Tumor Immune Microenvironment in Lymphoma: Focus on Epigenetics
Abstract
Simple Summary
Abstract
1. Introduction
2. Lymphoma: An Overview
3. Tumor Microenvironment: Interaction of Immune and Lymphoma Tumor Cells
4. Epigenetic
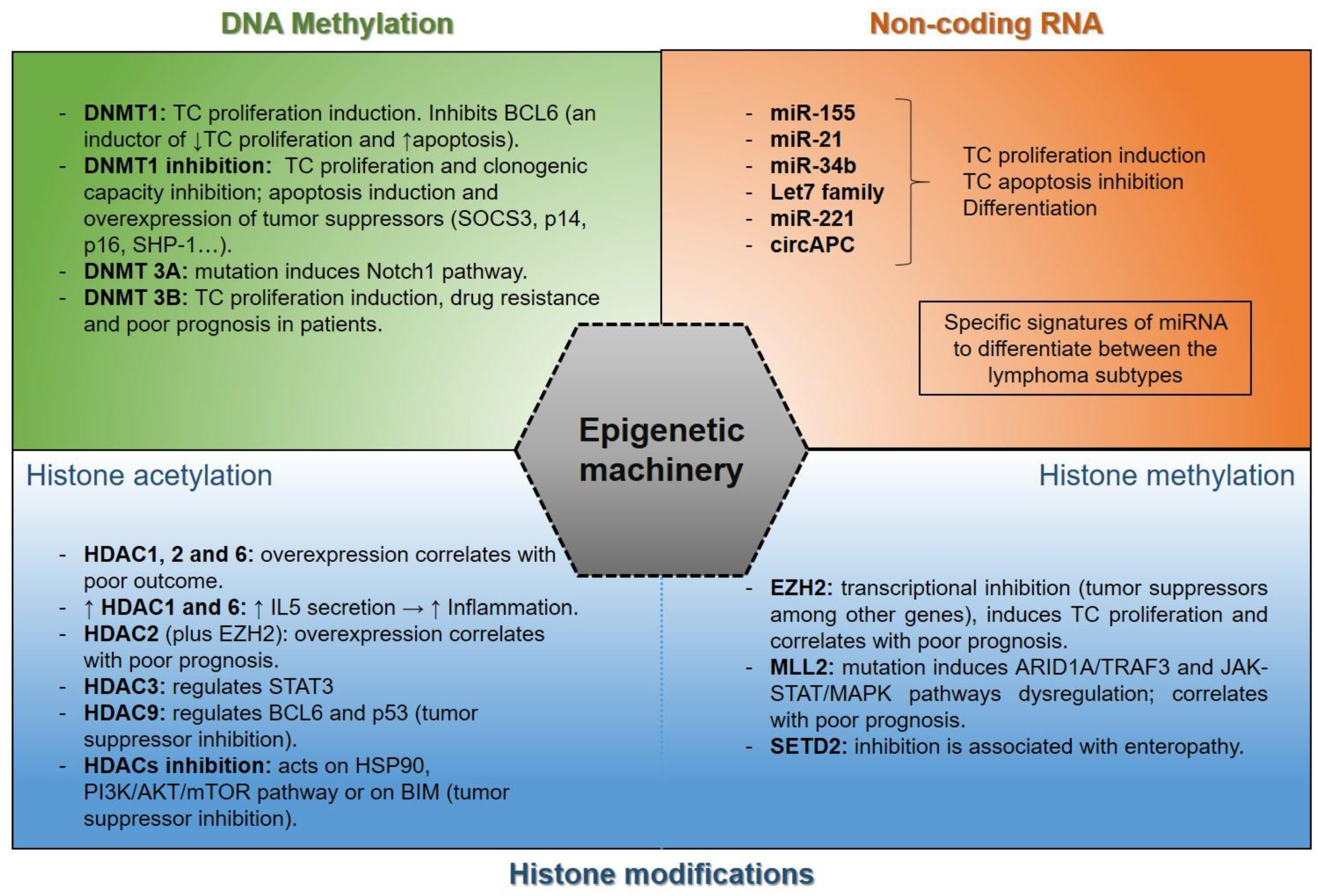
4.1. Epigenetic Alterations in Lymphoma
- (a)
- Histone acetylation: Chromosome structural modification and regulation of gene expression are regulated by the functional balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs), which contribute to acetylation and deacetylation of histones. Thus, an acetyl group is transferred from acetyl-CoA to the lysine residues NH2 group in proteins by HATs and is removed by HDACs [142,143].
- (b)
- Histone methylation: Histone methyltransferase enzymes are responsible for transferring methyl groups to lysine and arginine of histones. Mono-, di-, and trimethylation affects gene transcription and promotes lymphoma pathogenesis [152]. Several methyltransferases, such as EZH2, MML2, or SETD2, among others, are associated with lymphoma tumors. SETD2 (SET-domain-containing 2), a methyltransferase responsible for H3 lysine 36 trimethylation (H3K36me3), is mostly silenced in enteropathy-associated T-cell lymphoma (EATL) [153], as well as monomorphic epitheliotropic intestinal T-cell lymphoma (MEITL) [154]. MLL2 (also known as KMT2D) is mutated in angioimmunoblastic T-cell lymphoma (AITL), PTCL not otherwise specified (PTCL-NOS) [155], DLBCL, FL [156], and NK/T-cell lymphoma (NK/TCL) [157,158], where approximately 91% of mutations result in silencing its histone methylation function [159]. The loss of MLL2 functions alters several genes such as ARID1A, TRAF3, or signaling pathways such as JAK-STAT [160] or MAPK. In fact, the combination of chidamide and decitabine improves the KMT2D-PU.1 transcription factor interaction, which inactivates the MAPK pathway [161], constitutively activated in T-cell lymphoma [162]. Likewise, the loss of MLL2 functions influences the poor prognosis in lymphoma patients [157,158,163].
4.2. Epigenetic Regulation of Immune Cells in the TME
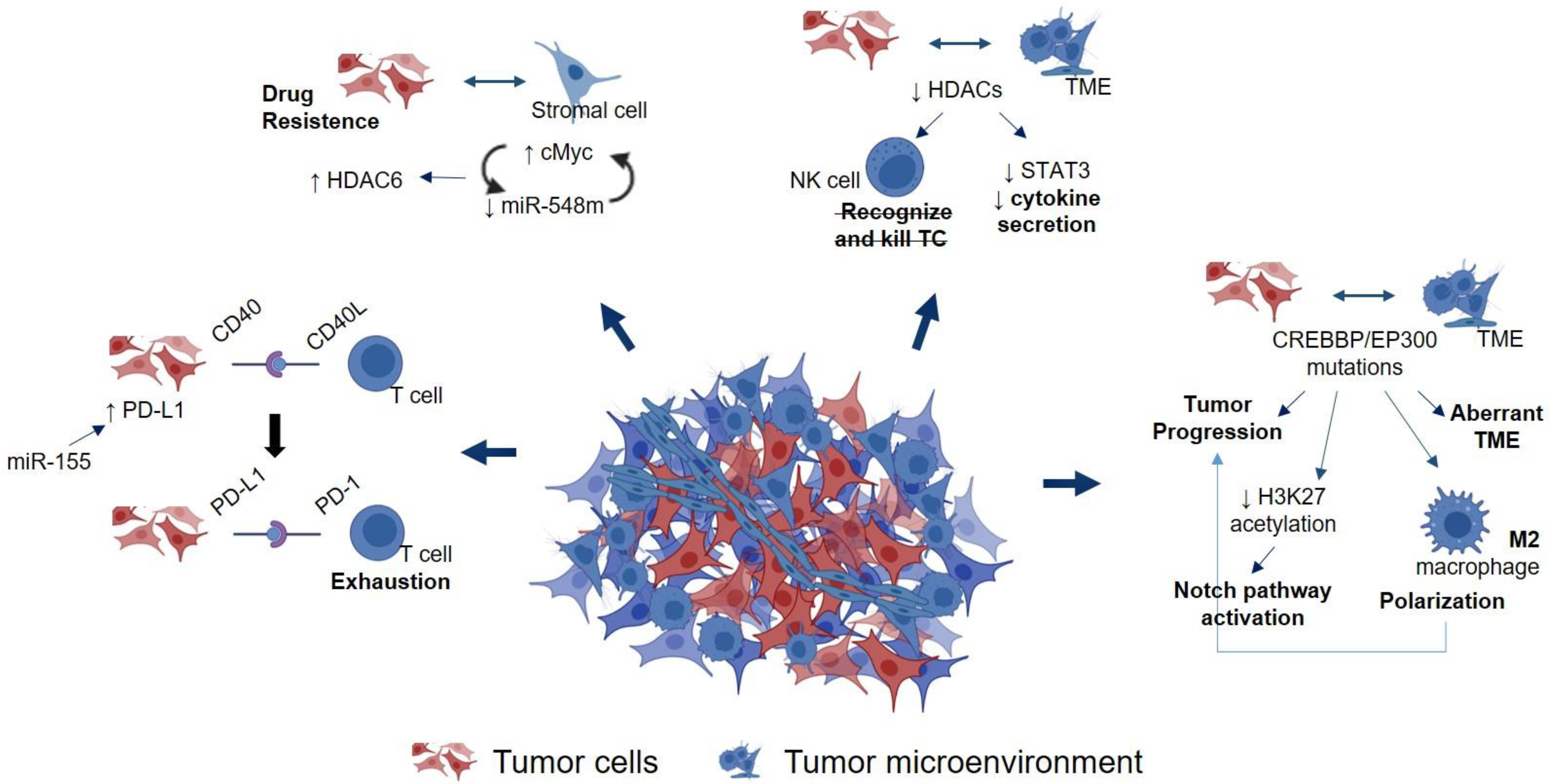
4.3. Epigenetic Cross-Talk between Lymphoma Tumor Cells and TME
4.4. Epigenetic Therapies: Clinical Trials in Lymphoma Disease
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.; Esteller, M. Cancer epigenomics: Beyond genomics. Curr. Opin. Genet. Dev. 2012, 22, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Boedtkjer, E.; Pedersen, S.F. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu. Rev. Physiol. 2020, 82, 103–126. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Gascoyne, R.D. The tumour microenvironment in B cell lymphomas. Nat. Rev. Cancer 2014, 14, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Craig, V.J.; Cogliatti, S.B.; Arnold, I.; Gerke, C.; Balandat, J.E.; Wundisch, T.; Muller, A. B-cell receptor signaling and CD40 ligand-independent T cell help cooperate in Helicobacter-induced MALT lymphomagenesis. Leukemia 2010, 24, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Dalai, S.K.; Mirshahidi, S.; Morrot, A.; Zavala, F.; Sadegh-Nasseri, S. Anergy in memory CD4+ T cells is induced by B cells. J. Immunol. 2008, 181, 3221–3231. [Google Scholar] [CrossRef]
- Brusa, D.; Serra, S.; Coscia, M.; Rossi, D.; D’Arena, G.; Laurenti, L.; Jaksic, O.; Fedele, G.; Inghirami, G.; Gaidano, G.; et al. The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica 2013, 98, 953–963. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Zhou, J.; Young, K.H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood 2018, 131, 68–83. [Google Scholar] [CrossRef]
- Lwin, T.; Zhao, X.; Cheng, F.; Zhang, X.; Huang, A.; Shah, B.; Zhang, Y.; Moscinski, L.C.; Choi, Y.S.; Kozikowski, A.P.; et al. A microenvironment-mediated c-Myc/miR-548m/HDAC6 amplification loop in non-Hodgkin B cell lymphomas. J. Clin. Investig. 2013, 123, 4612–4626. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Matasar, M.J.; Zelenetz, A.D. Overview of lymphoma diagnosis and management. Radiol. Clin. N. Am. 2008, 46, 175–198. [Google Scholar] [CrossRef] [PubMed]
- Mugnaini, E.N.; Ghosh, N. Lymphoma. Prim. Care 2016, 43, 661–675. [Google Scholar] [CrossRef] [PubMed]
- de Leval, L.; Jaffe, E.S. Lymphoma Classification. Cancer J. 2020, 26, 176–185. [Google Scholar] [CrossRef] [PubMed]
- The Non-Hodgkin’s Lymphoma Pathologic Classification Project. National Cancer Institute sponsored study of classifications of non-Hodgkin’s lymphomas: Summary and description of a working formulation for clinical usage. Cancer 1982, 49, 2112–2135. [Google Scholar] [CrossRef]
- Pfreundschuh, M.; Kuhnt, E.; Trumper, L.; Osterborg, A.; Trneny, M.; Shepherd, L.; Gill, D.S.; Walewski, J.; Pettengell, R.; Jaeger, U.; et al. CHOP-like chemotherapy with or without rituximab in young patients with good-prognosis diffuse large-B-cell lymphoma: 6-year results of an open-label randomised study of the MabThera International Trial (MInT) Group. Lancet. Oncol. 2011, 12, 1013–1022. [Google Scholar] [CrossRef]
- Scott, D.W.; Mottok, A.; Ennishi, D.; Wright, G.W.; Farinha, P.; Ben-Neriah, S.; Kridel, R.; Barry, G.S.; Hother, C.; Abrisqueta, P.; et al. Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin Determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies. J. Clin. Oncol. 2015, 33, 2848–2856. [Google Scholar] [CrossRef]
- Pfreundschuh, M.; Schubert, J.; Ziepert, M.; Schmits, R.; Mohren, M.; Lengfelder, E.; Reiser, M.; Nickenig, C.; Clemens, M.; Peter, N.; et al. Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: A randomised controlled trial (RICOVER-60). Lancet. Oncol. 2008, 9, 105–116. [Google Scholar] [CrossRef]
- Recher, C.; Coiffier, B.; Haioun, C.; Molina, T.J.; Ferme, C.; Casasnovas, O.; Thieblemont, C.; Bosly, A.; Laurent, G.; Morschhauser, F.; et al. Intensified chemotherapy with ACVBP plus rituximab versus standard CHOP plus rituximab for the treatment of diffuse large B-cell lymphoma (LNH03-2B): An open-label randomised phase 3 trial. Lancet 2011, 378, 1858–1867. [Google Scholar] [CrossRef]
- Landsburg, D.J.; Falkiewicz, M.K.; Maly, J.; Blum, K.A.; Howlett, C.; Feldman, T.; Mato, A.R.; Hill, B.T.; Li, S.; Medeiros, L.J.; et al. Outcomes of Patients With Double-Hit Lymphoma Who Achieve First Complete Remission. J. Clin. Oncol. 2017, 35, 2260–2267. [Google Scholar] [CrossRef]
- Philip, T.; Guglielmi, C.; Hagenbeek, A.; Somers, R.; Van der Lelie, H.; Bron, D.; Sonneveld, P.; Gisselbrecht, C.; Cahn, J.Y.; Harousseau, J.L.; et al. Autologous bone marrow transplantation as compared with salvage chemotherapy in relapses of chemotherapy-sensitive non-Hodgkin’s lymphoma. N. Engl. J. Med. 1995, 333, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Cazelles, C.; Belhadj, K.; Vellemans, H.; Camus, V.; Poullot, E.; Gaulard, P.; Veresezan, L.; Itti, E.; Becker, S.; Carvalho, M.; et al. Rituximab plus gemcitabine and oxaliplatin (R-GemOx) in refractory/relapsed diffuse large B-cell lymphoma: A real-life study in patients ineligible for autologous stem-cell transplantation. Leuk. Lymphoma 2021, 62, 2161–2168. [Google Scholar] [CrossRef] [PubMed]
- Sehn, L.H.; Herrera, A.F.; Flowers, C.R.; Kamdar, M.K.; McMillan, A.; Hertzberg, M.; Assouline, S.; Kim, T.M.; Kim, W.S.; Ozcan, M.; et al. Polatuzumab Vedotin in Relapsed or Refractory Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2020, 38, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Salles, G.; Duell, J.; Gonzalez Barca, E.; Tournilhac, O.; Jurczak, W.; Liberati, A.M.; Nagy, Z.; Obr, A.; Gaidano, G.; Andre, M.; et al. Tafasitamab plus lenalidomide in relapsed or refractory diffuse large B-cell lymphoma (L-MIND): A multicentre, prospective, single-arm, phase 2 study. Lancet. Oncol. 2020, 21, 978–988. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jager, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Bonadonna, G.; Zucali, R.; Monfardini, S.; De Lena, M.; Uslenghi, C. Combination chemotherapy of Hodgkin’s disease with adriamycin, bleomycin, vinblastine, and imidazole carboxamide versus MOPP. Cancer 1975, 36, 252–259. [Google Scholar] [CrossRef]
- Merli, F.; Luminari, S.; Gobbi, P.G.; Cascavilla, N.; Mammi, C.; Ilariucci, F.; Stelitano, C.; Musso, M.; Baldini, L.; Galimberti, S.; et al. Long-Term Results of the HD2000 Trial Comparing ABVD Versus BEACOPP Versus COPP-EBV-CAD in Untreated Patients With Advanced Hodgkin Lymphoma: A Study by Fondazione Italiana Linfomi. J. Clin. Oncol. 2016, 34, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Viviani, S.; Zinzani, P.L.; Rambaldi, A.; Brusamolino, E.; Levis, A.; Bonfante, V.; Vitolo, U.; Pulsoni, A.; Liberati, A.M.; Specchia, G.; et al. ABVD versus BEACOPP for Hodgkin’s lymphoma when high-dose salvage is planned. N. Engl. J. Med. 2011, 365, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.M.; Jurczak, W.; Straus, D.J.; Ansell, S.M.; Kim, W.S.; Gallamini, A.; Younes, A.; Alekseev, S.; Illes, A.; Picardi, M.; et al. Brentuximab Vedotin with Chemotherapy for Stage III or IV Hodgkin’s Lymphoma. N. Engl. J. Med. 2018, 378, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, C.H.; Nimer, S.D.; Zelenetz, A.D.; Trippett, T.; Hedrick, E.E.; Filippa, D.A.; Louie, D.; Gonzales, M.; Walits, J.; Coady-Lyons, N.; et al. A 2-step comprehensive high-dose chemoradiotherapy second-line program for relapsed and refractory Hodgkin disease: Analysis by intent to treat and development of a prognostic model. Blood 2001, 97, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Ansell, S.M.; Lin, Y. Immunotherapy of lymphomas. J. Clin. Investig. 2020, 130, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Nieto, Y.; Gruschkus, S.; Valdez, B.C.; Jones, R.B.; Anderlini, P.; Hosing, C.; Popat, U.; Qazilbash, M.; Kebriaei, P.; Alousi, A.; et al. Improved outcomes of high-risk relapsed Hodgkin lymphoma patients after high-dose chemotherapy: A 15-year analysis. Haematologica 2021. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.P.; Akhtari, M.; Smith, G.L.; Pinnix, C.; Osborne, E.M.; Gunther, J.R.; Allen, P.K.; Yehia, Z.A.; Fanale, M.; Rodriguez, M.A.; et al. Outcomes After Chemotherapy Followed by Radiation for Stage IIB Hodgkin Lymphoma With Bulky Disease. Clin. Lymphoma Myeloma Leuk. 2015, 15, 664–670.e2. [Google Scholar] [CrossRef] [PubMed]
- Armand, P.; Engert, A.; Younes, A.; Fanale, M.; Santoro, A.; Zinzani, P.L.; Timmerman, J.M.; Collins, G.P.; Ramchandren, R.; Cohen, J.B.; et al. Nivolumab for Relapsed/Refractory Classic Hodgkin Lymphoma After Failure of Autologous Hematopoietic Cell Transplantation: Extended Follow-Up of the Multicohort Single-Arm Phase II CheckMate 205 Trial. J. Clin. Oncol. 2018, 36, 1428–1439. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zinzani, P.L.; Fanale, M.A.; Armand, P.; Johnson, N.A.; Brice, P.; Radford, J.; Ribrag, V.; Molin, D.; Vassilakopoulos, T.P.; et al. Phase II Study of the Efficacy and Safety of Pembrolizumab for Relapsed/Refractory Classic Hodgkin Lymphoma. J. Clin. Oncol. 2017, 35, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zinzani, P.L.; Lee, H.J.; Armand, P.; Johnson, N.A.; Brice, P.; Radford, J.; Ribrag, V.; Molin, D.; Vassilakopoulos, T.P.; et al. Pembrolizumab in relapsed or refractory Hodgkin lymphoma: 2-year follow-up of KEYNOTE-087. Blood 2019, 134, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, E.; d’Amore, F.; Carlo-Stella, C. Immune and Inflammatory Cells of the Tumor Microenvironment Represent Novel Therapeutic Targets in Classical Hodgkin Lymphoma. Int. J. Mol. Sci. 2019, 20, 5503. [Google Scholar] [CrossRef]
- Joshi, M.; Ansell, S.M. Activating the Antitumor Immune Response in Non-Hodgkin Lymphoma Using Immune Checkpoint Inhibitors. J. Immunol. Res. 2020, 2020, 8820377. [Google Scholar] [CrossRef]
- Autio, M.; Leivonen, S.K.; Bruck, O.; Mustjoki, S.; Jorgensen, J.M.; Karjalainen-Lindsberg, M.L.; Beiske, K.; Holte, H.; Pellinen, T.; Leppa, S. Immune cell constitution in the tumor microenvironment predicts the outcome in diffuse large B-cell lymphoma. Haematologica 2021, 106, 718–729. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Tzankov, A. Lymphomas and Their Microenvironment: A Multifaceted Relationship. Pathobiology 2019, 86, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Gatti-Mays, M.E.; Balko, J.M.; Gameiro, S.R.; Bear, H.D.; Prabhakaran, S.; Fukui, J.; Disis, M.L.; Nanda, R.; Gulley, J.L.; Kalinsky, K.; et al. If we build it they will come: Targeting the immune response to breast cancer. NPJ Breast Cancer 2019, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Sanz, L.; Munoz, P. Tumor-Infiltrating Immunosuppressive Cells in Cancer-Cell Plasticity, Tumor Progression and Therapy Response. Cancer Microenviron. 2019, 12, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Mulder, T.A.; Wahlin, B.E.; Osterborg, A.; Palma, M. Targeting the Immune Microenvironment in Lymphomas of B-Cell Origin: From Biology to Clinical Application. Cancers 2019, 11, 915. [Google Scholar] [CrossRef] [PubMed]
- Wein, F.; Kuppers, R. The role of T cells in the microenvironment of Hodgkin lymphoma. J. Leukoc. Biol. 2016, 99, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Connors, J.M.; Gascoyne, R.D. Molecular pathogenesis of Hodgkin’s lymphoma: Increasing evidence of the importance of the microenvironment. J. Clin. Oncol. 2011, 29, 1812–1826. [Google Scholar] [CrossRef]
- Aldinucci, D.; Gloghini, A.; Pinto, A.; Colombatti, A.; Carbone, A. The role of CD40/CD40L and interferon regulatory factor 4 in Hodgkin lymp.phoma microenvironment. Leuk. Lymphoma 2012, 53, 195–201. [Google Scholar] [CrossRef]
- Ohshima, K.; Karube, K.; Hamasaki, M.; Suefuji, H.; Tutiya, T.; Yamaguchi, T.; Suzumiya, J.; Kikuchi, M. Imbalances of chemokines, chemokine receptors and cytokines in Hodgkin lymphoma: Classical Hodgkin lymphoma vs. Hodgkin-like ATLL. Int. J. Cancer 2003, 106, 706–712. [Google Scholar] [CrossRef]
- Beguelin, W.; Sawh, S.; Chambwe, N.; Chan, F.C.; Jiang, Y.; Choo, J.W.; Scott, D.W.; Chalmers, A.; Geng, H.; Tsikitas, L.; et al. IL10 receptor is a novel therapeutic target in DLBCLs. Leukemia 2015, 29, 1684–1694. [Google Scholar] [CrossRef]
- Cha, Z.; Qian, G.; Zang, Y.; Gu, H.; Huang, Y.; Zhu, L.; Li, J.; Liu, Y.; Tu, X.; Song, H.; et al. Circulating CXCR5+CD4+ T cells assist in the survival and growth of primary diffuse large B cell lymphoma cells through interleukin 10 pathway. Exp. Cell Res. 2017, 350, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Ame-Thomas, P.; Le Priol, J.; Yssel, H.; Caron, G.; Pangault, C.; Jean, R.; Martin, N.; Marafioti, T.; Gaulard, P.; Lamy, T.; et al. Characterization of intratumoral follicular helper T cells in follicular lymphoma: Role in the survival of malignant B cells. Leukemia 2012, 26, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.V.; Jones, D.B.; Wright, D.H. Reed-Sternberg-cell/lymphocyte interaction. Lancet 1977, 2, 768–769. [Google Scholar] [CrossRef]
- Goodman, A.; Patel, S.P.; Kurzrock, R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 2017, 14, 203–220. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Tzankov, A. Mechanisms of Immune Evasion and Immune Modulation by Lymphoma Cells. Front. Oncol. 2018, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef]
- Spasevska, I.; Josefsson, S.; Blaker, Y.N.; Steen, C.B.; Sharma, A.; Kushekhar, K.; Meyer, S.; Kolstad, A.; Beiske, K.; Holte, H.; et al. Heterogeneity of Regulatory T Cells in B-Cell Non-Hodgkin Lymphoma. Blood 2020, 136, 27–28. [Google Scholar] [CrossRef]
- Zhong, W.; Liu, X.; Zhu, Z.; Li, Q.; Li, K. High levels of Tim-3(+)Foxp3(+)Treg cells in the tumor microenvironment is a prognostic indicator of poor survival of diffuse large B cell lymphoma patients. Int. Immunopharmacol. 2021, 96, 107662. [Google Scholar] [CrossRef]
- Farinha, P.; Al-Tourah, A.; Gill, K.; Klasa, R.; Connors, J.M.; Gascoyne, R.D. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood 2010, 115, 289–295. [Google Scholar] [CrossRef]
- Tzankov, A.; Meier, C.; Hirschmann, P.; Went, P.; Pileri, S.A.; Dirnhofer, S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica 2008, 93, 193–200. [Google Scholar] [CrossRef]
- Muenst, S.; Hoeller, S.; Dirnhofer, S.; Tzankov, A. Increased programmed death-1+ tumor-infiltrating lymphocytes in classical Hodgkin lymphoma substantiate reduced overall survival. Hum. Pathol. 2009, 40, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Tzankov, A.; Matter, M.S.; Dirnhofer, S. Refined prognostic role of CD68-positive tumor macrophages in the context of the cellular micromilieu of classical Hodgkin lymphoma. Pathobiology 2010, 77, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Chen, Y.P.; Medeiros, L.J.; Chen, T.Y.; Chang, K.C. Higher infiltration of intratumoral CD25+ FOXP3+ lymphocytes correlates with a favorable prognosis in patients with diffuse large B-cell lymphoma. Leuk. Lymphoma 2021, 62, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Durgeau, A.; Virk, Y.; Corgnac, S.; Mami-Chouaib, F. Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy. Front. Immunol. 2018, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef] [PubMed]
- Duffield, A.S.; Ascierto, M.L.; Anders, R.A.; Taube, J.M.; Meeker, A.K.; Chen, S.; McMiller, T.L.; Phillips, N.A.; Xu, H.; Ogurtsova, A.; et al. Th17 immune microenvironment in Epstein-Barr virus-negative Hodgkin lymphoma: Implications for immunotherapy. Blood Adv. 2017, 1, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Willenbrock, K.; Roers, A.; Blohbaum, B.; Rajewsky, K.; Hansmann, M.L. CD8(+) T cells in Hodgkin’s disease tumor tissue are a polyclonal population with limited clonal expansion but little evidence of selection by antigen. Am. J. Pathol. 2000, 157, 171–175. [Google Scholar] [CrossRef]
- Mukai, H.Y.; Hasegawa, Y.; Kojima, H.; Okoshi, Y.; Takei, N.; Yamashita, Y.; Nagasawa, T.; Mori, N. Nodal CD8 positive cytotoxic T-cell lymphoma: A distinct clinicopathological entity. Mod. Pathol. 2002, 15, 1131–1139. [Google Scholar] [CrossRef][Green Version]
- Wobser, M.; Reinartz, T.; Roth, S.; Goebeler, M.; Rosenwald, A.; Geissinger, E. Cutaneous CD8+ Cytotoxic T-Cell Lymphoma Infiltrates: Clinicopathological Correlation and Outcome of 35 Cases. Oncol. Ther. 2016, 4, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Chatzitolios, A.; Venizelos, I.; Tripsiannis.s, G.; Anastassopoulos, G.; Papadopoulos, N. Prognostic significance of CD95, P53, and BCL2 expression in extranodal non-Hodgkin’s lymphoma. Ann. Hematol. 2010, 89, 889–896. [Google Scholar] [CrossRef][Green Version]
- Kondo, E.; Yoshino, T.; Yamadori, I.; Matsuo, Y.; Kawasaki, N.; Minowada, J.; Akagi, T. Expression of Bcl-2 protein and Fas antigen in non-Hodgkin’s lymphomas. Am. J. Pathol. 1994, 145, 330–337. [Google Scholar] [PubMed]
- Zoi-Toli, O.; Meijer, C.J.; Oudejans, J.J.; de Vries, E.; van Beek, P.; Willemze, R. Expression of Fas and Fas ligand in cutaneous B-cell lymphomas. J. Pathol. 1999, 189, 533–538. [Google Scholar] [CrossRef]
- Chao, M.P.; Alizadeh, A.A.; Tang, C.; Myklebust, J.H.; Varghese, B.; Gill, S.; Jan, M.; Cha, A.C.; Chan, C.K.; Tan, B.T.; et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010, 142, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Leavenworth, J.W.; Li, Y.; Luo, Q.; Xie, H.; Liu, X.; Huang, S.; Yan, H.; Fu, Z.; Zhang, L.Y.; et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl. Acad. Sci. USA 2015, 112, 15988–15993. [Google Scholar] [CrossRef] [PubMed]
- Gattringer, G.; Greil, R.; Radaszkiewicz, T.; Huber, H. In situ quantification of T-cell subsets, NK-like cells and macrophages in Hodgkin’s disease: Quantity and quality of infiltration density depends on histopathological subtypes. Blut 1986, 53, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Alvaro-Naranjo, T.; Lejeune, M.; Salvado-Usach, M.T.; Bosch-Princep, R.; Reverter-Branchat, G.; Jaen-Martinez, J.; Pons-Ferre, L.E. Tumor-infiltrating cells as a prognostic factor in Hodgkin’s lymphoma: A quantitative tissue microarray study in a large retrospective cohort of 267 patients. Leuk. Lymphoma 2005, 46, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Reiners, K.S.; Kessler, J.; Sauer, M.; Rothe, A.; Hansen, H.P.; Reusch, U.; Hucke, C.; Kohl, U.; Durkop, H.; Engert, A.; et al. Rescue of impaired NK cell activity in hodgkin lymphoma with bispecific antibodies in vitro and in patients. Mol. Ther. 2013, 21, 895–903. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Werner, L.; Dreyer, J.H.; Hartmann, D.; Barros, M.H.M.; Buttner-Herold, M.; Grittner, U.; Niedobitek, G. Tumor-associated macrophages in classical Hodgkin lymphoma: Hormetic relationship to outcome. Sci. Rep. 2020, 10, 9410. [Google Scholar] [CrossRef]
- Tudor, C.S.; Bruns, H.; Daniel, C.; Distel, L.V.; Hartmann, A.; Gerbitz, A.; Buettner, M.J. Macrophages and dendritic cells as actors in the immune reaction of classical Hodgkin lymphoma. PLoS ONE 2014, 9, e114345. [Google Scholar] [CrossRef]
- Wang, J.; Gao, K.; Lei, W.; Dong, L.; Xuan, Q.; Feng, M.; Wang, J.; Ye, X.; Jin, T.; Zhang, Z.; et al. Lymphocyte-to-monocyte ratio is associated with prognosis of diffuse large B-cell lymphoma: Correlation with CD163 positive M2 type tumor-associated macrophages, not PD-1 positive tumor-infiltrating lymphocytes. Oncotarget 2017, 8, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Shi, Z.H.; Wang, X.; Gu, K.S.; Zhai, Z.M. Tumor-associated macrophages predict prognosis in diffuse large B-cell lymphoma and correlation with peripheral absolute monocyte count. BMC Cancer 2019, 19, 1049. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Hiwatashi, K.; Ueno, S.; Sakoda, M.; Iino, S.; Okumura, H.; Hashiguchi, M.; Kawasaki, Y.; Kurahara, H.; Mataki, Y.; et al. Prognostic significance of CD68, CD163 and Folate receptor-beta positive macrophages in hepatocellular carcinoma. Exp. Med. 2018, 15, 4465–4476. [Google Scholar] [CrossRef]
- Guo, B.; Cen, H.; Tan, X.; Ke, Q. Meta-analysis of the prognostic and clinical value of tumor-associated macrophages in adult classical Hodgkin lymphoma. BMC Med. 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, F.; Cirillo, M.; Bianchi, A.; Gately, M.; Olimpieri, O.M.; Cerchiara, E.; Renzi, D.; Micera, A.; Balzamino, B.O.; Bonini, S.; et al. High density of CD68+/CD163+ tumour-associated macrophages (M2-TAM) at diagnosis is significantly correlated to unfavorable prognostic factors and to poor clinical outcomes in patients with diffuse large B-cell lymphoma. Hematol. Oncol. 2015, 33, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.J.; Go, H.; Paik, J.H.; Kim, T.M.; Heo, D.S.; Kim, C.W.; Jeon, Y.K. An increase of M2 macrophages predicts poor prognosis in patients with diffuse large B-cell lymphoma treated with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone. Leuk. Lymphoma 2014, 55, 2466–2476. [Google Scholar] [CrossRef] [PubMed]
- Pello, O.M.; Chevre, R.; Laoui, D.; De Juan, A.; Lolo, F.; Andres-Manzano, M.J.; Serrano, M.; Van Ginderachter, J.A.; Andres, V. In vivo inhibition of c-MYC in myeloid cells impairs tumor-associated macrophage maturation and pro-tumoral activities. PLoS ONE 2012, 7, e45399. [Google Scholar] [CrossRef] [PubMed]
- Palazon-Carrion, N.; Jimenez-Cortegana, C.; Sanchez-Leon, M.L.; Henao-Carrasco, F.; Nogales-Fernandez, E.; Chiesa, M.; Caballero, R.; Rojo, F.; Nieto-Garcia, M.A.; Sanchez-Margalet, V.; et al. Circulating immune biomarkers in peripheral blood correlate with clinical outcomes in advanced breast cancer. Sci. Rep. 2021, 11, 14426. [Google Scholar] [CrossRef]
- Ma, P.; Beatty, P.L.; McKolanis, J.; Brand, R.; Schoen, R.E.; Finn, O.J. Circulating Myeloid Derived Suppressor Cells (MDSC) That Accumulate in Premalignancy Share Phenotypic and Functional Characteristics With MDSC in Cancer. Front. Immunol. 2019, 10, 1401. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Cortegana, C.; Liro, J.; Palazon-Carrion, N.; Salamanca, E.; Sojo-Dorado, J.; de la Cruz-Merino, L.; Pascual, A.; Rodriguez-Bano, J.; Sanchez-Margalet, V. Increased Blood Monocytic Myeloid Derived Suppressor Cells but Low Regulatory T Lymphocytes in Patients with Mild COVID-19. Viral Immunol. 2021, 34, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Cortegana, C.; Palazon-Carrion, N.; Martin Garcia-Sancho, A.; Nogales-Fernandez, E.; Carnicero-Gonzalez, F.; Rios-Herranz, E.; de la Cruz-Vicente, F.; Rodriguez-Garcia, G.; Fernandez-Alvarez, R.; Rueda Dominguez, A.; et al. Circulating myeloid-derived suppressor cells and regulatory T cells as immunological biomarkers in refractory/relapsed diffuse large B-cell lymphoma: Translational results from the R2-GDP-GOTEL trial. J. Immunother. Cancer 2021, 9, e002323. [Google Scholar] [CrossRef] [PubMed]
- de Haas, N.; de Koning, C.; Spilgies, L.; de Vries, I.J.; Hato, S.V. Improving cancer immunotherapy by targeting the STATe of MDSCs. Oncoimmunology 2016, 5, e1196312. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Wang, L.Z.; Lin, P.C. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016, 380, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Dashnamoorthy, R.; Galera, P.; Makarenko, V.; Chang, H.; Ghosh, S.; Evens, A.M. The immune checkpoint molecules PD-1, PD-L1, TIM-3 and LAG-3 in diffuse large B-cell lymphoma. Oncotarget 2019, 10, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Marini, O.; Spina, C.; Mimiola, E.; Cassaro, A.; Malerba, G.; Todeschini, G.; Perbellini, O.; Scupoli, M.; Carli, G.; Facchinelli, D.; et al. Identification of granulocytic myeloid-derived suppressor cells (G-MDSCs) in the peripheral blood of Hodgkin and non-Hodgkin lymphoma patients. Oncotarget 2016, 7, 27676–27688. [Google Scholar] [CrossRef] [PubMed]
- Novosad, O.; Gorbach, O.; Skachkova, O.; Khranovska, N.; Kryachok, P.I. Role of Circulating Myeloid-Derived Suppressor Cell (MDSC) in Hodgkin Lymphoma (HL) Progression: Updated Prospective Study. Blood 2020, 136, 31. [Google Scholar] [CrossRef]
- Khalifa, K.A.; Badawy, H.M.; Radwan, W.M.; Shehata, M.A.; Bassuoni, M.A. CD14(+) HLA-DR low/(-) monocytes as indicator of disease aggressiveness in B-cell non-Hodgkin lymphoma. Int. J. Lab. Hematol. 2014, 36, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Xiu, B.; Lin, Y.; Grote, D.M.; Ziesmer, S.C.; Gustafson, M.P.; Maas, M.L.; Zhang, Z.; Dietz, A.B.; Porrata, L.F.; Novak, A.J.; et al. IL-10 induces the development of immunosuppressive CD14(+)HLA-DR(low/-) monocytes in B-cell non-Hodgkin lymphoma. Blood Cancer J. 2015, 5, e328. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Gustafson, M.P.; Bulur, P.A.; Gastineau, D.A.; Witzig, T.E.; Dietz, A.B. Immunosuppressive CD14+HLA-DR(low)/- monocytes in B-cell non-Hodgkin lymphoma. Blood 2011, 117, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Tadmor, T.; Fell, R.; Polliack, A.; Attias, D. Absolute monocytosis at diagnosis correlates with survival in diffuse large B-cell lymphoma-possible link with monocytic myeloid-derived suppressor cells. Hematol. Oncol. 2013, 31, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, R.A.; Feldman, A.L.; Wada, D.A.; Yang, Z.Z.; Comfere, N.I.; Dong, H.; Kwon, E.D.; Novak, A.J.; Markovic, S.N.; Pittelkow, M.R.; et al. B7-H1 (PD-L1, CD274) suppresses host immunity in T-cell lymphoproliferative disorders. Blood 2009, 114, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Azzaoui, I.; Uhel, F.; Rossille, D.; Pangault, C.; Dulong, J.; Le Priol, J.; Lamy, T.; Houot, R.; Le Gouill, S.; Cartron, G.; et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood 2016, 128, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Bontkes, H.J.; Jordanova, E.S.; Nijeboer, P.; Neefjes-Borst, E.A.; Cillessen, S.; Hayat, A.; Mulder, C.J.; Bouma, G.; von Blomberg, B.M.; de Gruijl, T.D. High myeloid-derived suppressor cell frequencies in the duodenum are associated with enteropathy associated T-cell lymphoma and its precursor lesions. Br. J. Haematol. 2017, 178, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Morris, J.R. Genes, genetics, and epigenetics: A correspondence. Science 2001, 293, 1103–1105. [Google Scholar] [CrossRef]
- Rodriguez-Paredes, M.; Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011, 17, 330–339. [Google Scholar] [CrossRef]
- Ashraf, N.; Zino, S.; Macintyre, A.; Kingsmore, D.; Payne, A.P.; George, W.D.; Shiels, P.G. Altered sirtuin expression is associated with node-positive breast cancer. Br. J. Cancer 2006, 95, 1056–1061. [Google Scholar] [CrossRef]
- Adams, H.; Fritzsche, F.R.; Dirnhofer, S.; Kristiansen, G.; Tzankov, A. Class I histone deacetylases 1, 2 and 3 are highly expressed in classical Hodgkin’s lymphoma. Expert Opin. Ther. Targets 2010, 14, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kwon, H.J.; Yoon, B.I.; Kim, J.H.; Han, S.U.; Joo, H.J.; Kim, D.Y. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn. J. Cancer Res. 2001, 92, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Baxter, E.; Windloch, K.; Gannon, F.; Lee, J.S. Epigenetic regulation in cancer progression. Cell Biosci. 2014, 4, 45. [Google Scholar] [CrossRef]
- Kovar, H.; Amatruda, J.; Brunet, E.; Burdach, S.; Cidre-Aranaz, F.; de Alava, E.; Dirksen, U.; van der Ent, W.; Grohar, P.; Grunewald, T.G.; et al. The second European interdisciplinary Ewing sarcoma research summit--A joint effort to deconstructing the multiple layers of a complex disease. Oncotarget 2016, 7, 8613–8624. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Perez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef]
- Thomas, R.M.; Gamper, C.J.; Ladle, B.H.; Powell, J.D.; Wells, A.D. De novo DNA methylation is required to restrict T helper lineage plasticity. J. Biol. Chem. 2012, 287, 22900–22909. [Google Scholar] [CrossRef]
- Zhang, J.A.; Mortazavi, A.; Williams, B.A.; Wold, B.J.; Rothenberg, E.V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 2012, 149, 467–482. [Google Scholar] [CrossRef]
- Queiros, A.C.; Beekman, R.; Vilarrasa-Blasi, R.; Duran-Ferrer, M.; Clot, G.; Merkel, A.; Raineri, E.; Russinol, N.; Castellano, G.; Bea, S.; et al. Decoding the DNA Methylome of Mantle Cell Lymphoma in the Light of the Entire B Cell Lineage. Cancer Cell 2016, 30, 806–821. [Google Scholar] [CrossRef]
- Chambwe, N.; Kormaksson, M.; Geng, H.; De, S.; Michor, F.; Johnson, N.A.; Morin, R.D.; Scott, D.W.; Godley, L.A.; Gascoyne, R.D.; et al. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood 2014, 123, 1699–1708. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef] [PubMed]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Mav, D.; Shah, R.; Grimm, S.A.; Phadke, D.; Hatzi, K.; Melnick, A.; Geigerman, C.; Sobol, S.E.; Jaye, D.L.; et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013, 23, 2030–2041. [Google Scholar] [CrossRef]
- Shaknovich, R.; Cerchietti, L.; Tsikitas, L.; Kormaksson, M.; De, S.; Figueroa, M.E.; Ballon, G.; Yang, S.N.; Weinhold, N.; Reimers, M.; et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 2011, 118, 3559–3569. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.K.; Ab Hamid, S.S.; Musa, M.; Wong, K.K. DNMT1 is associated with cell cycle and DNA replication gene sets in diffuse large B-cell lymphoma. Pathol. Res. Pr. 2018, 214, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Lv, Z.; Zhang, P.; Wang, Y.; Yuan, M.; Yu, X.; Otieno Odhiambo, W.; Zheng, M.; Zhang, H.; Ma, Y.; et al. AID assists DNMT1 to attenuate BCL6 expression through DNA methylation in diffuse large B-cell lymphoma cell lines. Neoplasia 2020, 22, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.L.; Hlady, R.A.; Opavska, J.; Klinkebiel, D.; Novakova, S.; Smith, L.M.; Lewis, R.E.; Karpf, A.R.; Simpson, M.A.; Wu, L.; et al. Essential role for Dnmt1 in the prevention and maintenance of MYC-induced T-cell lymphomas. Mol. Cell Biol. 2013, 33, 4321–4333. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Peng, Z.D.; Xu, S.H.; Gong, Y. Downregulation of miR-152 contributes to DNMT1-mediated silencing of SOCS3/SHP-1 in non-Hodgkin lymphoma. Cancer Gene 2019, 26, 195–207. [Google Scholar] [CrossRef]
- Robaina, M.C.; Mazzoccoli, L.; Arruda, V.O.; Reis, F.R.; Apa, A.G.; de Rezende, L.M.; Klumb, C.E. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015, 98, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920. [Google Scholar] [CrossRef] [PubMed]
- Amara, K.; Ziadi, S.; Hachana, M.; Soltani, N.; Korbi, S.; Trimeche, M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010, 101, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Couronne, L.; Bastard, C.; Bernard, O.A. TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl. J. Med. 2012, 366, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, T.; Singh, A.K.; Chen, T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics 2015, 7, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.A.; Kuo, Y.Y.; Liu, C.Y.; Chou, W.C.; Lee, M.C.; Chen, C.Y.; Lin, L.I.; Tseng, M.H.; Huang, C.F.; Chiang, Y.C.; et al. DNMT3A mutations in acute myeloid leukemia: Stability during disease evolution and clinical implications. Blood 2012, 119, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, K.; Nguyen, T.B.; Chiba, S.; Sakata-Yanagimoto, M. Review of the biologic and clinical significance of genetic mutations in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018, 109, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Scourzic, L.; Couronne, L.; Pedersen, M.T.; Della Valle, V.; Diop, M.; Mylonas, E.; Calvo, J.; Mouly, E.; Lopez, C.K.; Martin, N.; et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia 2016, 30, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Sakata-Yanagimoto, M.; Nishizawa, S.; Komori, D.; Gershon, P.; Kiryu, M.; Tanzima, S.; Fukumoto, K.; Enami, T.; Muratani, M.; et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia 2018, 32, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Vallois, D.; Dobay, M.P.; Morin, R.D.; Lemonnier, F.; Missiaglia, E.; Juilland, M.; Iwaszkiewicz, J.; Fataccioli, V.; Bisig, B.; Roberti, A.; et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016, 128, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Hajji, N.; Garcia-Dominguez, D.J.; Hontecillas-Prieto, L.; O’Neill, K.; de Alava, E.; Syed, N. The bitter side of epigenetics: Variability and resistance to chemotherapy. Epigenomics 2018, 13, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Hontecillas-Prieto, L.; Flores-Campos, R.; Silver, A.; de Alava, E.; Hajji, N.; Garcia-Dominguez, D.J. Synergistic Enhancement of Cancer Therapy Using HDAC Inhibitors: Opportunity for Clinical Trials. Front. Genet. 2020, 11, 578011. [Google Scholar] [CrossRef] [PubMed]
- Marquard, L.; Poulsen, C.B.; Gjerdrum, L.M.; de Nully Brown, P.; Christensen, I.J.; Jensen, P.B.; Sehested, M.; Johansen, P.; Ralfkiaer, E. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology 2009, 54, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, H.; Jia, X.; Hu, G.; Kong, L.; Zhang, T.; Li, L.; Pan, Y.; Zhai, Q.; Meng, B.; et al. Clinical significance of enhancer of zeste homolog 2 and histone deacetylases 1 and 2 expression in peripheral T-cell lymphoma. Oncol. Lett. 2019, 18, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Han, J.J.; Stenson, M.; Wellik, L.; Witzig, T.E. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: Implications for therapy. Leukemia 2012, 26, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; La Perle, K.; Kwiatkowski, S.; Sullivan, L.A.; Sams, G.H.; Johns, J.; Curphey, D.P.; Wen, J.; McConnell, K.; Qi, J.; et al. Mechanism, Consequences, and Therapeutic Targeting of Abnormal IL15 Signaling in Cutaneous T-cell Lymphoma. Cancer Discov. 2016, 6, 986–1005. [Google Scholar] [CrossRef]
- Gil, V.S.; Bhagat, G.; Howell, L.; Zhang, J.; Kim, C.H.; Stengel, S.; Vega, F.; Zelent, A.; Petrie, K. Deregulated expression of HDAC9 in B cells promotes development of lymphoproliferative disease and lymphoma in mice. Dis. Model. Mech. 2016, 9, 1483–1495. [Google Scholar] [CrossRef]
- Ding, H.; Peterson, K.L.; Correia, C.; Koh, B.; Schneider, P.A.; Nowakowski, G.S.; Kaufmann, S.H. Histone deacetylase inhibitors interrupt HSP90*RASGRP1 and HSP90*CRAF interactions to upregulate BIM and circumvent drug resistance in lymphoma cells. Leukemia 2017, 31, 1593–1602. [Google Scholar] [CrossRef]
- Valdez, B.C.; Brammer, J.E.; Li, Y.; Murray, D.; Liu, Y.; Hosing, C.; Nieto, Y.; Champlin, R.E.; Andersson, B.S. Romidepsin targets multiple survival signaling pathways in malignant T cells. Blood Cancer J. 2015, 5, e357. [Google Scholar] [CrossRef]
- Piazza, R.; Magistroni, V.; Mogavero, A.; Andreoni, F.; Ambrogio, C.; Chiarle, R.; Mologni, L.; Bachmann, P.S.; Lock, R.B.; Collini, P.; et al. Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex. Neoplasia 2013, 15, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Melnick, A. The epigenetic basis of diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, A.B.; Ondrejka, S.L.; McKinney, M.; Rempel, R.E.; Goodlad, J.R.; Teh, C.H.; Leppa, S.; Mannisto, S.; Kovanen, P.E.; Tse, E.; et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J. Exp. Med. 2017, 214, 1371.1–1386. [Google Scholar] [CrossRef] [PubMed]
- Tomita, S.; Kikuti, Y.Y.; Carreras, J.; Sakai, R.; Takata, K.; Yoshino, T.; Bea, S.; Campo, E.; Missiaglia, E.; Bouilly, J.; et al. Monomorphic Epitheliotropic Intestinal T-Cell Lymphoma in Asia Frequently Shows SETD2 Alterations. Cancers 2020, 12, 3539. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pol, S.; Ma, L.; Joshi, R.P.; Arber, D.A. A Survey of Somatic Mutations in 41 Genes in a Cohort of T-Cell Lymphomas Identifies Frequent Mutations in Genes Involved in Epigenetic Modification. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.C.; Dou, Y. Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 2015, 15, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Park, H.Y.; Kang, S.Y.; Kim, S.J.; Hwang, J.; Lee, S.; Kwak, S.H.; Park, K.S.; Yoo, H.Y.; Kim, W.S.; et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget 2015, 6, 17764–17776. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, W.; Li, J.; Xiong, J.; Liu, J.; Chen, T.; Wen, Q.; Zeng, Y.; Gao, L.; Zhang, C.; et al. Plasma circulating tumor DNA assessment reveals KMT2D as a potential poor prognostic factor in extranodal NK/T-cell lymphoma. Biomark. Res. 2020, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef]
- Ortega-Molina, A.; Boss, I.W.; Canela, A.; Pan, H.; Jiang, Y.; Zhao, C.; Jiang, M.; Hu, D.; Agirre, X.; Niesvizky, I.; et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 2015, 21, 1199–1208. [Google Scholar] [CrossRef]
- Jiang, L.; Gu, Z.H.; Yan, Z.X.; Zhao, X.; Xie, Y.Y.; Zhang, Z.G.; Pan, C.M.; Hu, Y.; Cai, C.P.; Dong, Y.; et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 2015, 47, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.M.; Huang, Y.H.; Huang, J.Y.; Wang, Z.F.; Fu, D.; Liu, H.; Liu, F.; Leboeuf, C.; Wang, L.; Ye, J.; et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica 2018, 103, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, S.; Rossi, D.; Rinaldi, A.; Bruscaggin, A.; Spina, V.; Eskelund, C.W.; Evangelista, A.; Moia, R.; Kwee, I.; Dahl, C.; et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: A FIL study. Haematologica 2020, 105, 1604–1612. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Helin, K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 2009, 9, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stutzer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 2014, 53, 49–62. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Graves, A.P.; Ganji, G.; Diaz, E.; Halsey, W.S.; Jiang, Y.; Smitheman, K.N.; Ott, H.M.; Pappalardi, M.B.; Allen, K.E.; et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA 2012, 109, 2989–2994. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Pelton, A.; Shahsafaei, A.; Dorfman, D.M. Differential expression of enhancer of zeste homolog 2 (EZH2) protein in small cell and aggressive B-cell non-Hodgkin lymphomas and differential regulation of EZH2 expression by p-ERK1/2 and MYC in aggressive B-cell lymphomas. Mod. Pathol. 2016, 29, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Ng, S.B.; Tay, J.L.; Lin, B.; Koh, T.L.; Tan, J.; Selvarajan, V.; Liu, S.C.; Bi, C.; Wang, S.; et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013, 121, 4512–4520. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Van Vlierberghe, P.; Nedjic, J.; Trimarchi, T.; Flaherty, M.S.; Ferres-Marco, D.; da Ros, V.; Tang, Z.; Siegle, J.; et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012, 18, 298–301. [Google Scholar] [CrossRef]
- Simon, C.; Chagraoui, J.; Krosl, J.; Gendron, P.; Wilhelm, B.; Lemieux, S.; Boucher, G.; Chagnon, P.; Drouin, S.; Lambert, R.; et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012, 26, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liang, L.; Huang, S.; Nong, L.; Li, D.; Zhang, B.; Li, T. Aberrant differential expression of EZH2 and H3K27me3 in extranodal NK/T-cell lymphoma, nasal type, is associated with disease progression and prognosis. Hum. Pathol. 2019, 83, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Soneji, S.; Marafioti, T.; Cooper, C.D.; Palazzo, S.; Paterson, J.C.; Cattan, H.; Enver, T.; Mager, R.; Boultwood, J.; et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer 2007, 121, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Roehle, A.; Hoefig, K.P.; Repsilber, D.; Thorns, C.; Ziepert, M.; Wesche, K.O.; Thiere, M.; Loeffler, M.; Klapper, W.; Pfreundschuh, M.; et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br. J. Haematol. 2008, 142, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shao, Q.; Li, C.; Liu, H.; Li, J.; Wang, Y.; Song, W.; Li, L.; Wang, G.; Shao, Z.; et al. Effects of microRNA-21 on apoptosis by regulating the expression of PTEN in diffuse large B-cell lymphoma. Medicine 2017, 96, e7952. [Google Scholar] [CrossRef] [PubMed]
- Leucci, E.; Cocco, M.; Onnis, A.; De Falco, G.; van Cleef, P.; Bellan, C.; van Rijk, A.; Nyagol, J.; Byakika, B.; Lazzi, S.; et al. MYC translocation-negative classical Burkitt lymphoma cases: An alternative pathogenetic mechanism involving miRNA deregulation. J. Pathol. 2008, 216, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.L.; Trinh, D.L.; Scott, D.W.; Chu, A.; Krzywinski, M.; Zhao, Y.; Robertson, A.G.; Mungall, A.J.; Schein, J.; Boyle, M.; et al. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol. 2015, 16, 18. [Google Scholar] [CrossRef]
- Iqbal, J.; Shen, Y.; Huang, X.; Liu, Y.; Wake, L.; Liu, C.; Deffenbacher, K.; Lachel, C.M.; Wang, C.; Rohr, J.; et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood 2015, 125, 1137–1145. [Google Scholar] [CrossRef]
- Lenze, D.; Leoncini, L.; Hummel, M.; Volinia, S.; Liu, C.G.; Amato, T.; De Falco, G.; Githanga, J.; Horn, H.; Nyagol, J.; et al. The different epidemiologic subtypes of Burkitt lymphoma share a homogenous micro RNA profile distinct from diffuse large B-cell lymphoma. Leukemia 2011, 25, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Leich, E.; Zamo, A.; Horn, H.; Haralambieva, E.; Puppe, B.; Gascoyne, R.D.; Chan, W.C.; Braziel, R.M.; Rimsza, L.M.; Weisenburger, D.D.; et al. MicroRNA profiles of t(14;18)-negative follicular lymphoma support a late germinal center B-cell phenotype. Blood 2011, 118, 5550–5558. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Jiang, Y.; Du, W.; Fairchild, L.L.; Melnick, A.; Elemento, O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhao, H.; Xu, W.; Bao, S.; Cheng, L.; Sun, J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, H.; Guo, Y.; Luo, Y.; Li, H.; Xu, Y.; Deng, J.; Sun, B. A pilot study of long noncoding RNA expression profiling by microarray in follicular lymphoma. Gene 2016, 577, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Calin, G.; Deng, M.D.; Au-Yeung, R.K.H.; Wang, L.Q.; Chim, C.S. Epigenetic Silencing of Tumor Suppressor lncRNA NKILA: Implication on NF-kappaB Signaling in Non-Hodgkin’s Lymphoma. Genes 2022, 13, 128. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Gronbaek, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhao, Y.; Shi, C.; Ren, P.; Wei, B.; Guo, Y.; Ma, J. A circular RNA from APC inhibits the proliferation of diffuse large B-cell lymphoma by inactivating Wnt/beta-catenin signaling via interacting with TET1 and miR-888. Aging 2019, 11, 8068–8084. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Cheng, S.; Luo, N.; Gao, R.; Yu, K.; Kang, B.; Wang, L.; Zhang, Q.; Fang, Q.; Zhang, L.; et al. Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol. 2019, 21, 2. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Vasilatos, S.N.; Chen, L.; Wu, H.; Cao, Z.; Fu, Y.; Huang, M.; Vlad, A.M.; Lu, B.; Oesterreich, S.; et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 2019, 38, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.J.; McCuaig, R.D.; Tan, A.H.Y.; Hardy, K.; Seddiki, N.; Ali, S.; Dahlstrom, J.E.; Bean, E.G.; Dunn, J.; Forwood, J.; et al. Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch. Front. Immunol. 2020, 11, 1228. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, N.; Hamaguchi, M.; Morikawa, H.; Sugimura, K.; Tanaka, A.; Ito, Y.; Osaki, M.; Tanaka, Y.; Yamashita, R.; Nakano, N.; et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 2012, 37, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M.; et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef] [PubMed]
- Schenk, A.; Bloch, W.; Zimmer, P. Natural Killer Cells—An Epigenetic Perspective of Development and Regulation. Int. J. Mol. Sci. 2016, 17, 326. [Google Scholar] [CrossRef] [PubMed]
- Ogbomo, H.; Michaelis, M.; Kreuter, J.; Doerr, H.W.; Cinatl, J., Jr. Histone deacetylase inhibitors suppress natural killer cell cytolytic activity. FEBS Lett. 2007, 581, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sanchez, A.; Baragano Raneros, A.; Carvajal Palao, R.; Sanz, A.B.; Ortiz, A.; Ortega, F.; Suarez-Alvarez, B.; Lopez-Larrea, C. DNA demethylation and histone H3K9 acetylation determine the active transcription of the NKG2D gene in human CD8+ T and NK cells. Epigenetics 2013, 8, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Liu, D.; Yu, L.; Xue, B.; Shi, H. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 2014, 28, 565–574. [Google Scholar] [CrossRef]
- Ishii, M.; Wen, H.; Corsa, C.A.; Liu, T.; Coelho, A.L.; Allen, R.M.; Carson, W.F.T.; Cavassani, K.A.; Li, X.; Lukacs, N.W.; et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009, 114, 3244–3254. [Google Scholar] [CrossRef]
- Kittan, N.A.; Allen, R.M.; Dhaliwal, A.; Cavassani, K.A.; Schaller, M.; Gallagher, K.A.; Carson, W.F.T.; Mukherjee, S.; Grembecka, J.; Cierpicki, T.; et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS ONE 2013, 8, e78045. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Takeuchi, O.; Vandenbon, A.; Yasuda, K.; Tanaka, Y.; Kumagai, Y.; Miyake, T.; Matsushita, K.; Okazaki, T.; Saitoh, T.; et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010, 11, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Lo Sasso, G.; Menzies, K.J.; Mottis, A.; Piersigilli, A.; Perino, A.; Yamamoto, H.; Schoonjans, K.; Auwerx, J. SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS ONE 2014, 9, e103573. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wei, J.; Zhong, L.; Shi, M.; Zhou, P.; Zuo, S.; Wu, K.; Zhu, M.; Huang, X.; Yu, Y.; et al. Cross talk between histone deacetylase 4 and STAT6 in the transcriptional regulation of arginase 1 during mouse dendritic cell differentiation. Mol. Cell Biol. 2015, 35, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Barozzi, I.; Termanini, A.; Prosperini, E.; Recchiuti, A.; Dalli, J.; Mietton, F.; Matteoli, G.; Hiebert, S.; Natoli, G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. USA 2012, 109, E2865–E2874. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Rong, S.; Repa, J.J.; St Clair, R.; Parks, J.S.; Mishra, N. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arter. Thromb. Vasc. Biol. 2014, 34, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cheng, F.; Wang, M.; Sahakian, E.; Powers, J.J.; Ibarz, J.P.; Smith, M.R.; Sotomayor, E. Loss of HDAC11 Promotes Myeloid-Derived Suppressor Cells Inhibition of T Cell Function in a Murine Lymphoma Microenvironment. Blood 2018, 132, 1105. [Google Scholar] [CrossRef]
- Orillion, A.; Hashimoto, A.; Damayanti, N.; Shen, L.; Adelaiye-Ogala, R.; Arisa, S.; Chintala, S.; Ordentlich, P.; Kao, C.; Elzey, B.; et al. Entinostat Neutralizes Myeloid-Derived Suppressor Cells and Enhances the Antitumor Effect of PD-1 Inhibition in Murine Models of Lung and Renal Cell Carcinoma. Clin. Cancer Res. 2017, 23, 5187–5201. [Google Scholar] [CrossRef] [PubMed]
- Sahakian, E.; Powers, J.J.; Chen, J.; Deng, S.L.; Cheng, F.; Distler, A.; Woods, D.M.; Rock-Klotz, J.; Sodre, A.L.; Youn, J.I.; et al. Histone deacetylase 11: A novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol. Immunol. 2015, 63, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Lienlaf, M.; Perez-Villarroel, P.; Wang, H.W.; Lee, C.; Woan, K.; Woods, D.; Knox, T.; Bergman, J.; Pinilla-Ibarz, J.; et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol. Immunol. 2014, 60, 44–53. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, L.; Chen, Q.; Song, Y.; Xu, S.; Ma, F.; Wang, X.; Wang, J.; Yu, H.; Cao, X.; et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J. Immunol. 2012, 188, 5500–5510. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.; Mascarenhas, J.; Telford, W.; Chadburn, A.; Friedman, S.M.; Schattner, E.J. Proliferative response of mantle cell lymphoma cells stimulated by CD40 ligation and IL-4. Leukemia 2000, 14, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Harrington, B.K.; Wheeler, E.; Hornbuckle, K.; Shana’ah, A.Y.; Youssef, Y.; Smith, L.; Hassan, Q., II; Klamer, B.; Zhang, X.; Long, M.; et al. Modulation of immune checkpoint molecule expression in mantle cell lymphoma. Leuk. Lymphoma 2019, 60, 2498–2507. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharawat, S.K. Epigenetic regulators of programmed death-ligand 1 expression in human cancers. Transl. Res. J. Lab. Clin. Med. 2018, 202, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, R.; Zhao, H.J.; Fu, D.; Zhong, H.J.; Weng, X.Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer 2019, 18, 54. [Google Scholar] [CrossRef] [PubMed]
- Tiper, I.V.; Webb, T.J. Histone deacetylase inhibitors enhance CD1d-dependent NKT cell responses to lymphoma. Cancer Immunol. Immunother. CII 2016, 65, 1411–1421. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Cai, K.; Xu, P.P.; Wang, L.; Huang, C.X.; Fang, Y.; Cheng, S.; Sun, X.J.; Liu, F.; Huang, J.Y.; et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal. Transduct. Target. 2021, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Li, L.H.; Olin, E.J.; Buskirk, H.H.; Reineke, L.M. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 1970, 30, 2760–2769. [Google Scholar]
- Vogler, W.R.; Miller, D.S.; Keller, J.W. 5-Azacytidine (NSC 102816): A new drug for the treatment of myeloblastic leukemia. Blood 1976, 48, 331–337. [Google Scholar] [CrossRef]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.J.; Issa, J.P.; Kurzrock, R.; Nunez, M.I.; Jelinek, J.; Hong, D.; Oki, Y.; Guo, Z.; Gupta, S.; Wistuba, I.I. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin. Cancer Res. 2009, 15, 3881–3888. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.A.; Liu, Z.; Lucas, D.M.; Chen, P.; Xie, Z.; Baiocchi, R.; Benson, D.M.; Devine, S.M.; Jones, J.; Andritsos, L.; et al. Phase I trial of low dose decitabine targeting DNA hypermethylation in patients with chronic lymphocytic leukaemia and non-Hodgkin lymphoma: Dose-limiting myelosuppression without evidence of DNA hypomethylation. Br. J. Haematol. 2010, 150, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kato, H.; Kobayashi, Y.; Yamasaki, S.; Morita-Hoshi, Y.; Yokoyama, H.; Morishima, Y.; Ricker, J.L.; Otsuki, T.; Miyagi-Maesima, A.; et al. Potential efficacy of the oral histone deacetylase inhibitor vorinostat in a phase I trial in follicular and mantle cell lymphoma. Cancer Sci. 2010, 101, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Ando, K.; Suzuki, T.; Ishizawa, K.; Oh, S.Y.; Itoh, K.; Yamamoto, K.; Au, W.Y.; Tien, H.F.; Matsuno, Y.; et al. A multicentre phase II study of vorinostat in patients with relapsed or refractory indolent B-cell non-Hodgkin lymphoma and mantle cell lymphoma. Br. J. Haematol. 2014, 165, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Frankel, P.; Popplewell, L.; Siddiqi, T.; Ruel, N.; Rotter, A.; Thomas, S.H.; Mott, M.; Nathwani, N.; Htut, M.; et al. A phase II study of vorinostat and rituximab for treatment of newly diagnosed and relapsed/refractory indolent non-Hodgkin lymphoma. Haematologica 2015, 100, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Persky, D.O.; Li, H.; Rimsza, L.M.; Barr, P.M.; Popplewell, L.L.; Bane, C.L.; Von Gehr, A.; LeBlanc, M.; Fisher, R.I.; Smith, S.M.; et al. A phase I/II trial of vorinostat (SAHA) in combination with rituximab-CHOP in patients with newly diagnosed advanced stage diffuse large B-cell lymphoma (DLBCL): SWOG S0806. Am. J. Hematol. 2018, 93, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, R.L.; Frye, R.; Prince, H.M.; Kirsch.hbaum, M.H.; Zain, J.; Allen, S.L.; Jaffe, E.S.; Ling, A.; Turner, M.; Peer, C.J.; et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. Blood 2011, 117, 5827–5834. [Google Scholar] [CrossRef]
- Piekarz, R.L.; Frye, R.; Turner, M.; Wright, J.J.; Allen, S.L.; Kirschbaum, M.H.; Zain, J.; Prince, H.M.; Leonard, J.P.; Geskin, L.J.; et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J. Clin. Oncol. 2009, 27, 5410–5417. [Google Scholar] [CrossRef]
- Puvvada, S.D.; Li, H.; Rimsza, L.M.; Bernstein, S.H.; Fisher, R.I.; LeBlanc, M.; Schmelz, M.; Glinsmann-Gibson, B.; Miller, T.P.; Maddox, A.M.; et al. A phase II study of belinostat (PXD101) in relapsed and refractory aggressive B-cell lymphomas: SWOG S0520. Leuk. Lymphoma 2016, 57, 2359–2369. [Google Scholar] [CrossRef] [PubMed]
- Foss, F.; Advani, R.; Duvic, M.; Hymes, K.B.; Intragumtornchai, T.; Lekhakula, A.; Shpilberg, O.; Lerner, A.; Belt, R.J.; Jacobsen, E.D.; et al. A Phase II trial of Belinostat (PXD101) in patients with relapsed or refractory peripheral or cutaneous T-cell lymphoma. Br. J. Haematol. 2015, 168, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Oki, Y.; Buglio, D.; Fanale, M.; Fayad, L.; Copeland, A.; Romaguera, J.; Kwak, L.W.; Pro, B.; de Castro Faria, S.; Neelapu, S.; et al. Phase I study of panobinostat plus everolimus in patients with relapsed or refractory lymphoma. Clin. Cancer Res. 2013, 19, 6882–6890. [Google Scholar] [CrossRef] [PubMed]
| Immune Cells in TME | Function | Mechanism | References |
|---|---|---|---|
| Th1 cells | Anti-tumor cells | Produce IL-2, IFN-gamma, and TNF-beta. Activate CTLs, antigen-presenting cells (APCs), and NK cells | [47] |
| Th2 cells | Pro-tumor cells | Express IL-4, IL-5, IL-6, and IL-10 and support tumor cell growth through CD40–CD40L interactions | [6,48] |
| Tregs | Pro- and anti-tumor cells | Express co-inhibitory and co-stimulatory markers. Three different Treg subsets have been identified in NHL. High levels of PD-1, OX40, ICOS, TIGIT, CTLA-4 and CD28, CD69, and CD95/Fas (activation markers). The third subset of Tregs had a lack of FOXP3 and a high expression of LAG3 and CTLA-4, IL10, CD38, KLRB1 (immunosuppression-associated genes) | [57] |
| CTL cells | Pro- and anti-tumor cells | Suppression of the progression of B-cell NHL [42]. Induce the progression in nodal cytotoxic T-cell lymphoma and cutaneous T-cell lymphoma [43,44] | [67,68,69] |
| B cells | Pro- and anti-tumor | Contribute to immune evasion [48], B cells allowing the progression of the disease inducing anergy in CD4+ T cells by CTLA-4 up-regulation [49] | [7,73] |
| NK cells | Anti-tumor cells | IL-2 and NKp30 and NKG2D surface receptors | [77] |
| M2 macrophages | Pro-tumor cells | High number of CD163+/MYC+ cells in HL is associated with worst outcome | [80] |
| MDSCs | Pro-tumor cells | Inhibit T cell immune responses by reactive oxygen species or nitrogen oxide production. Inhibit the maturation and functions of NK cells and to induce macrophages polarization into M2 phenotype by secreting IL-10 and transforming growth factor (TGF)-β secretion | [93,95,96] |
| Epigenetic Subgroup | Epigenetic Modifications/Mechanism | Function |
|---|---|---|
| DNA methylation | DNMT1 overexpression: hipermethylation of tumor supressors promoters/impaired T-cell infiltration in the TME | Pro-tumor cells |
| DNMT 3A mutation: hypomethylation and Notch1 pathway activation | Anti-tumor cells (mutation pro-tumor cells) | |
| DNMT 3B overexpression: hipermethylation MYC promoter; downregulation: increases M2 macrophage expression markers (Arg1) | Pro-tumor cells | |
| Histone acetylation | HDAC1 overexpression: inhibits the tumor supressor BIM and increase in IL-15 secretion | Pro-tumor cells |
| HDAC2 and EZH2 overexpression: inhibits the tumor supressor BIM/mpaired T-cell infiltration in the TME | Pro-tumor cells | |
| HDAC3 overexpression: downregulation: downregulation of inflammatory genes | Pro-tumor cells (downregulation anti-tumor cells) | |
| HDAC6 overexpression: increase in IL-15 secretion | Pro-tumor cells (PTCL) and anti-tumor cells (DLBCL) | |
| HDAC9 overexpression: modulates BCL6 activity and inhibits the tumor supressor p53; downregulation: downregulation of inflammatory genes | Pro-tumor cells (downregulation pro-tumor cells) | |
| HDAC11: impaires immunosuppressive–MDSC capacity | Anti-tumor cells | |
| Histone methylation | EZH2 (catalyzes H3K27me3) overexpression: regulates MYC and induces high proliferation of/increases in NKG2D receptor | Pro-tumor cells |
| JMJD3 (promotes di- and tri-demethylation of H3K27) induces activation of Arg1, among other M2 markers | Pro-tumor cells | |
| LSD1 (mono- or di-demethylase for H3K4) regulates infiltration of CD8+ T-cells into tumors | Anti-tumor cells (downregulation pro-tumor cells) | |
| MLL2 (catalyzes H3K4 monomethyltransferase) mutation: alters genes expression (ARID1A, TRAF3) or signaling pathways (JAK-STAT or MAPK) | Anti-tumor cells (mutation pro-tumor cells) | |
| SETD2 (catalyzes H3K36me3) mutation: enteropathy-associated T-cell lymphoma | Anti-tumor cells (mutation pro-tumor cells) | |
| SMYD3 (catalyzes H3K4 and H4K5 methyltransferase) induces activation of Arg1, among other M2 markers | Pro-tumor cells | |
| Non-coding RNA | circ-APC regulates tumor cell proliferation | Anti-tumor cells (downregulation pro-tumor cells) |
| let-7 family inhibits MYC expression | Anti-tumor cells | |
| miR-34b inhibits MYC expression | Anti-tumor cells | |
| miR494 promotes the accumulation and activity of MDSCs by Akt pathway activation | Pro-tumor cells | |
| miRNA-21 downregulation: increases PTEN expression and induces apoptosis | Anti-tumor cells |
| NCT Number | Title | Conditions | Phase |
|---|---|---|---|
| SAHA/Vorinostat (HDACi) | |||
| NCT00972478 | Vorinostat, Rituximab, and Combination Chemotherapy in Treating Patients With Newly Diagnosed Stage II, Stage III, or Stage IV Diffuse Large B-Cell Lymphoma |
|
|
| NCT00336063 | Vorinostat and Azacitidine in Treating Patients with Locally Recurrent or Metastatic Nasopharyngeal Cancer or Nasal Natural Killer T-Cell Lymphoma |
| Phase 1 |
| NCT03150329 | Pembrolizumab and Vorinostat in Treating Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, or Hodgkin Lymphoma |
| Phase 1 |
| NCT01193842 | Vorinostat and Combination Chemotherapy with Rituximab in Treating Patients With HIV-Related Diffuse Large B-Cell Non-Hodgkin Lymphoma or Other Aggressive B-Cell Lymphomas |
|
|
| Romidepsin (HDACi) | |||
| NCT03547700 | Study of Ixazomib and Romidepsin in Peripheral T-cellLymphoma (PTCL) |
|
|
| NCT03141203 | Evaluation of the Combination of Romidepsin and Carfilzomib in Relapsed/Refractory Peripheral T Cell Lymphoma Patients |
|
|
| NCT01796002 | Efficacy and Safety of Romidepsin CHOP vs. CHOP inPatients with Untreated Peripheral T-Cell Lymphoma |
| Phase 3 |
| NCT02616965 | A Study to Assess the Feasibility of Romidepsin Combined with Brentuximab Vedotin in Cutaneous T-cell Lymphoma |
| Phase 1 |
| NCT01908777 | A Phase 2 Multicenter Study of High Dose Chemotherapy with Autologous Stem Cell Transplant Followed by Maintenance Therapy with Romidepsin for the Treatment of T-Cell Non-Hodgkin Lymphoma |
| Phase 2 |
| NCT01755975 | Romidepsin in Combination with Lenalidomide in Adultswith Relapsed or Refractory Lymphomas and Myeloma |
|
|
| NCT03534180 | Venetoclax and Romidepsin in Treating Patients withRecurrent or Refractory Mature T-Cell Lymphoma |
| Phase 2 |
| Belinostat (HDACi) | |||
| NCT02737046 | Belinostat Therapy With Zidovudine for Adult T-CellLeukemia-Lymphoma |
| Phase 2 |
| Panobinostat (HDACi) | |||
| NCT01261247 | Panobinostat in Treating Patients with Relapsed orRefractory Non-Hodgkin Lymphoma |
| Phase 2 |
| 5-azacytidine (DNMTi) | |||
| NCT03450343 | Oral Azacitidine Plus Salvage Chemotherapy inRelapsed/Refractory Diffuse Large B-Cell Lymphoma |
| Phase 1 |
| NCT03703375 | Efficacy and Safety of Oral Azacitidine (CC-486) Compared to Investigator’s Choice Therapy in Patients with Relapsed or Refractory Angioimmunoblastic T-Cell Lymphoma |
| Phase 3 |
| NCT04897477 | Azacytidine, Bendamustine, Piamprizumab inRefractory/Relapsed B-Cell Non-Hodgkin Lymphoma |
|
|
| NCT04480125 | Azacitidine Combined with Chidamide in the Treatment of Newly Diagnosed PTCL Unfit for Conventional Chemotherapy |
| Phase 2 |
| NCT03593018 | Efficacy and Safety of Oral Azacitidine Compared to Investigator’s Choice Therapy in Patients with Relapsed or Refractory AITL |
| Phase 3 |
| NCT04747236 | A Randomized, Phase IIB, Multicenter, Trial of Oral Azacytidine Plus Romidepsin Versus Investigator’s Choice in Patients with Relapse or Refractory Peripheral T-Cell Lymphoma (PTCL) |
| Phase 2 |
| NCT05162976 | CC-486 and Nivolumab for the Treatment of HodgkinLymphoma Refractory to PD-1 Therapy or Relapsed |
| Phase 1 |
| NCT03542266 | CC486-CHOP in Patients with Previously UntreatedPeripheral T-Cell Lymphoma |
| Phase 2 |
| NCT04578600 | CC-486, Lenalidomide, and Obinutuzumab for the Treatment of Recurrent or Refractory CD20 Positive B- Cell Lymphoma |
| Phase 1 |
| NCT02828358 | Azacitidine and Combination Chemotherapy in Treating Infants With Acute Lymphoblastic Leukemia and KMT2A Gene Rearrangement |
| Phase 2 |
| 5-Aza-2-deoxycytidine (DNMTi) | |||
| NCT02951728 | Decitabine Plus R-CHOP in Diffuse Large B-CellLymphoma |
|
|
| NCT04697940 | Decitabine-primed Tandem CD19/CD20 CAR T-Cells’Treatment in r/r B-NHL |
|
|
| NCT04850560 | Sequential Low-dose Decitabine with PD-1/CD28 CD19CAR-T in Relapsed or Refractory B-Cell Lymphoma |
| Phase 1 |
| NCT03494296 | A Prospective Study of Low-dose Decitabine Combined with COP Regimen in the Treatment of Relapsed and Refractory DLBCL |
| |
| NCT04446130 | Study of Decitabine Combined with HAAG Regimen in Newly Diagnosed ETP-ALL/LBL, T/M-MPAL, and ALL/ LBL with Myeloid or Stem Cell Markers Patients |
| Phase 3 |
| NCT04553393 | Decitabine-primed Tandem CD19/CD20 CAR T-Cells Plus Epigenetic Agents in Aggressive r/r B-NHL with Huge Tumor Burden |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Domínguez, D.J.; Hontecillas-Prieto, L.; Palazón-Carrión, N.; Jiménez-Cortegana, C.; Sánchez-Margalet, V.; de la Cruz-Merino, L. Tumor Immune Microenvironment in Lymphoma: Focus on Epigenetics. Cancers 2022, 14, 1469. https://doi.org/10.3390/cancers14061469
García-Domínguez DJ, Hontecillas-Prieto L, Palazón-Carrión N, Jiménez-Cortegana C, Sánchez-Margalet V, de la Cruz-Merino L. Tumor Immune Microenvironment in Lymphoma: Focus on Epigenetics. Cancers. 2022; 14(6):1469. https://doi.org/10.3390/cancers14061469
Chicago/Turabian StyleGarcía-Domínguez, Daniel J., Lourdes Hontecillas-Prieto, Natalia Palazón-Carrión, Carlos Jiménez-Cortegana, Víctor Sánchez-Margalet, and Luis de la Cruz-Merino. 2022. "Tumor Immune Microenvironment in Lymphoma: Focus on Epigenetics" Cancers 14, no. 6: 1469. https://doi.org/10.3390/cancers14061469
APA StyleGarcía-Domínguez, D. J., Hontecillas-Prieto, L., Palazón-Carrión, N., Jiménez-Cortegana, C., Sánchez-Margalet, V., & de la Cruz-Merino, L. (2022). Tumor Immune Microenvironment in Lymphoma: Focus on Epigenetics. Cancers, 14(6), 1469. https://doi.org/10.3390/cancers14061469






