In Silico Analysis of Ion Channels and Their Correlation with Epithelial to Mesenchymal Transition in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
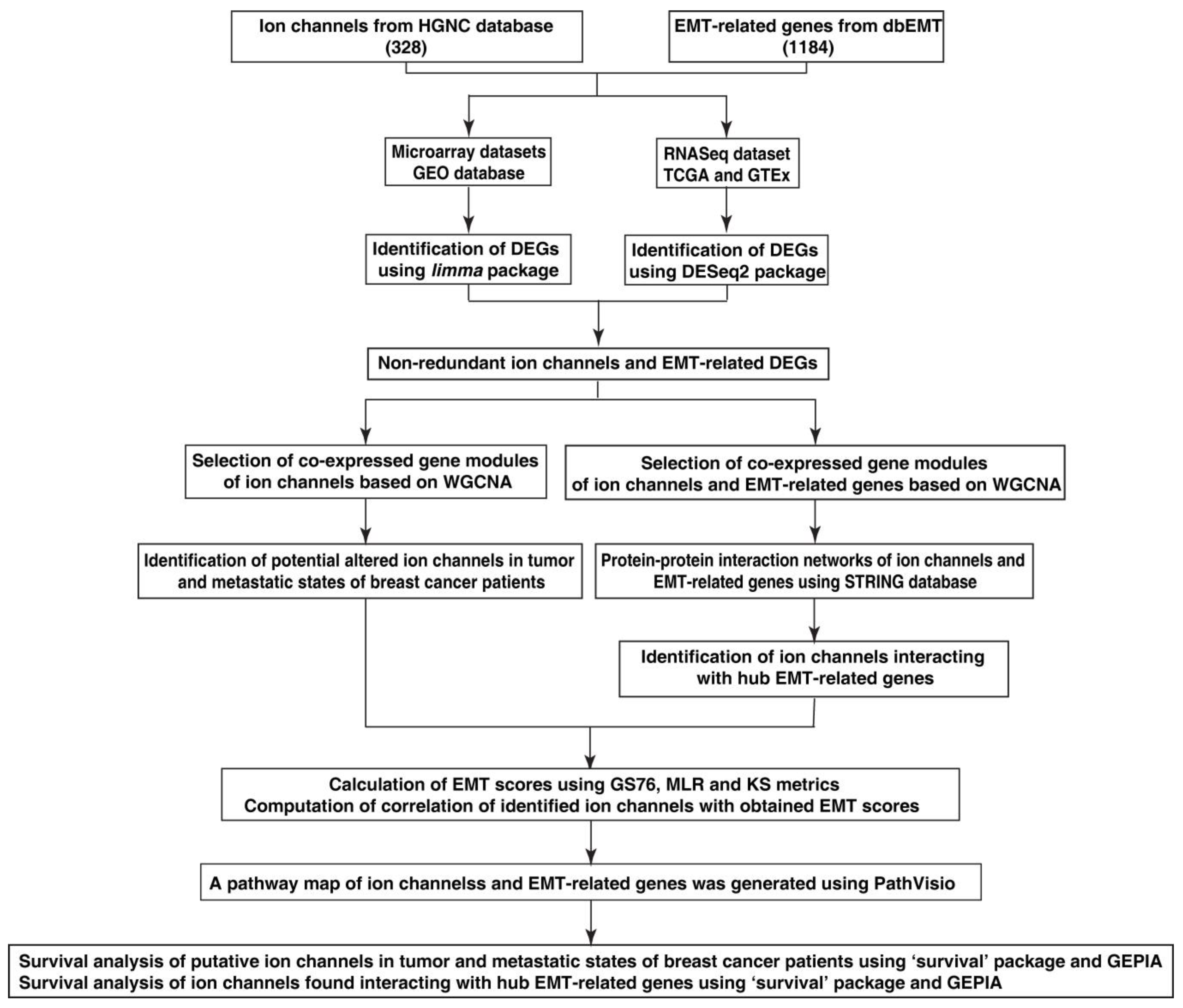
2. Material and Methods
2.1. Data Collection
2.2. Data Pre-Processing and Identification of Differentially Expressed Genes
2.3. Construction of Co-Expression Networks of Altered Ion Channels and EMT-Related Genes
2.4. Identification of Altered Ion Channels in Tumor and Metastatic States
2.5. EMT Score Calculation
2.6. Protein–Protein Interaction Networks of Altered Ion Channels Co-Expressed with EMT-Related Genes
2.7. Computation of Correlation of EMT Scores with Identified Ion Channels
2.8. Generation of Pathway Map of Altered Ion Channels and EMT-Related Genes
2.9. Survival Analysis of Identified Altered Ion Channels
3. Results
3.1. Differentially Expressed Ion Channels and EMT-Related Genes
3.2. Altered Ion Channels in Tumor and Metastatic States of Breast Cancer
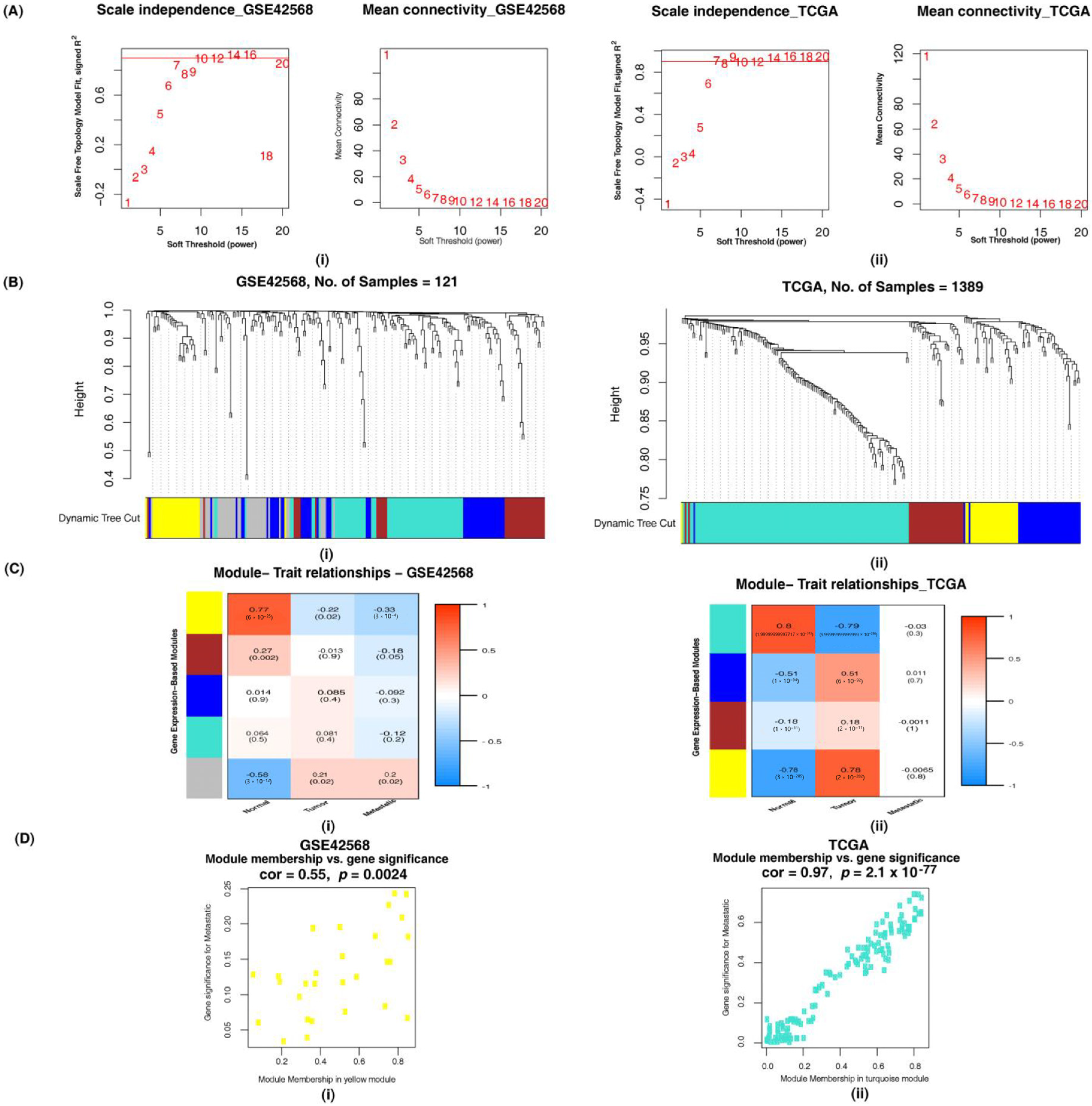
3.2.1. Identification of Altered Ion Channel Modules by Weighted Gene Co-Expression Network Analysis
3.2.2. Overlapping Ion Channels in GSE42568, GSE52604 and TCGA Datasets
3.3. Identification of Altered Ion Channels Interacting with EMT-Related Genes
3.3.1. Weighted Gene Co-Expression Network Analysis for the Identification of Altered Combined Gene Set Modules
3.3.2. Network Analysis of Altered Ion Channels Co-Expressed with EMT-Related Genes
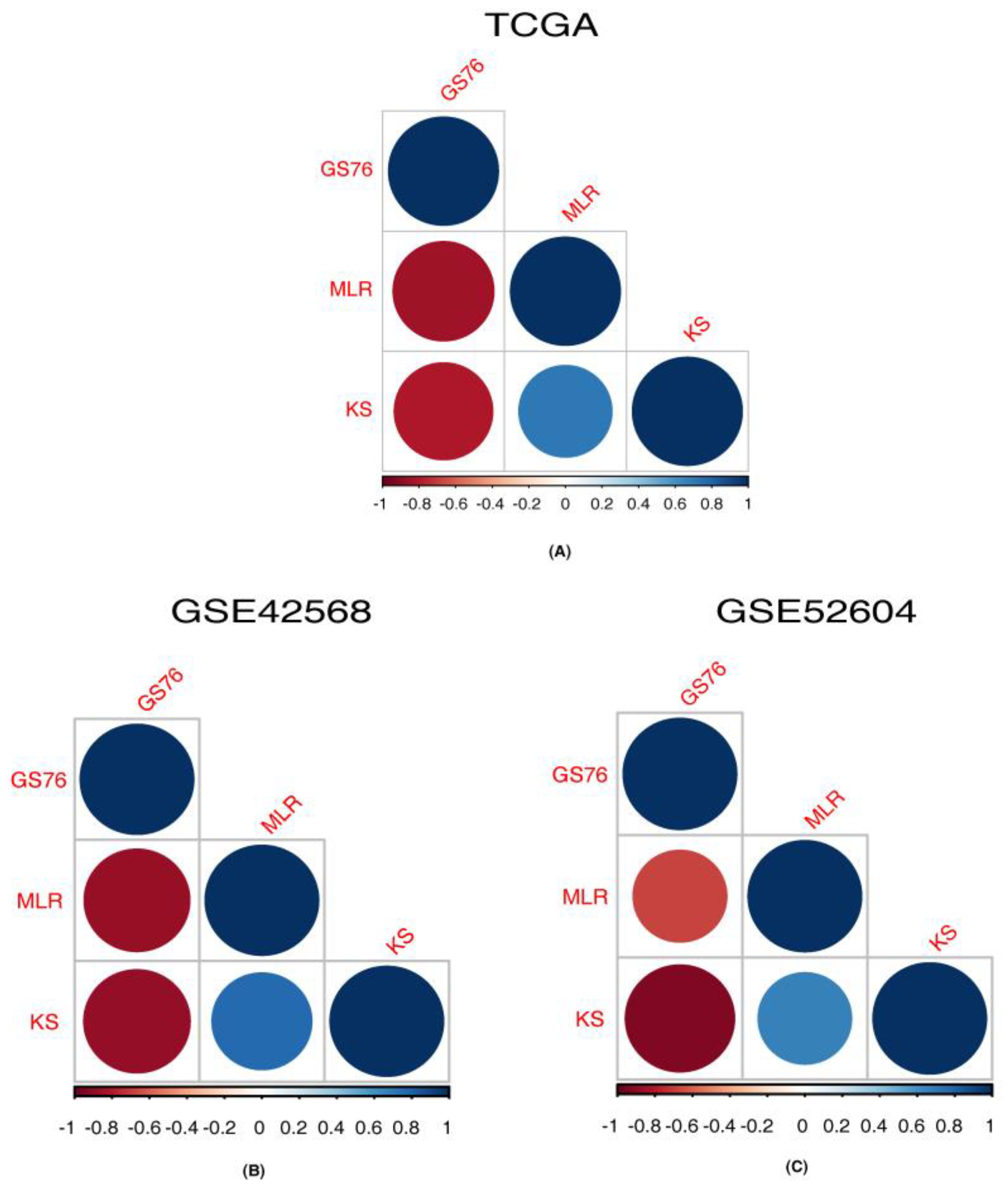
3.4. Correlation of Identified Ion Channels with EMT Scores
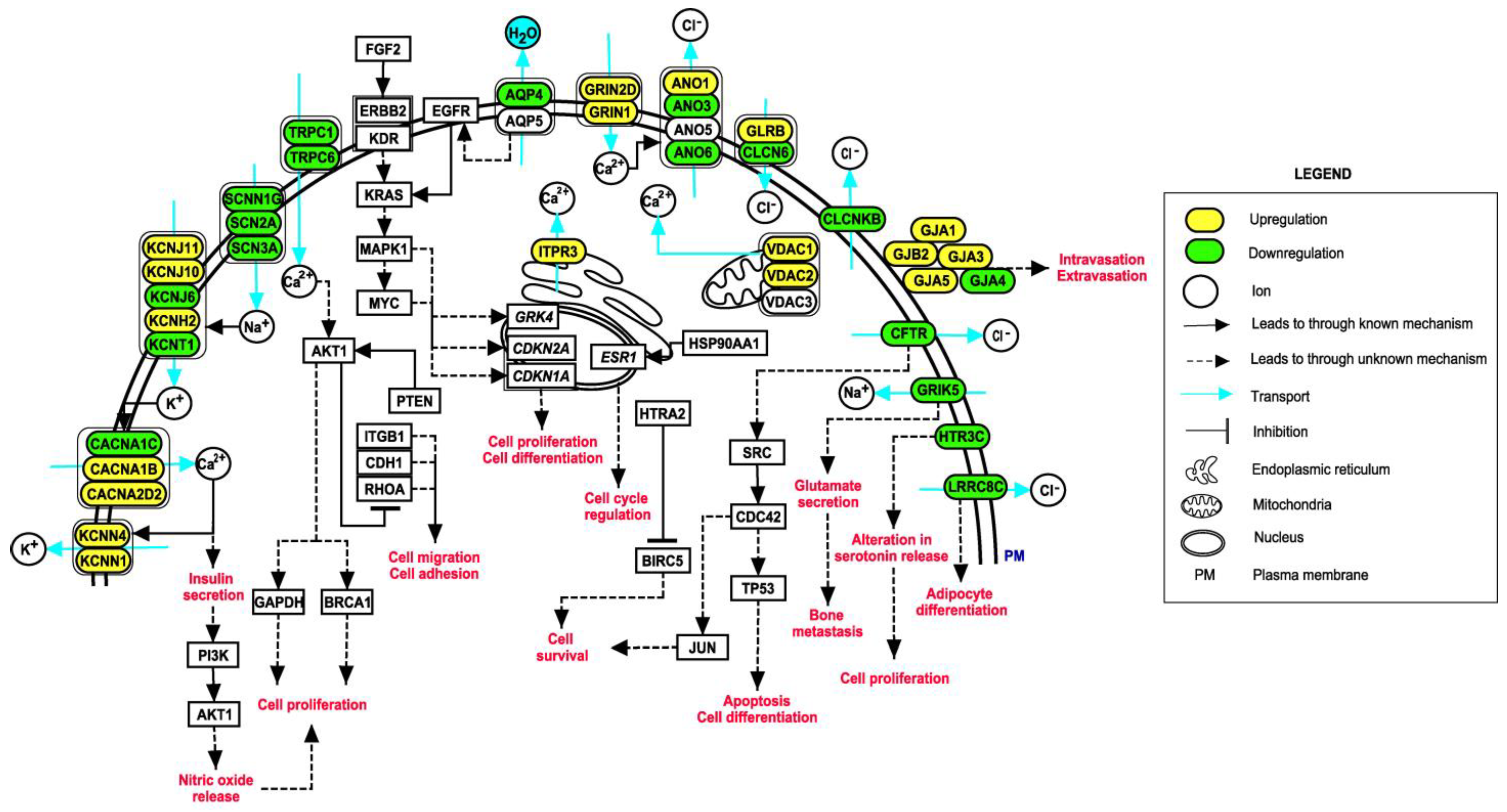
3.5. Pathway Map of Identified Ion Channels and Their Associations with EMT
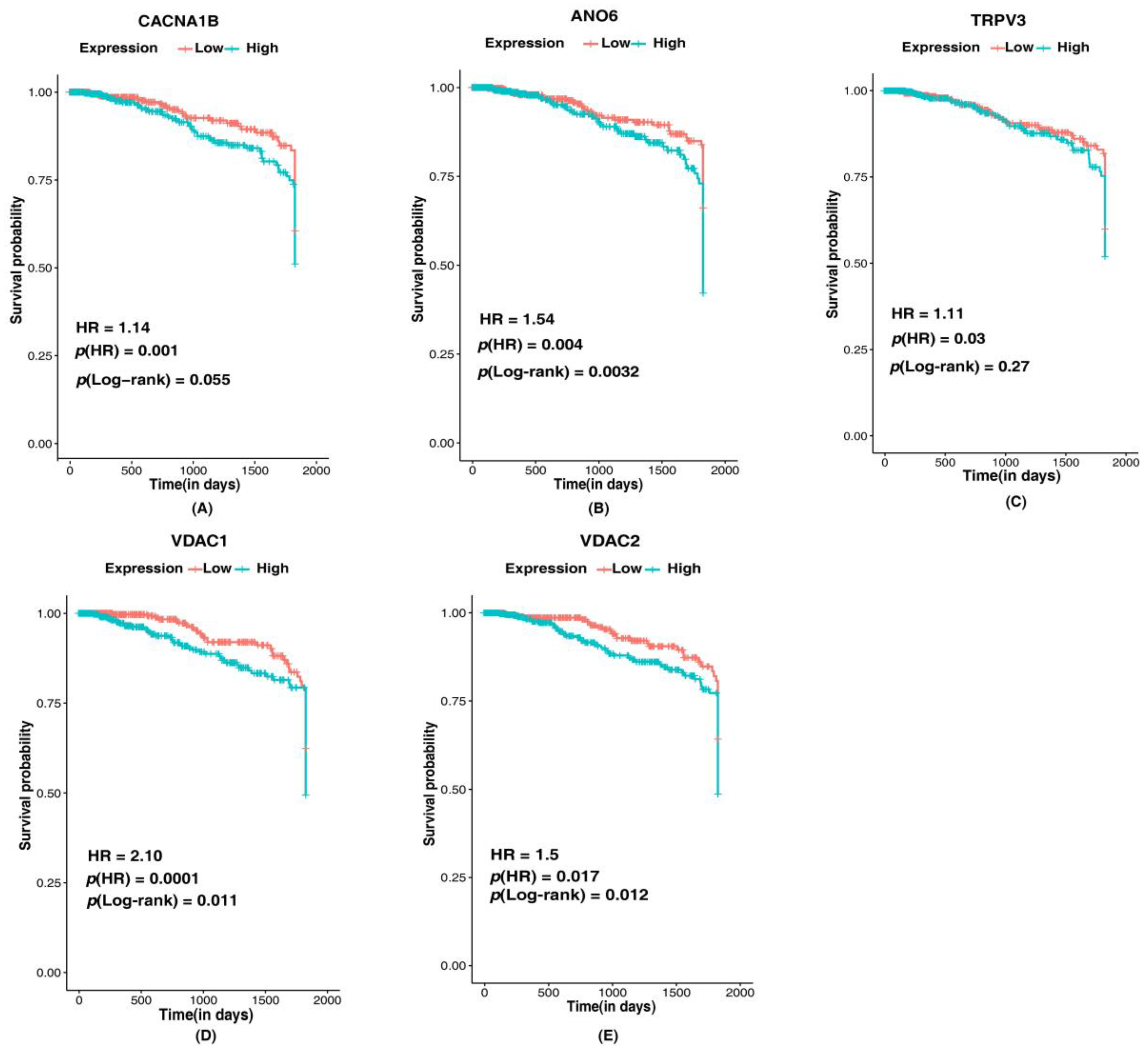
3.6. Association of Identified Putative Ion Channels in Survival of Breast Cancer Patients
4. Discussion
4.1. Ion Channels Identified as Putative Ion Channels in the Tumor State of Breast Cancer
- HTR3C, a ligand-gated ion channel, is one of the receptors for serotonin, a hormone that functions as a neurotransmitter and mitogen. Activation of this receptor causes fast, depolarizing responses. Additionally, SCN3A, a voltage gated sodium channel, is known to maintain depolarization in the enterochromaffin cells, which results in the regulation of serotonin release [57]. Serotonin functions as a tumor-suppressant in non-transformed breast cells and early-stage breast cancers. During tumor progression, cells acquire genetic or epigenetic alterations in serotonin signaling. This makes them resistant to suppressive actions of serotonin and favors tumor-promoting actions [58].
- CLCN6 is a voltage-dependent chloride channel and has a role in regulating blood pressure levels and hypertension [59]. cMyc a protooncogene responsible for cell proliferation in various cancers, transcriptionally regulates GRK4 protein that was reported to be overexpressed in breast cancer tissues [60]. GRK4 has been demonstrated to be associated with an increased risk of hypertension, indicating hypertension as an important factor in breast cancer [60].
- GLRB, a glycine receptor, is a ligand-gated ion channel that mediates the inhibitory effects of glycine. Vascular endothelial growth factor (VEGF) has a crucial role in cancer progression as it promotes the formation of new blood vessels. Activation of VEGF receptor results in activation of phospholipase C-gamma and increases intracellular Ca2+ concentration. VEGF-induced cell proliferation is dependent on intracellular Ca2+ concentration. Hyperpolarization of the cell membrane due to glycine-gated chloride channels blocks the influx of Ca2+, thereby minimizing VEGF-mediated signaling. Thus, changes in the functioning of GLRB may promote tumor growth [61].
- Anoctamins (ANOs) are Ca2+ activated chloride channels. The activation of receptors of growth hormone signaling takes place through RAS-RAF-ERK, PI3K-AKT and DAG-IP3 pathways. It is known that anoctamin-controlled calcium channels are relevant for the activation of ERK-1,2. Additionally, the rise in intracellular Ca2+ together with activation of RAS-RAF/ERK pathway is a major aspect of cell proliferation [62]. ANO1 has been widely studied in various tumors in this respect. However, very few studies have reported the involvement of other members of anoctamins. ANO3, ANO5 and ANO6 were identified in this study.
- LRRC8C is a volume-regulated anion channel (VRAC). VRACs are activated on cell swelling and play a critical role in cell volume regulation [63]. The Fad158 gene, which has a crucial role in adipocyte differentiation, belongs to the LRRC8 family and is also known as LRRC8C [64]. As adipocytes are the major components of breast tissue, abnormal adipocyte metabolism leads to accumulation of tumor-supporting cells and other effects, such as insulin resistance, dyslipidemia and oxidative stress [65]. These effects may further lead to the aggressive nature of the tumor. Additionally, VRACs are known to mediate cellular uptake of drugs, such as cisplatin and carboplatin, which are widely used in the treatment of cancer [66]. However, LRRC8C was not reported to have a direct effect on the uptake of these drugs but was stated to have a possible role in mediating the action of LRRC8A [66].
4.2. Ion Channels Identified as Putative Ion Channels in the Metastatic State of Breast Cancer
- TRPV3 is not a well-understood ion channel and can be activated by temperature. Although it has been reported as overexpressed in non-small cell lung cancer [84].
- GABRG3 belongs to the GABA-A receptor gene family of hetero-pentameric ligand-gated ion channels. GABA-A receptor GABRP is required for maintaining basal-like cytokeratin expression, ERK1/2 phosphorylation and pro-migratory phenotype of breast cancer cells [85]. GABA-B receptors promote metastasis by enhancing ERK phosphorylation and thus activating metalloproteins that enable tumor cells to penetrate the basement membrane [86]. These studies indicate the importance of GABA receptors in the metastatic progression of breast cancer.
- KCNT1 is a Na+ activated K+ channel with diverse functions, including insulin secretion and cell volume regulation. With the help of insulin receptors, insulin regulates endothelial cell migration, proliferation and production of VEGF. It also activates PI3K/Akt signaling that promotes nitric oxide (NO) release. NO increases endothelial survival, migration, proliferation and vascular permeability [87].
- KCNN1 is a Ca2+ dependent potassium channel involved in regulating cell volume. Increased permeability of K+ due to activation of these channels results in membrane hyperpolarization. This enhances Ca2+ entry and thus contributes to a decrease in the regulatory volume of a cell [88].
- CLCNKB is a voltage-gated chloride channel. Chloride channels participate in cell volume regulation, membrane potential stabilization, signal transduction and transepithelial transport. CLCNKB has been widely studied in Bartter syndrome. Its role in breast cancer is not well established. These channels aid in maintaining intracellular ion concentration lower than extracellular ion concentration, preventing osmotic cell swelling. Altered expression in these channels may result in osmotic stress contributing to cell migration [89].
- GJA5 is a member of the connexin family. Connexins are involved in the formation of heterologous gap junctions between tumor and endothelial cells, which facilitate intravasation and extravasation [90]. Various gap junctions have been reported in this regard. Very few studies have reported GJA5 [91].
- GRIK5 belongs to the glutamate-gated ionic family. Cells with breast cancer phenotype secrete high levels of glutamate and metastasize to bones. These cells with excess glutamate result in cancer-induced bone pain, which is a significant co-morbidity in advanced stage breast cancer patients [92].
4.3. Ion Channels Interacting with Multiple Hub EMT-Related Genes in Breast Cancer
- GJA1 has roles in the cAMP pathway, Wnt signaling pathway and activation of p38. It was also reported as a tumor suppressor and a potent molecule in the acceleration of cancer progression [93].
- TRPC6 is present ubiquitously in human tissues and is involved in Ca2+ dependent pathways. It is also known to be upregulated in various other disease conditions [94].
- TRPC1, another ion channel involved in modulation of Ca2+, is an established biomarker in certain cancers [95].
- VDAC1 is a multifunctional channel involved in controlling the communications between mitochondria and the rest of the cell [96].
- ANO1 channel is involved in cell proliferation, survival, migration, contraction, secretion and neuronal excitation [97].
- AQP5, an aquaporin, is involved in the movement of water across cellular membranes. This process is one of the most important phenomena resulting in cell movement, cellular viscosity and signal transductions [39].
- CLIC1 was identified in maintenance of cell volume, ion homeostasis, trans-epithelial transport, pH regulation and cell cycle regulation [98].
- GJB2 regulates cell migration and colonization and thus aggressive phenotype in breast cancer [99].
- KCNH2 was reported to be involved in cell proliferation and cell migration processes implicating MAP kinase and c-fos pathways through a cell line study [100].
- CFTR, an anion channel, is known for its role in ion and acid-base homeostasis. It was also reported as a tumor suppressor [101].
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Scully, O.J.; Bay, B.H.; Yip, G.; Yu, Y. Breast cancer metastasis. Cancer Genom. Proteom. 2012, 9, 311–320. [Google Scholar]
- Ko, J.H.; Ko, E.A.; Gu, W.; Lim, I.; Bang, H.; Zhou, T. Expression profiling of ion channel genes predicts clinical outcome in breast cancer. Mol. Cancer 2013, 12, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, J.; Deb, B.; George, I.A.; Kapil, S.; Coral, K.; Kakkar, N.; Pattanaik, S.; Mandal, A.K.; Mavuduru, R.S.; Kumar, P. Somatic Mutations Profile of a Young Patient With Metastatic Urothelial Carcinoma Reveals Mutations in Genes Involved in Ion Channels. Front. Oncol. 2019, 9, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Liu, C.; Zhang, D.; Men, H.; Huo, L.; Geng, Q.; Wang, S.; Gao, Y.; Zhang, W.; Zhang, Y.; et al. Mechanosensitive ion channel Piezo1 promotes prostate cancer development through the activation of the Akt/mTOR pathway and acceleration of cell cycle. Int. J. Oncol. 2019, 55, 629–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, P.H.; Kourghi, M.; Pei, J.V.; Nourmohammadi, S.; Yool, A.J. 5-Hydroxymethyl-Furfural and Structurally Related Compounds Block the Ion Conductance in Human Aquaporin-1 Channels and Slow Cancer Cell Migration and Invasion. Mol. Pharm. 2020, 98, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhong, D.; Wei, Y.; Wu, X.; Kang, L.; Ding, Z. Potassium channel ether a go-go1 is aberrantly expressed in human liposarcoma and promotes tumorigenesis. Biomed. Res. Int. 2014, 2014, 345678. [Google Scholar] [CrossRef]
- Li, M.; Xiong, Z.G. Ion channels as targets for cancer therapy. Int. J. Physiol. Pathophysiol. Pharm. 2011, 3, 156–166. [Google Scholar]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Azimi, I.; Monteith, G.R. Plasma membrane ion channels and epithelial to mesenchymal transition in cancer cells. Endocr. Relat. Cancer 2016, 23, R517–R525. [Google Scholar] [CrossRef] [Green Version]
- Fortunato, A. The role of hERG1 ion channels in epithelial-mesenchymal transition and the capacity of riluzole to reduce cisplatin resistance in colorectal cancer cells. Cell. Oncol. 2017, 40, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, G.; Li, M.; Fang, Y.; Qian, K.; Tang, Y.; Wu, X.; Lei, X.; Li, X.; Liu, Q.; et al. Targeting TRPV1 on cellular plasticity regulated by Ovol 2 and Zeb 1 in hepatocellular carcinoma. Biomed. Pharm. 2019, 118, 109270. [Google Scholar] [CrossRef]
- Zhu, S.; Zhou, H.Y.; Deng, S.C.; Deng, S.J.; He, C.; Li, X.; Chen, J.Y.; Jin, Y.; Hu, Z.L.; Wang, F.; et al. ASIC1 and ASIC3 contribute to acidity-induced EMT of pancreatic cancer through activating Ca(2+)/RhoA pathway. Cell Death Dis. 2017, 8, e2806. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef]
- Chakraborty, P.; George, J.T.; Tripathi, S.; Levine, H.; Jolly, M.K. Comparative Study of Transcriptomics-Based Scoring Metrics for the Epithelial-Hybrid-Mesenchymal Spectrum. Front. Bioeng. Biotechnol. 2020, 8, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Kong, L.; Liu, Y.; Qu, H. dbEMT: An epithelial-mesenchymal transition associated gene resource. Sci. Rep. 2015, 5, 11459. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- George, J.T.; Jolly, M.K.; Xu, S.; Somarelli, J.A.; Levine, H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017, 77, 6415–6428. [Google Scholar] [CrossRef] [Green Version]
- Mandal, S.; Tejaswi, T.; Janivara, R.; Srikrishnan, S.; Thakur, P.; Sahoo, S.; Chakraborty, P.; Sohal, S.S.; Levine, H.; George, J.T.; et al. Transcriptomic-based quantification of the epithelial-hybrid-mesenchymal spectrum across biological contexts. Biomolecules 2022, 12, 29. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [Green Version]
- Sharma, J.; Balakrishnan, L.; Datta, K.K.; Sahasrabuddhe, N.A.; Khan, A.A.; Sahu, A.; Singhal, A.; Getnet, D.; Raju, R.; Chatterjee, A.; et al. A knowledgebase resource for interleukin-17 family mediated signaling. J. Cell Commun. Signal. 2015, 9, 291–296. [Google Scholar] [CrossRef] [Green Version]
- Parthasarathi, K.T.S.; Munjal, N.S.; Dey, G.; Kumar, A.; Pandey, A.; Balakrishnan, L.; Sharma, J. A pathway map of signaling events triggered upon SARS-CoV infection. J. Cell Commun. Signal. 2021, 15, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Sharma, J.; Raju, R.; Palapetta, S.M.; Prasad, T.S.; Huang, T.C.; Yoda, A.; Tyner, J.W.; van Bodegom, D.; Weinstock, D.M.; et al. TSLP signaling pathway map: A platform for analysis of TSLP-mediated signaling. Database 2014, 2014, bau007. [Google Scholar] [CrossRef] [PubMed]
- Jyoti Sharma, L.B. Sandeep Kaushik and Manoj Kumar Kashyap. Editorial: Multi-Omics approaches to study signaling pathways. Front. Bioeng. Biotechnol. 2020, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Kutmon, M.; van Iersel, M.P.; Bohler, A.; Kelder, T.; Nunes, N.; Pico, A.R.; Evelo, C.T. PathVisio 3: An extendable pathway analysis toolbox. PLoS Comput. Biol. 2015, 11, e1004085. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Davis, F.M.; Azimi, I.; Faville, R.A.; Peters, A.A.; Jalink, K.; Putney, J.W., Jr.; Goodhill, G.J.; Thompson, E.W.; Roberts-Thomson, S.J.; Monteith, G.R. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene 2014, 33, 2307–2316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.T.; Jiang, X.H.; Xie, C.; Cheng, H.; Da Dong, J.; Wang, Y.; Fok, K.L.; Zhang, X.H.; Sun, T.T.; Tsang, L.L.; et al. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochim. Biophys. Acta 2013, 1833, 2961–2969. [Google Scholar] [CrossRef] [Green Version]
- Fernandez Fernandez, E.; De Santi, C.; De Rose, V.; Greene, C.M. CFTR dysfunction in cystic fibrosis and chronic obstructive pulmonary disease. Expert Rev. Respir. Med. 2018, 12, 483–492. [Google Scholar] [CrossRef]
- Milkovic, L.; Cipak Gasparovic, A. AQP3 and AQP5-Potential Regulators of Redox Status in Breast Cancer. Molecules 2021, 26, 2613. [Google Scholar] [CrossRef]
- Kale, V.P.; Amin, S.G.; Pandey, M.K. Targeting ion channels for cancer therapy by repurposing the approved drugs. Biochim. Biophys. Acta 2015, 1848, 2747–2755. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Shao, Q.; Curtis, H.; Galipeau, J.; Belliveau, D.J.; Wang, T.; Alaoui-Jamali, M.A.; Laird, D.W. Retroviral delivery of connexin genes to human breast tumor cells inhibits in vivo tumor growth by a mechanism that is independent of significant gap junctional intercellular communication. J. Biol. Chem. 2002, 277, 29132–29138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva-Font, C.; Jin, B.J.; Verkman, A.S. Aquaporin-1 gene deletion reduces breast tumor growth and lung metastasis in tumor-producing MMTV-PyVT mice. FASEB J. 2014, 28, 1446–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bon, E.; Driffort, V.; Gradek, F.; Martinez-Caceres, C.; Anchelin, M.; Pelegrin, P.; Cayuela, M.L.; Marionneau-Lambot, S.; Oullier, T.; Guibon, R.; et al. SCN4B acts as a metastasis-suppressor gene preventing hyperactivation of cell migration in breast cancer. Nat. Commun. 2016, 7, 13648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, C.; Charlestin, V.; Wang, M.; Walker, Z.T.; Miranda-Vergara, M.C.; Facchine, B.A.; Wu, J.; Kaliney, W.J.; Dovichi, N.J.; Li, J.; et al. Aquaporin-7 Regulates the Response to Cellular Stress in Breast Cancer. Cancer Res. 2020, 80, 4071–4086. [Google Scholar] [CrossRef]
- Azoitei, N.; Kleger, A.; Schoo, N.; Thal, D.R.; Brunner, C.; Pusapati, G.V.; Filatova, A.; Genze, F.; Moller, P.; Acker, T.; et al. Protein kinase D2 is a novel regulator of glioblastoma growth and tumor formation. Neuro Oncol. 2011, 13, 710–724. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, C.; Zheng, M.; Sun, J. Prognostic value of the mRNA expression of gap junction alpha members in patients with gastric cancer. Oncol. Lett. 2019, 18, 1669–1678. [Google Scholar] [CrossRef]
- Warnecke-Eberz, U.; Metzger, R.; Holscher, A.H.; Drebber, U.; Bollschweiler, E. Diagnostic marker signature for esophageal cancer from transcriptome analysis. Tumour. Biol. 2016, 37, 6349–6358. [Google Scholar] [CrossRef]
- Zeng, Y.Z.; Zhang, Y.Q.; Chen, J.Y.; Zhang, L.Y.; Gao, W.L.; Lin, X.Q.; Huang, S.M.; Zhang, F.; Wei, X.L. TRPC1 Inhibits Cell Proliferation/Invasion and Is Predictive of a Better Prognosis of Esophageal Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 627713. [Google Scholar] [CrossRef]
- Tajeddine, N.; Gailly, P. TRPC1 protein channel is major regulator of epidermal growth factor receptor signaling. J. Biol. Chem. 2012, 287, 16146–16157. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Huang, J.; Peng, J.; Wu, X.; Zhang, Y.; Zhu, W.; Guo, L. Upregulation of the inwardly rectifying potassium channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell lung cancer under the regulation of miR-7 and the Ras/MAPK pathway. Mol. Cancer 2015, 14, 59. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Li, Y.; Du, Y.; Ma, J. KCND2 upregulation might be an independent indicator of poor survival in gastric cancer. Future Oncol. 2018, 14, 2811–2820. [Google Scholar] [CrossRef] [PubMed]
- Xuan, H.; Zhengjun, C.; Yang, H.; Guohan, C. Case report: Mutation analysis of primary pulmonary lymphoepithelioma-like carcinoma via whole-exome sequencing. Diagn. Pathol. 2019, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Neveu, B.; Spinella, J.F.; Richer, C.; Lagace, K.; Cassart, P.; Lajoie, M.; Jananji, S.; Drouin, S.; Healy, J.; Hickson, G.R.; et al. CLIC5: A novel ETV6 target gene in childhood acute lymphoblastic leukemia. Haematologica 2016, 101, 1534–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Tellez, T.N.; Lopez, T.V.; Vasquez Garzon, V.R.; Villa-Trevino, S. Co-Expression of Ezrin-CLIC5-Podocalyxin Is Associated with Migration and Invasiveness in Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0131605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, S.; Umakoshi, A.; Yano, H.; Ohsumi, S.; Sumida, Y.; Hayase, E.; Usa, E.; Islam, A.; Choudhury, M.E.; Nishi, Y.; et al. Chloride intracellular channel protein 2 is secreted and inhibits MMP14 activity, while preventing tumor cell invasion and metastasis. Neoplasia 2021, 23, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Gong, Y.Z.; Shao, M.N.; Ruan, G.T.; Xie, H.L.; Liao, X.W.; Wang, X.K.; Han, Q.F.; Zhou, X.; Zhu, L.C.; et al. Distinct diagnostic and prognostic values of gamma-aminobutyric acid type A receptor family genes in patients with colon adenocarcinoma. Oncol. Lett. 2020, 20, 275–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strege, P.R.; Knutson, K.; Eggers, S.J.; Li, J.H.; Wang, F.; Linden, D.; Szurszewski, J.H.; Milescu, L.; Leiter, A.B.; Farrugia, G.; et al. Sodium channel NaV1.3 is important for enterochromaffin cell excitability and serotonin release. Sci. Rep. 2017, 7, 15650. [Google Scholar] [CrossRef] [Green Version]
- Pai, V.P.; Marshall, A.M.; Hernandez, L.L.; Buckley, A.R.; Horseman, N.D. Altered serotonin physiology in human breast cancers favors paradoxical growth and cell survival. Breast Cancer Res. 2009, 11, R81. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Pulit, S.L.; Hwang, S.J.; Brody, J.A.; Amin, N.; Auer, P.L.; Bis, J.C.; Boerwinkle, E.; Burke, G.L.; Chakravarti, A.; et al. Rare Exome Sequence Variants in CLCN6 Reduce Blood Pressure Levels and Hypertension Risk. Circ. Cardiovasc. Genet. 2016, 9, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Yue, W.; Tran, H.T.; Wang, J.P.; Schiermeyer, K.; Gildea, J.J.; Xu, P.; Felder, R.A. The Hypertension Related Gene G-Protein Coupled Receptor Kinase 4 Contributes to Breast Cancer Proliferation. Breast Cancer 2021, 15, 11782234211015753. [Google Scholar] [CrossRef]
- Yamashina, S.; Ikejima, K.; Rusyn, I.; Sato, N. Glycine as a potent anti-angiogenic nutrient for tumor growth. J. Gastroenterol. Hepatol. 2007, 22, S62–S64. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, K.; Ousingsawat, J.; Benedetto, R.; Cabrita, I.; Schreiber, R. Contribution of Anoctamins to Cell Survival and Cell Death. Cancers 2019, 11, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osei-Owusu, J.; Yang, J.; Vitery, M.D.C.; Qiu, Z. Molecular Biology and Physiology of Volume-Regulated Anion Channel (VRAC). Curr. Top. Membr. 2018, 81, 177–203. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Nozaki, Y.; Nishizuka, M.; Ikawa, M.; Osada, S.; Imagawa, M. Factor for adipocyte differentiation 158 gene disruption prevents the body weight gain and insulin resistance induced by a high-fat diet. Biol. Pharm. Bull. 2011, 34, 1257–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Liu, S. Mechanistic insights of adipocyte metabolism in regulating breast cancer progression. Pharm. Res. 2020, 155, 104741. [Google Scholar] [CrossRef]
- Planells-Cases, R.; Lutter, D.; Guyader, C.; Gerhards, N.M.; Ullrich, F.; Elger, D.A.; Kucukosmanoglu, A.; Xu, G.; Voss, F.K.; Reincke, S.M.; et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J. 2015, 34, 2993–3008. [Google Scholar] [CrossRef]
- Yu, Y.P.; Liu, P.; Nelson, J.; Hamilton, R.L.; Bhargava, R.; Michalopoulos, G.; Chen, Q.; Zhang, J.; Ma, D.; Pennathur, A.; et al. Identification of recurrent fusion genes across multiple cancer types. Sci. Rep. 2019, 9, 1074. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.; Deng, Z.; Zhuang, X.; Ju, S.; Mu, J.; Jiang, H.; Zhang, L.; Yan, J.; Miller, D.; Zhang, H.G. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS ONE 2012, 7, e50781. [Google Scholar] [CrossRef]
- Cornen, S.; Guille, A.; Adelaide, J.; Addou-Klouche, L.; Finetti, P.; Saade, M.R.; Manai, M.; Carbuccia, N.; Bekhouche, I.; Letessier, A.; et al. Candidate luminal B breast cancer genes identified by genome, gene expression and DNA methylation profiling. PLoS ONE 2014, 9, e81843. [Google Scholar] [CrossRef]
- Lallet-Daher, H.; Wiel, C.; Gitenay, D.; Navaratnam, N.; Augert, A.; Le Calve, B.; Verbeke, S.; Carling, D.; Aubert, S.; Vindrieux, D.; et al. Potassium channel KCNA1 modulates oncogene-induced senescence and transformation. Cancer Res. 2013, 73, 5253–5265. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Song, L.; Li, J.; Wang, Y.; Yang, C.; Kou, X.; Xiao, B.; Zhang, W.; Li, L.; Liu, S.; et al. Bone sialoprotein-alphavbeta3 integrin axis promotes breast cancer metastasis to the bone. Cancer Sci. 2019, 110, 3157–3172. [Google Scholar] [CrossRef]
- Ahn, J.W.; Kim, H.S.; Yoon, J.K.; Jang, H.; Han, S.M.; Eun, S.; Shim, H.S.; Kim, H.J.; Kim, D.J.; Lee, J.G.; et al. Identification of somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers. Genome Med. 2014, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, X.; Zhao, Y.; Wang, J.; Li, W.; Xiang, Q.; Zhang, Z.; Wu, S.; Liu, A.; Du, G.; Cui, Y. Competing Endogenous RNA and Coexpression Network Analysis for Identification of Potential Biomarkers and Therapeutics in association with Metastasis Risk and Progression of Prostate Cancer. Oxid. Med. Cell. Longev. 2019, 2019, 8265958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, T.; Muhanhali, D.; Jia, X.; Wu, Z.; Cai, Z.; Ling, Y. Identification of gene co-expression modules and hub genes associated with lymph node metastasis of papillary thyroid cancer. Endocrine 2019, 66, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Jin, W.; Zheng, Z.; Chen, E.; Wang, Q.; Wang, Y.; Wang, O.; Zhang, X. GABRB2 plays an important role in the lymph node metastasis of papillary thyroid cancer. Biochem. Biophys. Res. Commun. 2017, 492, 323–330. [Google Scholar] [CrossRef]
- Hauptman, N.; Jevsinek Skok, D.; Spasovska, E.; Bostjancic, E.; Glavac, D. Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4, and KCNA5 as potential diagnostic biomarkers in colorectal cancer. BMC Med. Genom. 2019, 12, 54. [Google Scholar] [CrossRef] [Green Version]
- Palaniappan, A.; Ramar, K.; Ramalingam, S. Computational Identification of Novel Stage-Specific Biomarkers in Colorectal Cancer Progression. PLoS ONE 2016, 11, e0156665. [Google Scholar] [CrossRef]
- Wei, X.; Walia, V.; Lin, J.C.; Teer, J.K.; Prickett, T.D.; Gartner, J.; Davis, S.; Stemke-Hale, K.; Davies, M.A.; Gershenwald, J.E.; et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011, 43, 442–446. [Google Scholar] [CrossRef] [Green Version]
- Kanda, M.; Shimizu, D.; Nakamura, S.; Sawaki, K.; Umeda, S.; Miwa, T.; Tanaka, H.; Inokawa, Y.; Hattori, N.; Hayashi, M.; et al. Blockade of CHRNB2 signaling with a therapeutic monoclonal antibody attenuates the aggressiveness of gastric cancer cells. Oncogene 2021, 40, 5495–5504. [Google Scholar] [CrossRef]
- Gong, D.; Zhang, J.; Chen, Y.; Xu, Y.; Ma, J.; Hu, G.; Huang, Y.; Zheng, J.; Zhai, W.; Xue, W. The m(6)A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca(2+) influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 233. [Google Scholar] [CrossRef]
- Yu, D.; Holm, R.; Goscinski, M.A.; Trope, C.G.; Nesland, J.M.; Suo, Z. Prognostic and clinicopathological significance of Cacna2d1 expression in epithelial ovarian cancers: A retrospective study. Am. J. Cancer Res. 2016, 6, 2088–2097. [Google Scholar] [PubMed]
- Lu, A.; Shi, Y.; Liu, Y.; Lin, J.; Zhang, H.; Guo, Y.; Li, L.; Lin, Z.; Wu, J.; Ji, D.; et al. Integrative analyses identified ion channel genes GJB2 and SCNN1B as prognostic biomarkers and therapeutic targets for lung adenocarcinoma. Lung Cancer 2021, 158, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.C.; Huang, W.C.; Lin, Y.P.; Becker, E.B.E.; Ansorge, O.; Flockerzi, V.; Conti, D.; Cenacchi, G.; Glitsch, M.D. Functional expression of calcium-permeable canonical transient receptor potential 4-containing channels promotes migration of medulloblastoma cells. J. Physiol. 2017, 595, 5525–5544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, Q.; Fan, K.; Li, B.; Li, H.; Qi, H.; Guo, J.; Cao, Y.; Sun, H. Overexpression of TRPV3 Correlates with Tumor Progression in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2016, 17, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sizemore, G.M.; Sizemore, S.T.; Seachrist, D.D.; Keri, R.A. GABA(A) receptor pi (GABRP) stimulates basal-like breast cancer cell migration through activation of extracellular-regulated kinase 1/2 (ERK1/2). J. Biol. Chem. 2014, 289, 24102–24113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Li, X.; Yao, Z.; Wei, C.; Ning, N.; Li, J. GABAergic signaling facilitates breast cancer metastasis by promoting ERK1/2-dependent phosphorylation. Cancer Lett. 2014, 348, 100–108. [Google Scholar] [CrossRef]
- Yee, L.D.; Mortimer, J.E.; Natarajan, R.; Dietze, E.C.; Seewaldt, V.L. Metabolic Health, Insulin, and Breast Cancer: Why Oncologists Should Care About Insulin. Front. Endocrinol. 2020, 11, 58. [Google Scholar] [CrossRef] [Green Version]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol. Rev. 2018, 98, 559–621. [Google Scholar] [CrossRef] [Green Version]
- Morishita, K.; Watanabe, K.; Ichijo, H. Cell volume regulation in cancer cell migration driven by osmotic water flow. Cancer Sci. 2019, 110, 2337–2347. [Google Scholar] [CrossRef] [Green Version]
- Aasen, T.; Leithe, E.; Graham, S.V.; Kameritsch, P.; Mayan, M.D.; Mesnil, M.; Pogoda, K.; Tabernero, A. Connexins in cancer: Bridging the gap to the clinic. Oncogene 2019, 38, 4429–4451. [Google Scholar] [CrossRef] [Green Version]
- Alonso, F.; Domingos-Pereira, S.; Le Gal, L.; Derre, L.; Meda, P.; Jichlinski, P.; Nardelli-Haefliger, D.; Haefliger, J.A. Targeting endothelial connexin40 inhibits tumor growth by reducing angiogenesis and improving vessel perfusion. Oncotarget 2016, 7, 14015–14028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazzari, J.; Lin, H.; Murphy, C.; Ungard, R.; Singh, G. Inhibitors of glutamate release from breast cancer cells; new targets for cancer-induced bone-pain. Sci. Rep. 2015, 5, 8380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, S.; Fan, X.; Zhang, J.; An, R.; Li, S. GJA1 Expression and Its Prognostic Value in Cervical Cancer. Biomed. Res. Int. 2020, 2020, 8827920. [Google Scholar] [CrossRef] [PubMed]
- Jardin, I.; Nieto, J.; Salido, G.M.; Rosado, J.A. TRPC6 channel and its implications in breast cancer: An overview. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118828. [Google Scholar] [CrossRef]
- Elzamzamy, O.M.; Penner, R.; Hazlehurst, L.A. The Role of TRPC1 in Modulating Cancer Progression. Cells 2020, 9, 388. [Google Scholar] [CrossRef] [Green Version]
- Shoshan-Barmatz, V.; Shteinfer-Kuzmine, A.; Verma, A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020, 10, 1485. [Google Scholar] [CrossRef]
- Dulin, N.O. Calcium-Activated Chloride Channel ANO1/TMEM16A: Regulation of Expression and Signaling. Front. Physiol. 2020, 11, 590262. [Google Scholar] [CrossRef]
- Yu, W.; Cui, R.; Qu, H.; Liu, C.; Deng, H.; Zhang, Z. Expression and prognostic value of CLIC1 in epithelial ovarian cancer. Exp. Ther. Med. 2018, 15, 4943–4949. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Pandey, P.R.; Sharma, S.; Xing, F.; Wu, K.; Chittiboyina, A.; Wu, S.Y.; Tyagi, A.; Watabe, K. ID2 and GJB2 promote early-stage breast cancer progression by regulating cancer stemness. Breast Cancer Res. Treat. 2019, 175, 77–90. [Google Scholar] [CrossRef]
- Afrasiabi, E.; Hietamaki, M.; Viitanen, T.; Sukumaran, P.; Bergelin, N.; Tornquist, K. Expression and significance of HERG (KCNH2) potassium channels in the regulation of MDA-MB-435S melanoma cell proliferation and migration. Cell Signal. 2010, 22, 57–64. [Google Scholar] [CrossRef]
- Than, B.L.; Linnekamp, J.F.; Starr, T.K.; Largaespada, D.A.; Rod, A.; Zhang, Y.; Bruner, V.; Abrahante, J.; Schumann, A.; Luczak, T.; et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene 2016, 35, 4179–4187. [Google Scholar] [CrossRef] [PubMed]



| Dataset | Upregulated | Downregulated | ||||
|---|---|---|---|---|---|---|
| Ion Channels | EMT-Related Genes | Common Genes | Ion Channels | EMT-Related Genes | Common Genes | |
| TCGA | ||||||
| HT | 145 | 343 | 6 | 80 | 264 | - |
| TM | 31 | 192 | - | 26 | 144 | - |
| HM | 71 | 311 | 3 | 70 | 290 | - |
| GSE42568 | ||||||
| HT | 12 | 150 | 1 | 16 | 176 | - |
| TM | - | - | - | - | - | - |
| HM | 13 | 181 | 2 | 16 | 163 | - |
| GSE52604 | ||||||
| HT | - | - | - | - | - | |
| TM | - | - | - | - | - | |
| HM | 35 | 310 | 2 | 120 | 160 | 5 |
| Ion Channels | Description | Differential Expression |
|---|---|---|
| HTR3C | 5-hydroxytryptamine receptor 3C | Downregulated |
| CLCN6 | chloride voltage-gated channel 6 | Downregulated |
| GLRB | glycine receptor beta | Upregulated |
| SCN3A | sodium voltage-gated channel alpha subunit 3 | Downregulated |
| ANO3 | anoctamin 3 | Downregulated |
| ANO6 | anoctamin 6 | Downregulated |
| LRRC8C | leucine rich repeat containing 8 VRAC subunit C | Downregulated |
| Ion Channels | Description | Differential Expression |
|---|---|---|
| TRPV3 | transient receptor potential cation channel subfamily V member 6 | Downregulated |
| GRIK5 | glutamate ionotropic receptor kainate type subunit 5 | Downregulated |
| CLCNKB | chloride voltage-gated channel Kb | Downregulated |
| GABRG3 | gamma-aminobutyric acid type A receptor gamma3 subunit | Upregulated |
| KCNT1 | potassium sodium-activated channel subfamily T member 1 | Downregulated |
| KCNN1 | potassium calcium-activated channel subfamily N member 1 | Downregulated |
| GJA5 | gap junction protein alpha 5 | Upregulated |
| Dataset | Degree | EMT-Related Proteins | Description | Interacting Ion Channels |
|---|---|---|---|---|
| HT-GSE42568 | 54 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | TRPC6, GJA1 |
| 54 | MYC | MYC proto-oncogene, bHLH transcription factor | GJA1 | |
| 46 | KRAS | KRAS proto-oncogene, GTPase | GRIN1, CLCN6 | |
| 44 | FGF2 | Fibroblast growth factor 2 | GJA1 | |
| 44 | PTEN | phosphatase and tensin homolog | GJA1 | |
| 43 | KDR | kinase insert domain receptor | GJA1, SCN3A, SCN2A | |
| 40 | RHOA | ras homolog family member A | GJA1, TRPC6, TRPC1 | |
| 31 | CAV1 | caveolin 1 | CACNA1B, GJA1, CLIC2, GJA4, TRPC1 | |
| 30 | ITGB1 | integrin subunit beta 1 | GJA1 | |
| 29 | CXCL12 | C-X-C motif chemokine ligand 12 | GJA1 |
| Dataset | Degree | EMT-Related Proteins | Description | Interacting Ion Channels |
|---|---|---|---|---|
| HT-TCGA | 74 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | CLIC1, VDAC1, VDAC2, VDAC3 |
| 46 | SRC | SRC proto-oncogene, non-receptor tyrosine kinase | CLCN3, GJA3, KCNH2 | |
| 46 | HSP90AA1 | heat shock protein 90 alpha family class A member 1 | KCNH2, VDAC1 | |
| 42 | CDKN2A | cyclin dependent kinase inhibitor 2A | KCNJ11 | |
| 41 | KRAS | KRAS proto-oncogene, GTPase | GRIN2D | |
| 36 | DNMT1 | DNA methyltransferase 1 | KCNJ10 | |
| 36 | HSPA4 | heat shock protein family A (Hsp70) member 4 | KCNH2, VDAC1 | |
| 30 | CDC42 | cell division cycle 42 | CACNA2D2 |
| Dataset | Degree | EMT-Related Proteins | Description | Interacting Ion Channels |
|---|---|---|---|---|
| HM-GSE42568 | 99 | TP53 | tumor protein p53 | CFTR |
| 93 | AKT1 | AKT serine/threonine kinase 1 | CLIC1 | |
| 90 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | AQP4, AQP5, CACNA1C, CFTR, KCNN4, CLIC1 | |
| 90 | CDH1 | cadherin 1 | ANO1, AQP5 | |
| 61 | ERBB2 | erb-b2 receptor tyrosine kinase 2 | ANO1 | |
| 58 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit | CACNA1C | |
| 47 | BRCA1 | BRCA1, DNA repair associated | CFTR | |
| 42 | DNMT1 | DNA methyltransferase 1 | CFTR |
| Dataset | Degree | EMT-Related Proteins | Description | Interacting Ion Channels |
|---|---|---|---|---|
| HM-GSE52604 | 117 | AKT1 | AKT serine/threonine kinase 1 | ITPR3, CLIC1, GJB2 |
| 116 | CDH1 | cadherin 1 | GJB2 | |
| 84 | NOTCH1 | notch 1 | KCNT1 | |
| 75 | MAPK1 | mitogen-activated protein kinase 1 | SCNN1G |
| Dataset | Degree | EMT-Related Proteins | Description | Interacting Ion Channels |
|---|---|---|---|---|
| HM-TCGA | 59 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | CLIC1, VDAC1, VDAC3 |
| 41 | HSP90AA1 | heat shock protein 90 alpha family class A member 1 | KCNH2, VDAC1 | |
| 34 | CDKN2A | cyclin dependent kinase inhibitor 2A | KCNJ11 | |
| 33 | DNMT1 | DNA methyltransferase 1 | KCNJ10 | |
| 31 | HSPA4 | heat shock protein family A (Hsp70) member 4 | KCNH2, VDAC1 | |
| 25 | BIRC5 | baculoviral IAP repeat containing 5 | KCNJ6 | |
| 24 | CDC42 | cell division cycle 42 | CACNA2D2 |
| Dataset | GS76.MLR-Cor | GS76.MLR-pval | GS76.KS-Cor | GS76.KS-pval | KS.MLR-Cor | KS.MLR-pval |
|---|---|---|---|---|---|---|
| TCGA | −0.843467533 | <0.00001 | −0.803881144 | 0.00 | 0.718122546 | 6.1098 × 10−221 |
| GSE42568 | −0.863655242 | <0.00001 | −0.876536316 | <0.00001 | 0.770347822 | <0.00001 |
| GSE52604 | −0.677929378 | <0.00001 | −0.923135739 | <0.00001 | 0.673478229 | <0.00001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parthasarathi, K.T.S.; Mandal, S.; Singh, S.; Gundimeda, S.; Jolly, M.K.; Pandey, A.; Sharma, J. In Silico Analysis of Ion Channels and Their Correlation with Epithelial to Mesenchymal Transition in Breast Cancer. Cancers 2022, 14, 1444. https://doi.org/10.3390/cancers14061444
Parthasarathi KTS, Mandal S, Singh S, Gundimeda S, Jolly MK, Pandey A, Sharma J. In Silico Analysis of Ion Channels and Their Correlation with Epithelial to Mesenchymal Transition in Breast Cancer. Cancers. 2022; 14(6):1444. https://doi.org/10.3390/cancers14061444
Chicago/Turabian StyleParthasarathi, K. T. Shreya, Susmita Mandal, Smrita Singh, Seetaramanjaneyulu Gundimeda, Mohit Kumar Jolly, Akhilesh Pandey, and Jyoti Sharma. 2022. "In Silico Analysis of Ion Channels and Their Correlation with Epithelial to Mesenchymal Transition in Breast Cancer" Cancers 14, no. 6: 1444. https://doi.org/10.3390/cancers14061444
APA StyleParthasarathi, K. T. S., Mandal, S., Singh, S., Gundimeda, S., Jolly, M. K., Pandey, A., & Sharma, J. (2022). In Silico Analysis of Ion Channels and Their Correlation with Epithelial to Mesenchymal Transition in Breast Cancer. Cancers, 14(6), 1444. https://doi.org/10.3390/cancers14061444





