Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements
Abstract
Simple Summary
Abstract
1. Introduction
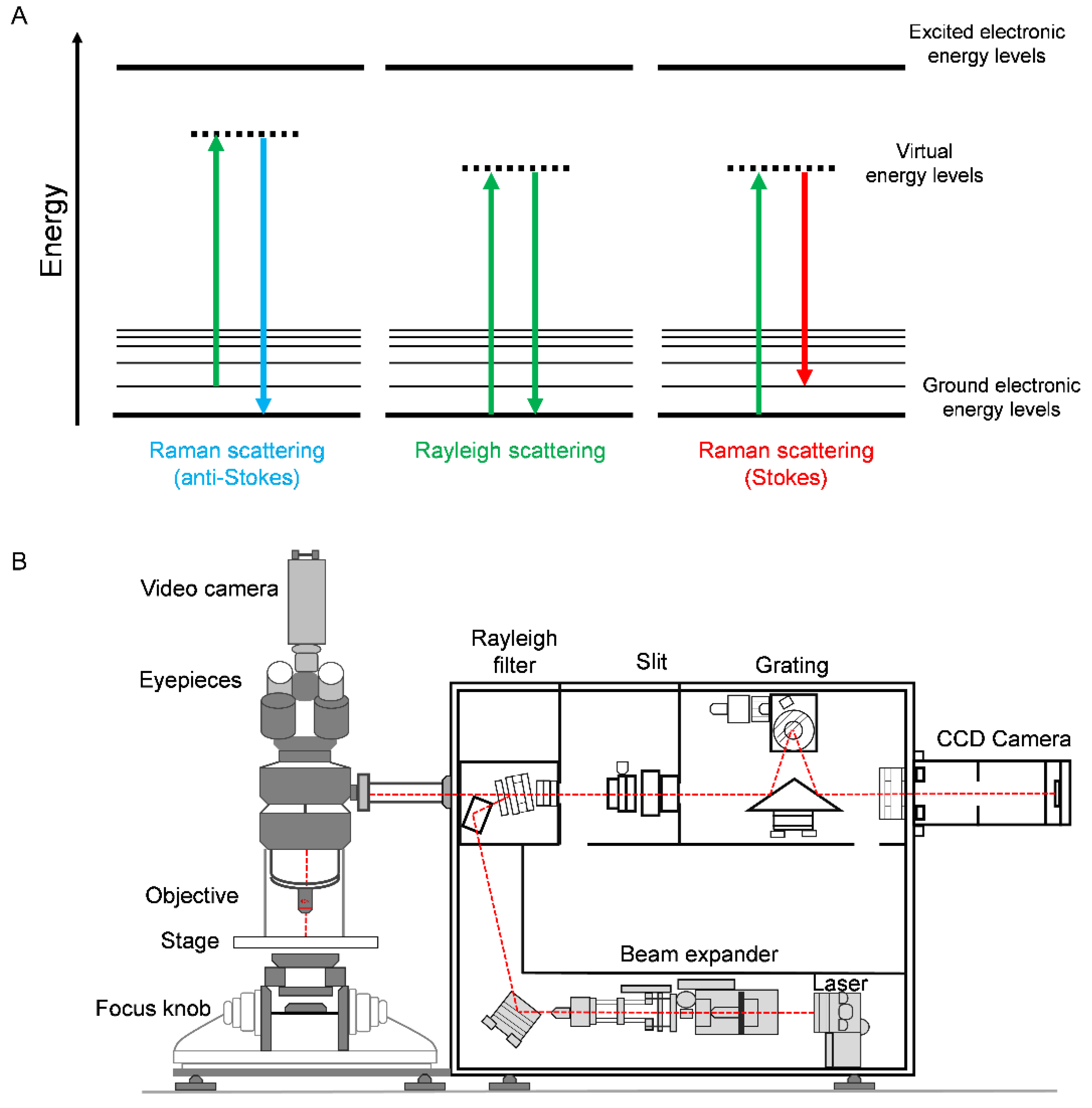
2. Principles of Raman Spectroscopy
3. Enhanced Raman Techniques
3.1. Resonance Raman Scattering (RRS)
3.2. Surface-Enhanced Raman Scattering (SERS)
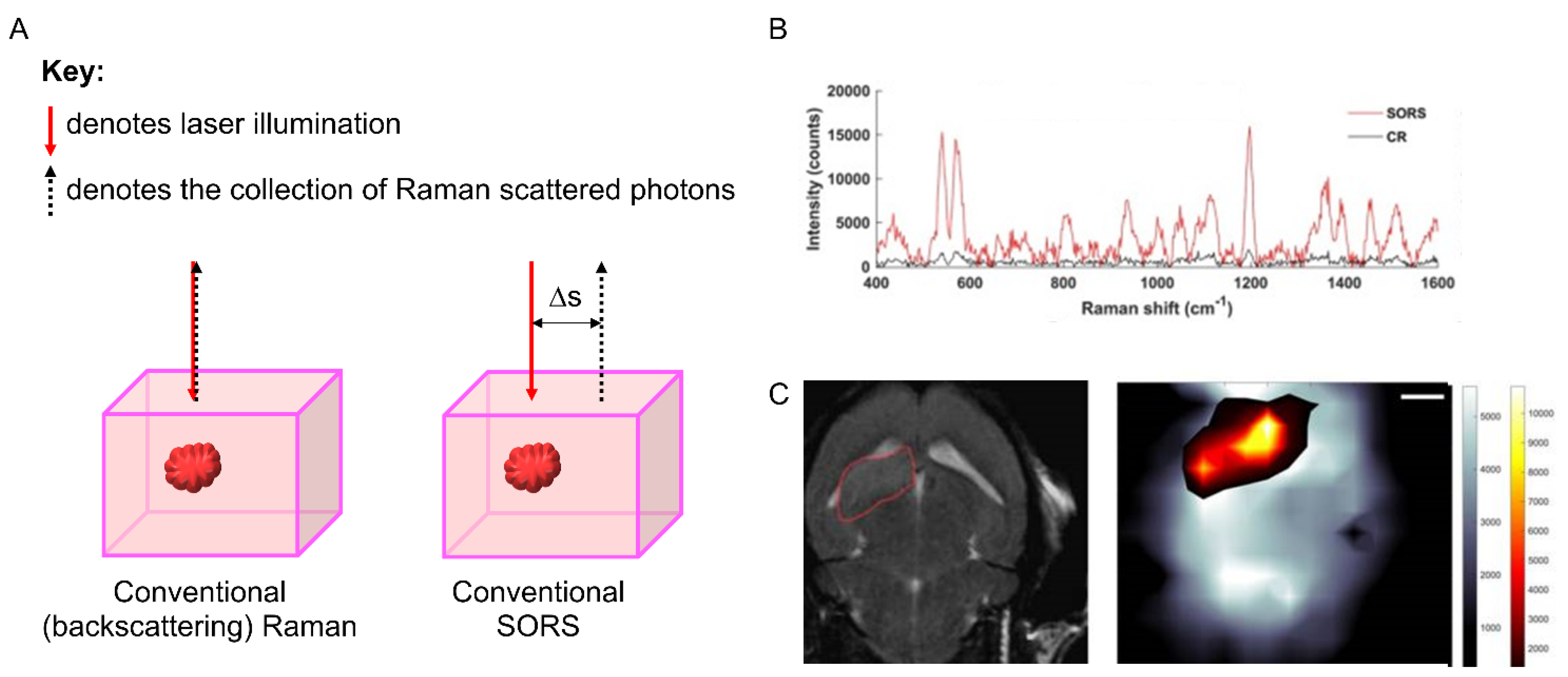
3.3. Spatially Offset Raman Scattering (SORS)
3.4. Coherent Raman Scattering (CRS): Coherent Anti-Stokes Raman Scattering (CARS) and Stimulated Raman Scattering (SRS)
4. Sample Preparation
5. Spectral Analysis and Multivariate Techniques
6. Raman Spectroscopy in Preclinical Prostate Cancer Models and Clinical Tumours
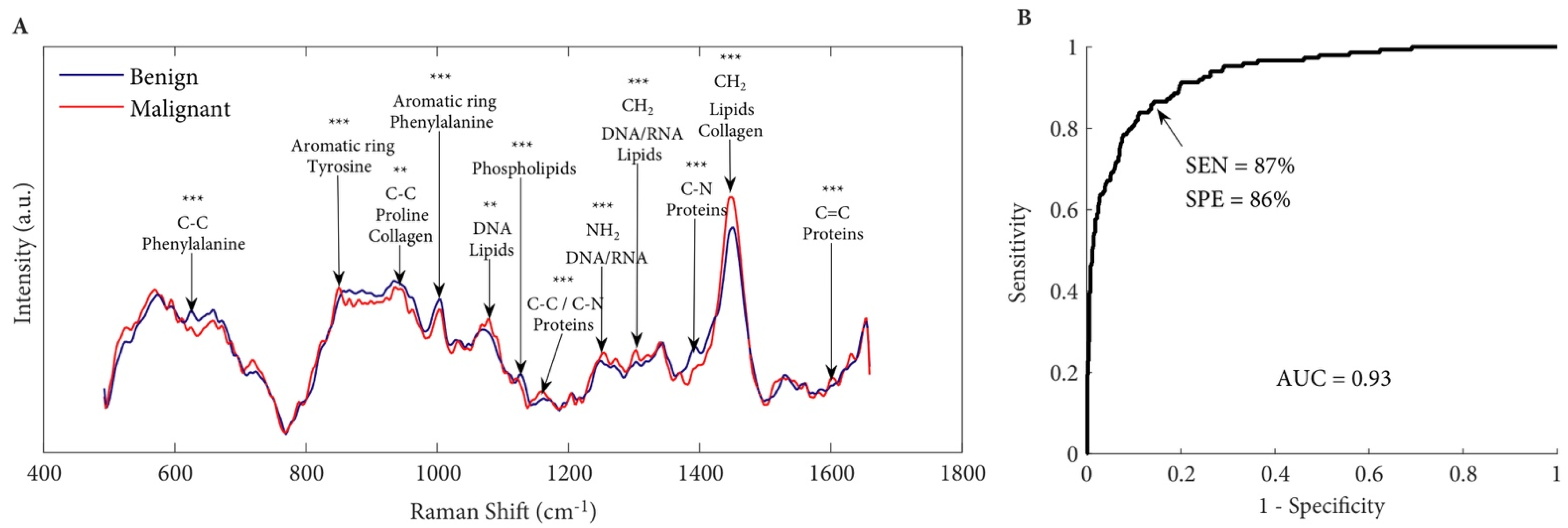
6.1. Detecting Prostate Cancer at the Tissue Level
6.2. Treating Residual or Recurrent Microscopic Disease
6.3. Raman-Based Analysis of Castration-Resistant Prostate Cancer (CRPC)
6.4. Analysing Tumour Microenvironment of Lymphatics and Bony Metastasis
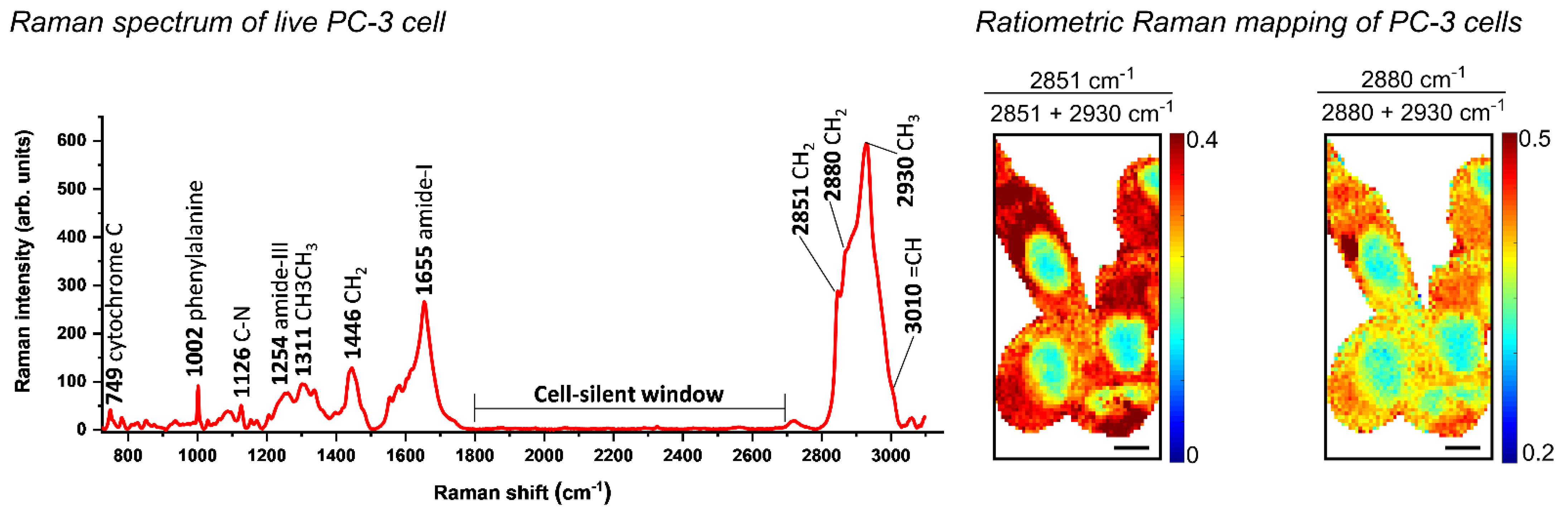
6.5. Raman-Based Analysis to Characterise Altered Lipid Metabolism in Prostate Carcinogenesis
6.6. Analysis of Biofluids as Liquid Biopsies in Detecting Prostate Cancer
7. Discussion
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADT | Androgen deprivation therapy |
| CRPC | Castration-resistant prostate cancer |
| cfDNA | Cell-free DNA |
| CTCs | Circulating tumour cells |
| CNN | Convolutional neural network |
| EVs | Extracellular vesicles |
| NIR | Near Infrared |
| PLND | Pelvic lymph node dissection |
| RRS | Resonance Raman scattering |
| SERS | Surface-enhanced Raman spectroscopy |
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Tipping, W.J.; Lee, M.; Serrels, A.; Brunton, V.G.; Hulme, A.N. Stimulated Raman scattering microscopy: An emerging tool for drug discovery. Chem. Soc. Rev. 2016, 45, 2075–2089. [Google Scholar] [CrossRef]
- Jamieson, L.E.; Wetherill, C.; Faulds, K.; Graham, D. Ratiometric Raman imaging reveals the new anti-cancer potential of lipid targeting drugs. Chem. Sci. 2018, 9, 6935–6943. [Google Scholar] [CrossRef]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA 2011, 109, 28–32. [Google Scholar] [CrossRef]
- Sloan-Dennison, S.; Laing, S.; Graham, D.; Faulds, K. From Raman to SESORRS: Moving deeper into cancer detection and treatment monitoring. Chem. Commun. 2021, 57, 12436–12451. [Google Scholar] [CrossRef]
- Phyo, J.B.; Woo, A.; Yu, H.J.; Lim, K.; Cho, B.H.; Jung, H.S.; Lee, M.Y. Label-Free Sers Analysis of Urine Using a 3d-Stacked Agnw-Glass Fiber Filter Sensor for the Diagnosis of Pancreatic Cancer and Prostate Cancer. Anal. Chem. 2021, 93, 3778–3785. [Google Scholar] [CrossRef]
- Gao, R.; Lv, Z.; Mao, Y.; Yu, L.; Bi, X.; Xu, S.; Cui, J.; Wu, Y.-C. SERS-Based Pump-Free Microfluidic Chip for Highly Sensitive Immunoassay of Prostate-Specific Antigen Biomarkers. ACS Sens. 2019, 4, 938–943. [Google Scholar] [CrossRef]
- Bhamidipati, M.; Lee, G.; Kim, I.; Fabris, L. SERS-Based Quantification of PSMA in Tissue Microarrays Allows Effective Stratification of Patients with Prostate Cancer. ACS Omega 2018, 3, 16784–16794. [Google Scholar] [CrossRef]
- Spaliviero, M.; Harmsen, S.; Huang, R.; Wall, M.A.; Andreou, C.; Eastham, J.A.; Touijer, K.A.; Scardino, P.T.; Kircher, M.F. Detection of Lymph Node Metastases with SERRS Nanoparticles. Mol. Imaging Biol. 2016, 18, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Thakor, A.S.; Luong, R.; Paulmurugan, R.; Lin, F.I.; Kempen, P.; Zavaleta, C.; Chu, P.; Massoud, T.F.; Sinclair, R.; Gambhir, S.S. The Fate and Toxicity of Raman-Active Silica-Gold Nanoparticles in Mice. Sci. Transl. Med. 2011, 3, 79ra33. [Google Scholar] [CrossRef]
- Harmsen, S.; Wall, M.A.; Huang, R.; Kircher, M.F. Cancer imaging using surface-enhanced resonance Raman scattering nanoparticles. Nat. Protoc. 2017, 12, 1400–1414. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-enhanced Raman spectroscopy for in vivo biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar] [CrossRef]
- Nicolson, F.; Kircher, M.F.; Stone, N.; Matousek, P. Spatially offset Raman spectroscopy for biomedical applications. Chem. Soc. Rev. 2021, 50, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, F.; Jamieson, L.E.; Mabbott, S.; Plakas, K.; Shand, N.C.; Detty, M.R.; Graham, D.; Faulds, K. Through tissue imaging of a live breast cancer tumour model using handheld surface enhanced spatially offset resonance Raman spectroscopy (SESORRS). Chem. Sci. 2018, 9, 3788–3792. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, F.; Andreiuk, B.; Andreou, C.; Hsu, H.-T.; Rudder, S.; Kircher, M.F. Non-invasive In Vivo Imaging of Cancer Using Surface-Enhanced Spatially Offset Raman Spectroscopy (SESORS). Theranostics 2019, 9, 5899–5913. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Miao, K.; Lin, L.-E.; Chen, X.; Du, J.; Wei, L. Super-resolution label-free volumetric vibrational imaging. Nat. Commun. 2021, 12, 3648. [Google Scholar] [CrossRef]
- Sepp, K.; Lee, M.; Bluntzer, M.T.J.; Helgason, G.V.; Hulme, A.N.; Brunton, V.G. Utilizing Stimulated Raman Scattering Microscopy To Study Intracellular Distribution of Label-Free Ponatinib in Live Cells. J. Med. Chem. 2019, 63, 2028–2034. [Google Scholar] [CrossRef]
- Aljakouch, K.; Lechtonen, T.; Yosef, H.K.; Hammoud, M.K.; Alsaidi, W.; Kötting, C.; Mügge, C.; Kourist, R.; El-Mashtoly, S.F.; Gerwert, K. Raman Microspectroscopic Evidence for the Metabolism of a Tyrosine Kinase Inhibitor, Neratinib, in Cancer Cells. Angew. Chem. Int. Ed. 2018, 57, 7250–7254. [Google Scholar] [CrossRef] [PubMed]
- Tipping, W.J.; Lee, M.; Serrels, A.; Brunton, V.G.; Hulme, A.N. Imaging drug uptake by bioorthogonal stimulated Raman scattering microscopy. Chem. Sci. 2017, 8, 5606–5615. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.; Miao, Y.; Porterfield, W.; Cai, W.; Zhu, X.; Kim, S.-J.; Hu, F.; Bhattarai-Kline, S.; Min, W.; Zhang, W. Structure–activity–distribution relationship study of anti-cancer antimycin-type depsipeptides. Chem. Commun. 2019, 55, 9379–9382. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Chen, Z.; Zhang, L.; Shen, Y.; Wei, L.; Min, W. Vibrational Imaging of Glucose Uptake Activity in Live Cells and Tissues by Stimulated Raman Scattering. Angew. Chem. Int. Ed. 2015, 54, 9821–9825. [Google Scholar] [CrossRef] [PubMed]
- De Moliner, F.; Knox, K.; Gordon, D.; Lee, M.; Tipping, W.J.; Geddis, A.; Reinders, A.; Ward, J.M.; Oparka, K.; Vendrell, M. A Palette of Minimally Tagged Sucrose Analogues for Real-Time Raman Imaging of Intracellular Plant Metabolism. Angew. Chem. Int. Ed. 2021, 60, 7637–7642. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; McWilliams, A.; Lam, S.; English, J.; McLean, D.I.; Lui, H.; Zeng, H. Effect of formalin fixation on the near-infrared Raman spectroscopy of normal and cancerous human bronchial tissues. Int. J. Oncol. 2003, 23, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.; Maguire, A.; Meade, A.D.; Flint, S.; Toner, M.; Byrne, H.J.; Lyng, F.M. Improved protocols for pre-processing Raman spectra of formalin fixed paraffin preserved tissue sections. Anal. Methods 2017, 9, 4709–4717. [Google Scholar] [CrossRef]
- Stiufiuc, R.; Iacovita, C.; Lucaciu, C.M.; Stiufiuc, G.; Dutu, A.G.; Braescu, C.; Leopold, N. SERS-active silver colloids prepared by reduction of silver nitrate with short-chain polyethylene glycol. Nanoscale Res. Lett. 2013, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Korbekandi, H.; Mirmohammadi, S.V.; Zolfaghari, B. Synthesis of silver nanoparticles: Chemical, physical and biological methods. Res. Pharm. Sci. 2014, 9, 385–406. [Google Scholar]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef]
- Dorney, J.; Bonnier, F.; Garcia, A.; Casey, A.; Chambers, G.; Byrne, H.J. Identifying and localizing intracellular nanoparticles using Raman spectroscopy. Analyst 2012, 137, 1111–1119. [Google Scholar] [CrossRef]
- Bonnier, F.; Byrne, H.J. Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems. Analyst 2012, 137, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, H.; Bonnier, F.; Knief, P.; Howe, O.; Lyng, F.M.; Meade, A.D.; Byrne, H.J. Evaluation of the potential of Raman microspectroscopy for prediction of chemotherapeutic response to cisplatin in lung adenocarcinoma. Analyst 2010, 135, 3070–3076. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Xie, X.S. Reliable Cell Segmentation Based on Spectral Phasor Analysis of Hyperspectral Stimulated Raman Scattering Imaging Data. Anal. Chem. 2014, 86, 4115–4119. [Google Scholar] [CrossRef] [PubMed]
- Tipping, W.J.; Wilson, L.T.; An, C.; Leventi, A.A.; Wark, A.W.; Wetherill, C.; Tomkinson, N.C.O.; Faulds, K.; Graham, D. Stimulated Raman scattering microscopy with spectral phasor analysis: Applications in assessing drug–cell interactions. Chem. Sci. 2022. [Google Scholar] [CrossRef]
- Byrne, H.J.; Knief, P.; Keating, M.E.; Bonnier, F. Spectral pre and post processing for infrared and Raman spectroscopy of biological tissues and cells. Chem. Soc. Rev. 2016, 45, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Aubertin, K.; Trinh, V.Q.; Jermyn, M.; Baksic, P.; Grosset, A.-A.; Desroches, J.; St-Arnaud, K.; Birlea, M.; Vladoiu, M.C.; Latour, M.; et al. Mesoscopic characterization of prostate cancer using Raman spectroscopy: Potential for diagnostics and therapeutics. Br. J. Urol. 2018, 122, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.; Zorn, K.C.; Tremblay, J.-P.; Desroches, J.; Dallaire, F.; Aubertin, K.; Marple, E.T.; Kent, C.; Leblond, F.; Trudel, D.; et al. Integration of a Raman spectroscopy system to a robotic-assisted surgical system for real-time tissue characterization during radical prostatectomy procedures. J. Biomed. Opt. 2019, 24, 025001. [Google Scholar] [CrossRef]
- Cookson, M.S.; Aus, G.; Burnett, A.L.; Canby-Hagino, E.D.; D’Amico, A.V.; Dmochowski, R.R.; Eton, D.; Forman, J.D.; Goldenberg, S.L.; Hernandez, J.; et al. Variation in the Definition of Biochemical Recurrence in Patients Treated for Localized Prostate Cancer: The American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel Report and Recommendations for a Standard in the Reporting of Surgical Outcomes. J. Urol. 2007, 177, 540–545. [Google Scholar]
- Qiu, Y.; Zhang, Y.; Li, M.; Chen, G.; Fan, C.; Cui, K.; Wan, J.-B.; Han, A.; Ye, J.; Xiao, Z. Intraoperative Detection and Eradication of Residual Microtumors with Gap-Enhanced Raman Tags. ACS Nano 2018, 12, 7974–7985. [Google Scholar] [CrossRef]
- Harmsen, S.; Huang, R.; Wall, M.A.; Karabeber, H.; Samii, J.M.; Spaliviero, M.; White, J.R.; Monette, S.; O’Connor, R.; Pitter, K.L.; et al. Surface-enhanced resonance Raman scattering nanostars for high-precision cancer imaging. Sci. Transl. Med. 2015, 7, 271ra7. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Nakamura, H.; Fang, J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Deliv. Rev. 2012, 65, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.A.; Arora, V.K.; Sawyers, C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Cancer 2015, 15, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Blomme, A.; Ford, C.A.; Mui, E.; Patel, R.; Ntala, C.; Jamieson, L.E.; Planque, M.; McGregor, G.H.; Peixoto, P.; Hervouet, E.; et al. 2,4-Dienoyl-Coa Reductase Regulates Lipid Homeostasis in Treatment-Resistant Prostate Cancer. Nat. Commun. 2020, 11, 2508. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, D.; Zeng, J.; Guan, Z.; Dang, Q.; Wang, X.; Wang, J.; Huang, L.; Cao, P.; Zhang, G.; et al. Raman spectroscopy, a potential tool in diagnosis and prognosis of castration-resistant prostate cancer. J. Biomed. Opt. 2013, 18, 087001. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cagiannos, I.; Karakiewicz, P.; Eastham, J.A.; Ohori, M.; Rabbani, F.; Gerigk, C.; Reuter, V.; Graefen, M.; Hammerer, P.G.; Erbersdobler, A.; et al. A Preoperative Nomogram Identifying Decreased Risk of Positive Pelvic Lymph Nodes in Patients with Prostate Cancer. J. Urol. 2003, 170, 1798–1803. [Google Scholar] [CrossRef] [PubMed]
- Abdollah, F.; Sun, M.; Suardi, N.; Gallina, A.; Capitanio, U.; Bianchi, M.; Tutolo, M.; Passoni, N.; Karakiewicz, P.I.; Rigatti, P.; et al. National Comprehensive Cancer Network Practice Guidelines 2011: Need for More Accurate Recommendations for Pelvic Lymph Node Dissection in Prostate Cancer. J. Urol. 2012, 188, 423–428. [Google Scholar] [CrossRef]
- Petros, J.A.; Catalona, W.J. Lower Incidence of Unsuspected Lymph Node Metastases in 521 Consecutive Patients with Clinically Localized Prostate Cancer. J. Urol. 1992, 147, 1574–1575. [Google Scholar] [CrossRef]
- Heidenreich, A.; Varga, Z.; Von Knobloch, R. Extended pelvic lymphadenectomy in patients undergoing radical prostatectomy: High incidence of lymph node metastasis. J. Urol. 2002, 167, 1681–1686. [Google Scholar] [CrossRef]
- Godoy, G.; Von Bodman, C.; Chade, D.C.; Dillioglugil, O.; Eastham, J.A.; Fine, S.W.; Scardino, P.T.; Laudone, V.P. Pelvic Lymph Node Dissection for Prostate Cancer: Frequency and Distribution of Nodal Metastases in a Contemporary Radical Prostatectomy Series. J. Urol. 2012, 187, 2082–2086. [Google Scholar] [CrossRef]
- Joung, J.Y.; Cho, I.-C.; Lee, K.H. Role of Pelvic Lymph Node Dissection in Prostate Cancer Treatment. Korean J. Urol. 2011, 52, 437–445. [Google Scholar] [CrossRef]
- Chien, T.-M.; Lu, Y.-M.; Geng, J.-H.; Huang, T.-Y.; Ke, H.-L.; Huang, C.-N.; Li, C.-C.; Chou, Y.-H.; Wu, W.J.; Huang, S.-P. Predictors of Positive Bone Metastasis in Newly Diagnosed Prostate Cancer Patients. Asian Pac. J. Cancer Prev. 2016, 17, 1187–1191. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Schöpfer, A.; Wagner, U.; Sauter, G.; Moch, H.; Willi, N.; Gasser, T.C.; Mihatsch, M.J. Metastatic patterns of prostate cancer: An autopsy study of 1589 patients. Hum. Pathol. 2000, 31, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Berruti, A.; Dogliotti, L.; Bitossi, R.; Fasolis, G.; Gorzegno, G.; Bellina, M.; Torta, M.; Porpiglia, F.; Fontana, D.; Angeli, A. Incidence of skeletal complications in patients with bone metastatic prostate cancer and hormone refractory disease: Predictive role of bone resorption and formation markers evaluated at baseline. J. Urol. 2000, 164, 1248–1253. [Google Scholar] [CrossRef]
- Merdan, S.; Womble, P.R.; Miller, D.C.; Barnett, C.; Ye, Z.; Linsell, S.M.; Montie, J.E.; Denton, B.T. Toward Better Use of Bone Scans Among Men with Early-stage Prostate Cancer. Urology 2014, 84, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Zacho, H.D.; Nielsen, J.B.; Haberkorn, U.; Stenholt, L.; Petersen, L.J. 68 Ga-PSMA PET/CT for the detection of bone metastases in prostate cancer: A systematic review of the published literature. Clin. Physiol. Funct. Imaging 2017, 38, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Zhang, H.; Wang, Y.; Qian, H.; Zhu, Y.; Dong, B.; Xu, F.; Chen, N.; Liu, S.; Pan, J.; et al. Deep convolutional neural networks combine Raman spectral signature of serum for prostate cancer bone metastases screening. Nanomed. Nanotechnol. Biol. Med. 2020, 29, 102245. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Sterling, J.A.; Merkel, A.R.; Perrien, D.; Nyman, J.S.; Mahadevan-Jansen, A. Prostate cancer metastases alter bone mineral and matrix composition independent of effects on bone architecture in mice—A quantitative study using microCT and Raman spectroscopy. Bone 2013, 56, 454–460. [Google Scholar] [CrossRef]
- Ahmad, F.; Cherukuri, M.K.; Choyke, P.L. Metabolic reprogramming in prostate cancer. Br. J. Cancer 2021, 125, 1185–1196. [Google Scholar] [CrossRef]
- Roman, M.; Wrobel, T.P.; Panek, A.; Paluszkiewicz, C.; Kwiatek, W.M. Lipid droplets in prostate cancer cells and effect of irradiation studied by Raman microspectroscopy. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2020, 1865, 158753. [Google Scholar] [CrossRef]
- O’Malley, J.; Kumar, R.; Kuzmin, A.; Pliss, A.; Yadav, N.; Balachandar, S.; Wang, J.; Attwood, K.; Prasad, P.N.; Chandra, D. Lipid quantification by Raman microspectroscopy as a potential biomarker in prostate cancer. Cancer Lett. 2017, 397, 52–60. [Google Scholar] [CrossRef]
- Farah, B.L.; Sinha, R.A.; Wu, Y.; Singh, B.K.; Zhou, J.; Bay, B.-H.; Yen, P.M. β-Adrenergic Agonist and Antagonist Regulation of Autophagy in HepG2 Cells, Primary Mouse Hepatocytes, and Mouse Liver. PLoS ONE 2014, 9, e98155. [Google Scholar] [CrossRef] [PubMed]
- Brohée, L.; Peulen, O.; Nusgens, B.; Castronovo, V.; Thiry, M.; Colige, A.C.; Deroanne, C.F. Propranolol sensitizes prostate cancer cells to glucose metabolism inhibition and prevents cancer progression. Sci. Rep. 2018, 8, 7050. [Google Scholar] [CrossRef] [PubMed]
- Medipally, D.K.R.; Cullen, D.; Untereiner, V.; Sockalingum, G.D.; Maguire, A.; Nguyen, T.N.Q.; Bryant, J.; Noone, E.; Bradshaw, S.; Finn, M.; et al. Vibrational spectroscopy of liquid biopsies for prostate cancer diagnosis. Ther. Adv. Med. Oncol. 2020, 12, 1758835920918499. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, J.; Lee, S.-Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Fleming, J.; Mui, E.; Loveridge, C.; Repiscak, P.; Blomme, A.; Harle, V.; Salji, M.; Ahmad, I.; Teo, K.; et al. Sprouty2 loss-induced IL 6 drives castration-resistant prostate cancer through scavenger receptor B1. EMBO Mol. Med. 2018, 10, e8347. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.-L.; Lin, C.-J.; Li, T.-K.; Shen, T.-L.; Hsieh, J.-T.; Chen, B.P.C. The role of extracellular vesicles in prostate cancer with clinical applications. Endocr.-Relat. Cancer 2020, 27, R133–R144. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Nanou, A.; Rikkert, L.; Coumans, F.A.W.; Otto, C.; Terstappen, L.W.M.M.; Offerhaus, H.L. Label-Free Prostate Cancer Detection by Characterization of Extracellular Vesicles Using Raman Spectroscopy. Anal. Chem. 2018, 90, 11290–11296. [Google Scholar] [CrossRef] [PubMed]
- Del Mistro, G.; Cervo, S.; Mansutti, E.; Spizzo, R.; Colombatti, A.; Belmonte, P.; Zucconelli, R.; Steffan, A.; Sergo, V.; Bonifacio, A. Surface-enhanced Raman spectroscopy of urine for prostate cancer detection: A preliminary study. Anal. Bioanal. Chem. 2015, 407, 3271–3275. [Google Scholar] [CrossRef]
- Ma, Y.; Chi, J.; Zheng, Z.; Attygalle, A.; Kim, I.Y.; Du, H. Therapeutic prognosis of prostate cancer using surface-enhanced Raman scattering of patient urine and multivariate statistical analysis. J. Biophoton. 2021, 14, 202000275. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, H.; Yang, X.; Shao, X.; Li, T.; Chen, N.; Chen, Z.; Xue, W.; Pan, J.; Liu, S. Raman Spectroscopy Reveals Abnormal Changes in the Urine Composition of Prostate Cancer: An Application of an Intelligent Diagnostic Model with a Deep Learning Algorithm. Adv. Intell. Syst. 2021, 3, 2000090. [Google Scholar] [CrossRef]
- Wolf, A.M.D.; Wender, R.C.; Etzioni, R.B.; Thompson, I.M.; D’Amico, A.V.; Volk, R.; Brooks, D.D.; Dash, C.; Guessous, I.; Andrews, K.; et al. American Cancer Society Guideline for the Early Detection of Prostate Cancer: Update 2010. CA Cancer J. Clin. 2010, 60, 70–98. [Google Scholar] [CrossRef] [PubMed]
- Tikkinen, K.; Dahm, P.; Lytvyn, L.; Heen, A.F.; Vernooij, R.W.M.; Siemieniuk, R.A.C.; Wheeler, R.; Vaughan, B.; Fobuzi, A.C.; Blanker, M.H.; et al. Prostate cancer screening with prostate-specific antigen (PSA) test: A clinical practice guideline. BMJ 2018, 362, k3581. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Rong, M.; Shao, X.; Zhang, H.; Liu, S.; Dong, B.; Xue, W.; Wang, T.; Li, T.; Pan, J. Surface-enhanced Raman spectroscopy of serum accurately detects prostate cancer in patients with prostate-specific antigen levels of 4–10 ng/mL. Int. J. Nanomed. 2017, 12, 5399–5407. [Google Scholar] [CrossRef] [PubMed]
- Medipally, D.K.R.; Maguire, A.; Bryant, J.; Armstrong, J.; Dunne, M.; Finn, M.; Lyng, F.M.; Meade, A.D. Development of a high throughput (HT) Raman spectroscopy method for rapid screening of liquid blood plasma from prostate cancer patients. Analyst 2017, 142, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer by using Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 12371–12376. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Sekine, R.; Kagoshima, H.; Kazama, K.; Kato, A.; Shiozawa, M.; Tanaka, J.-I. All-in-one Raman spectroscopy approach to diagnosis of colorectal cancer: Analysis of spectra in the fingerprint regions. J. Anus Rectum Colon 2019, 3, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Orringer, D.; Freudiger, C.W.; Ramkissoon, S.; Liu, X.; Lau, D.; Golby, A.J.; Norton, I.; Hayashi, M.; Agar, N.Y.R.; et al. Rapid, Label-Free Detection of Brain Tumors with Stimulated Raman Scattering Microscopy. Sci. Transl. Med. 2013, 5, 201ra119. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Arbel, M.; Zhang, L.; Freudiger, C.W.; Hou, S.S.; Lin, D.; Yang, X.; Bacskai, B.J.; Xie, X.S. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Sci. Adv. 2018, 4, eaat7715. [Google Scholar] [CrossRef] [PubMed]
- Pence, I.J.; Evans, C.L. Translational biophotonics with Raman imaging: Clinical applications and beyond. Analyst 2021, 146, 6379–6393. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.E.; Kearns, H.; Graham, D.; Faulds, K. Surface Enhanced Raman Scattering for the Multiplexed Detection of Pathogenic Microorganisms: Towards Point-of-Use Applications. Analyst 2021, 146, 6084–6101. [Google Scholar] [CrossRef]
- Sebba, D.; Lastovich, A.G.; Kuroda, M.; Fallows, E.; Johnson, J.; Ahouidi, A.; Honko, A.N.; Fu, H.; Nielson, R.; Carruthers, E.; et al. A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases. Sci. Transl. Med. 2018, 10, eaat0944. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Bando, K.; Ando, J.; Yamakoshi, H.; Terayama, N.; Dodo, K.; Smith, N.I.; Sodeoka, M.; Fujita, K. Quantitative Drug Dynamics Visualized by Alkyne-Tagged Plasmonic-Enhanced Raman Microscopy. ACS Nano 2020, 14, 15032–15041. [Google Scholar] [CrossRef] [PubMed]
- Saar, B.G.; Freudiger, C.W.; Reichman, J.; Stanley, C.M.; Holtom, G.R.; Xie, X.S. Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science 2010, 330, 1368–1370. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Fung, A.A.; Zhou, A. Advances in stimulated Raman scattering imaging for tissues and animals. Quant. Imaging Med. Surg. 2020, 11, 1078–1101. [Google Scholar] [CrossRef] [PubMed]


| Terms | Definitions |
|---|---|
| Rayleigh scattering | Rayleigh scattering is an elastic scattering process where there is no change in the energy of the photons upon interaction with the target sample. |
| Stokes/anti-Stokes Raman scattering | Raman scattering (or inelastic scattering of incident photons) may result in either anti-Stokes scattering when there is an energy gain or Stokes scattering when there is an intensity loss with respect to the incident photons following interaction with the molecular vibration. |
| Resonance Raman spectroscopy | The frequency of the excitation laser source matches (or approaches) that of an electronic transition within the target sample, thus enhancing the Raman scattering signal. |
| SERS—Surface-enhanced Raman scattering | A technique for molecular detection that relies on the enhanced Raman scattering of molecules that are adsorbed on, or in close proximity to, SERS-active metal surfaces, including gold or silver nanoparticles. |
| SORS—Spatially offset Raman scattering | Low-intensity laser excitation is directed onto the surface of the sample, and Raman spectra are obtained at a known spatial offset from the illumination spot. By applying a spatial offset to the detection, it enables the collection of Raman scattered photons that have been produced at greater (and variable) depths within the sample material. |
| Coherent Raman scattering | Raman-active vibrations can be selectively driven into coherence by exciting with two (or more) laser wavelengths. The frequency difference of the excitation sources is matched to a vibrational resonance of the target molecule, thus enhancing its detection. Coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS) are examples of coherent Raman scattering. |
| Advantages | Disadvantages |
|---|---|
| Non-destructive, non-invasive High specificity Simultaneous detection of biomolecules Compatible with physiological measurements due to minimal water interaction In vivo applications Suitable for chemical analysis, quantification, classification and imaging of biological materials | Weak Raman signals can lead to long acquisition times Not widely incorporated into current clinical workflows Sophisticated data analysis Autofluorescence can overwhelm the Raman signal (sample dependent) |
| Technique | Advantages | Disadvantages |
|---|---|---|
| Spontaneous Raman scattering | Minimal sample preparation and setup Minimal interference with water for biological investigation in live cells | Autofluorescence from cells and tissues can overwhelm Raman signals Image acquisition can be slow (seconds to minutes) for some applications |
| Resonance Raman scattering (RRS) | Improved sensitivity and selectivity over standard Raman spectroscopy | High-fluorescence background signal capable of obscuring true Raman signals |
| Surface-enhanced Raman scattering (SERS) | Improved sensitivity and selectivity over standard Raman spectroscopy | Requires coupling with nanoparticles SERS nanoparticles must be biocompatible if used in vivo |
| Spatially offset Raman scattering (SORS) | Delineation of spectral differences in composition at greater depths In vivo detection through tissue | Complex setup and hardware required |
| Surface-enhanced spatially offset Raman spectroscopy (SESORS) | Couples the sensitivity afforded by SERS with subsurface probing of SORS Detection at greater depths can be achieved | Requires active targeting of SERS nanoparticles for detection |
| Coherent anti-Stokes Raman scattering (CARS) | Fast image acquisition Typically uses biocompatible NIR laser excitation Minimal background fluorescence | Requires tuneable lasers to probe molecular structures, which are expensive Non-resonance signal distorts the CARS spectrum |
| Stimulated Raman scattering (SRS) | Enhanced signal strength compared to spontaneous Raman scattering Fast image acquisition (μs/pixel) Biocompatible NIR excitation SRS spectrum matches Raman spectrum for easy peak assignment and quantification | Requires tuneable lasers to probe molecular structures, which are expensive Some background signal can be detected from pump-probe-based effects Complex hardware makes it difficult to incorporate into a handheld device for intraoperative use |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaba, F.; Tipping, W.J.; Salji, M.; Faulds, K.; Graham, D.; Leung, H.Y. Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements. Cancers 2022, 14, 1535. https://doi.org/10.3390/cancers14061535
Gaba F, Tipping WJ, Salji M, Faulds K, Graham D, Leung HY. Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements. Cancers. 2022; 14(6):1535. https://doi.org/10.3390/cancers14061535
Chicago/Turabian StyleGaba, Fortis, William J. Tipping, Mark Salji, Karen Faulds, Duncan Graham, and Hing Y. Leung. 2022. "Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements" Cancers 14, no. 6: 1535. https://doi.org/10.3390/cancers14061535
APA StyleGaba, F., Tipping, W. J., Salji, M., Faulds, K., Graham, D., & Leung, H. Y. (2022). Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements. Cancers, 14(6), 1535. https://doi.org/10.3390/cancers14061535






