate metabolism | TCA cycle | 0.022 | 0.54 |
| Lactate | Pyruvate metabolism | Glycolysis | 0.029 | 0.14 |
| Glutamine | Glutamine/glutamate metabolism | Glutaminolysis | 0.032 | 0.50 |
| Key Metabolite(s) | KEGG Pathways | KEGG-ID | p-Value | Fold Enrichment | FDR | Genes * |
|---|---|---|---|---|---|---|
| Glutathione | Glutathione metabolism | hsa00480 | 1.3 × 10−9 | 25.3 | 1.9 × 10−8 | GSTK1, GCLC, GGT6, GPX4, ANPEP, IDH1, 2, MGST1, MGST2, LAP3 |
| Alanine | Alanine, aspartate and glutamate metabolism | hsa00250 | 1 × 10−7 | 28.7 | 1.1 × 10−11 | ALDH4A1, ADSL, FOLH1, GOT1, RIMKLA, NIT2, ASS1 |
| Proline | Arginine andproline metabolism | hsa00330 | 2.9 × 10−11 | 28.7 | 5.8 × 10−10 | ALDH4A1, GOT1, P4HA1, NOS2, NOS3, ARG1, PYCR1, LAP3, PRODH, ALDH9A1 |
| Glucose and lactic acid | Glycolysis/Gluconeogenesis | hsa00010 | 5.3 × 10−6 | 15 | 3.9 × 10−5 | HK3, PKM, ALDOA, FBP1, PCK2, HK1, ALDH9A1, SLC2A1, SLC16A3, SLC16A14 |
| Oxypurinol | PurineMetabolism ** | hsa00230 | 4.8 × 10−4 | 89.6 | 3.2 × 10−3 | NT5C3A, ADCY10, ADSL ***, PKM, PNP, IMPDH1, NUDT2, HPRT1, DCK, GART |
| Cysteine | Cysteine and methionine metabolism | hsa00270 | 2.1 × 10−3 | 15.1 | 1.1 × 10−3 | SDS, GOT1, MDH1, CBS, SLC1A5, SLC3A2 |
| Glycine | Glycine, serineand threonine metabolism | hsa00260 | 2.3 × 10−3 | 14.7 | 1.1 × 10−3 | SDS, SHMT2, CBS, PSPH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alwahsh, M.; Knitsch, R.; Marchan, R.; Lambert, J.; Hoerner, C.; Zhang, X.; Schalke, B.; Lee, D.-H.; Bulut, E.; Graeter, T.; et al. Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets. Cancers 2022, 14, 1564. https://doi.org/10.3390/cancers14061564
Alwahsh M, Knitsch R, Marchan R, Lambert J, Hoerner C, Zhang X, Schalke B, Lee D-H, Bulut E, Graeter T, et al. Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets. Cancers. 2022; 14(6):1564. https://doi.org/10.3390/cancers14061564
Chicago/Turabian StyleAlwahsh, Mohammad, Robert Knitsch, Rosemarie Marchan, Jörg Lambert, Christian Hoerner, Xiaonan Zhang, Berthold Schalke, De-Hyung Lee, Elena Bulut, Thomas Graeter, and et al. 2022. "Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets" Cancers 14, no. 6: 1564. https://doi.org/10.3390/cancers14061564
APA StyleAlwahsh, M., Knitsch, R., Marchan, R., Lambert, J., Hoerner, C., Zhang, X., Schalke, B., Lee, D.-H., Bulut, E., Graeter, T., Ott, G., Kurz, K. S., Preissler, G., Schölch, S., Farhat, J., Yao, Z., Sticht, C., Ströbel, P., Hergenröder, R., ... Belharazem, D. (2022). Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets. Cancers, 14(6), 1564. https://doi.org/10.3390/cancers14061564








