Advance of SOX Transcription Factors in Hepatocellular Carcinoma: From Role, Tumor Immune Relevance to Targeted Therapy
Abstract
Simple Summary
Abstract
1. Introduction
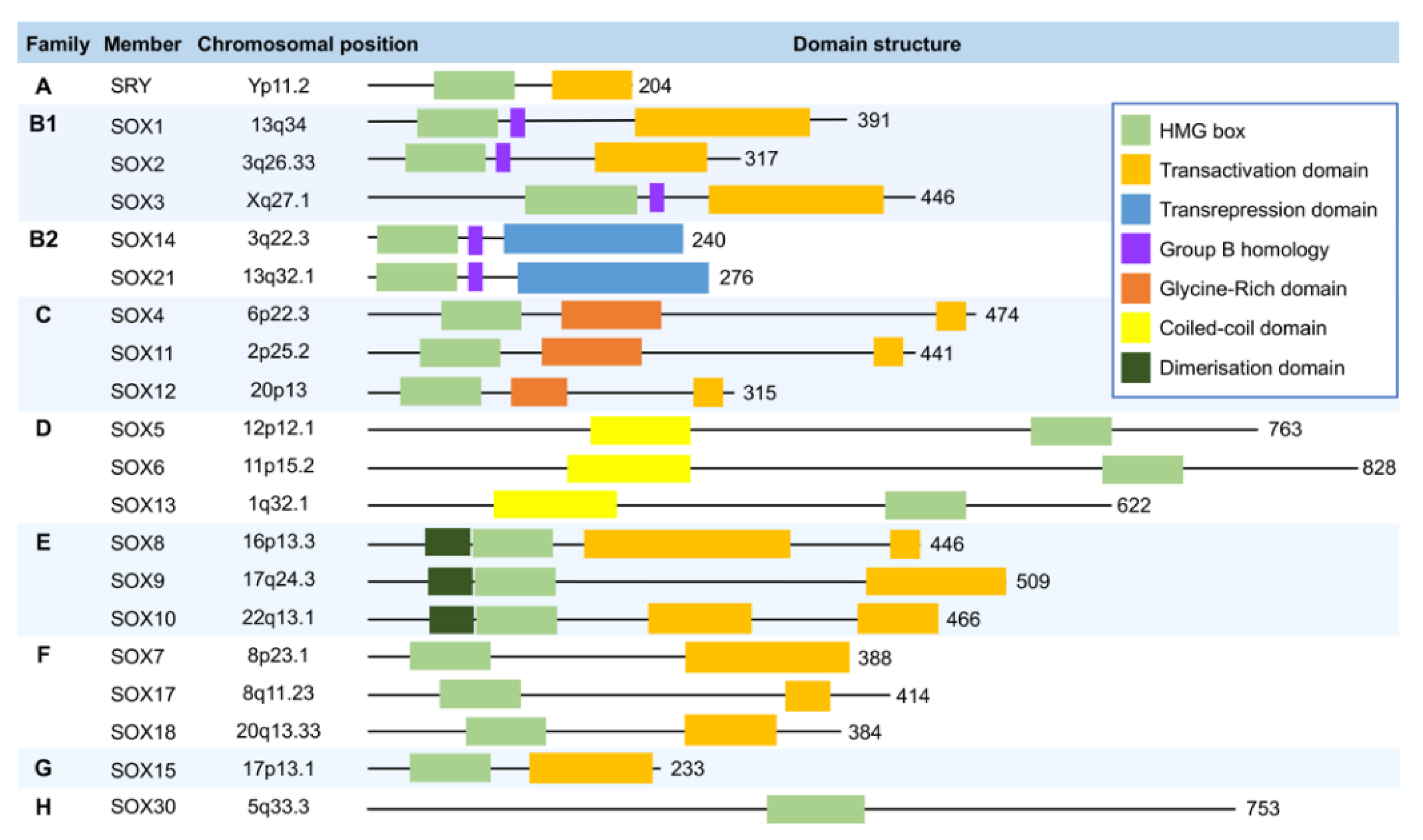
2. Overview of the SOX Transcription Factors
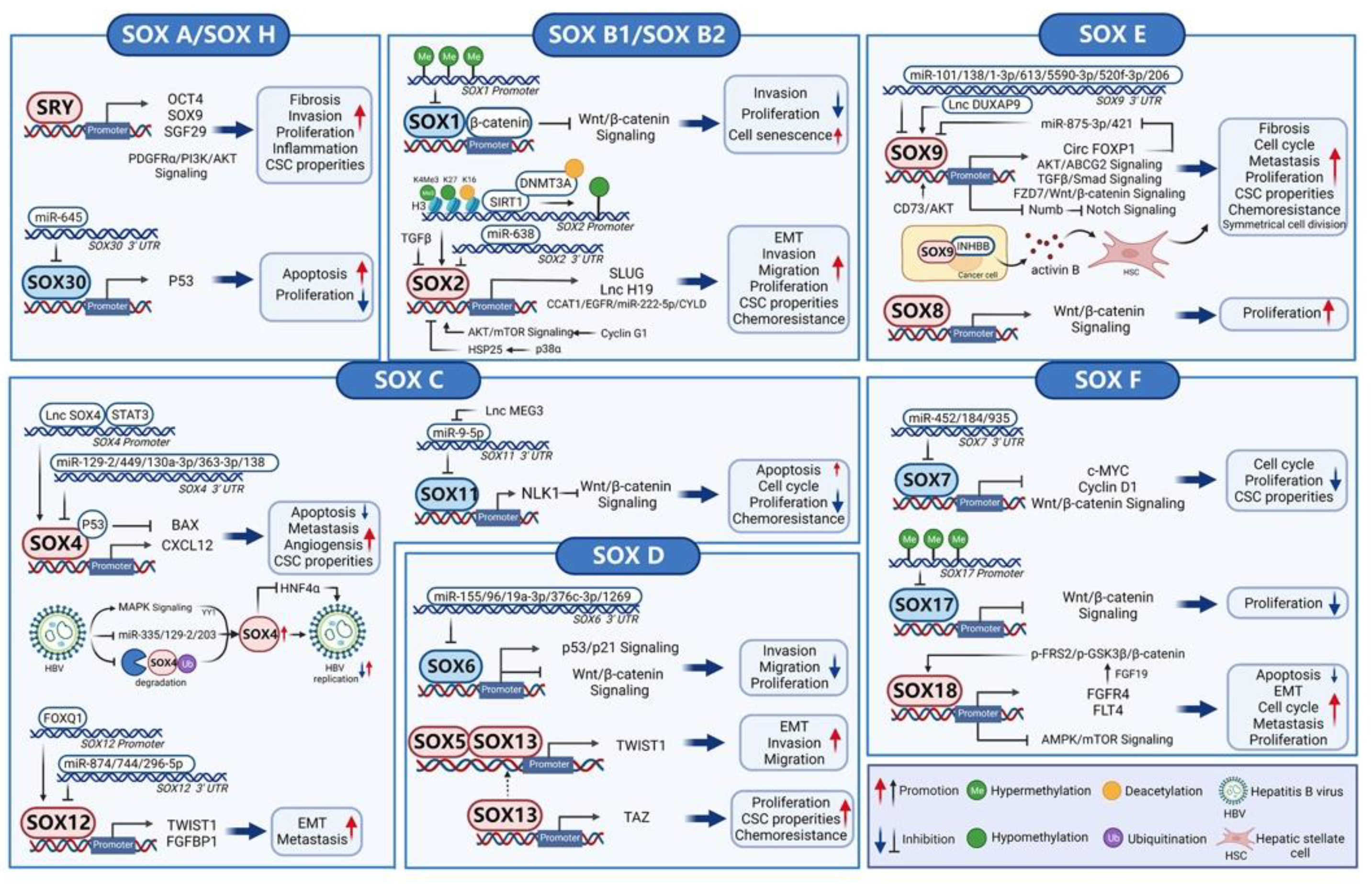
3. SOX Transcription Factors in Hepatocellular Carcinoma
3.1. SOX A (SRY)
3.2. SOX B1 and SOX B2 (SOX1, SOX2, SOX3, SOX14, and SOX21)
3.3. SOX C (SOX4, SOX11, and SOX12)
3.4. SOX D (SOX5, SOX6, SOX13, and SOX23)
3.5. SOX E (SOX8, SOX9, and SOX10)
3.6. SOX F (SOX7, SOX17, and SOX18) and SOX H (SOX30)
4. SOX Transcription Factors and Tumor Immune Microenvironment
4.1. The Role of SOX in Tumor Immune Microenvironment
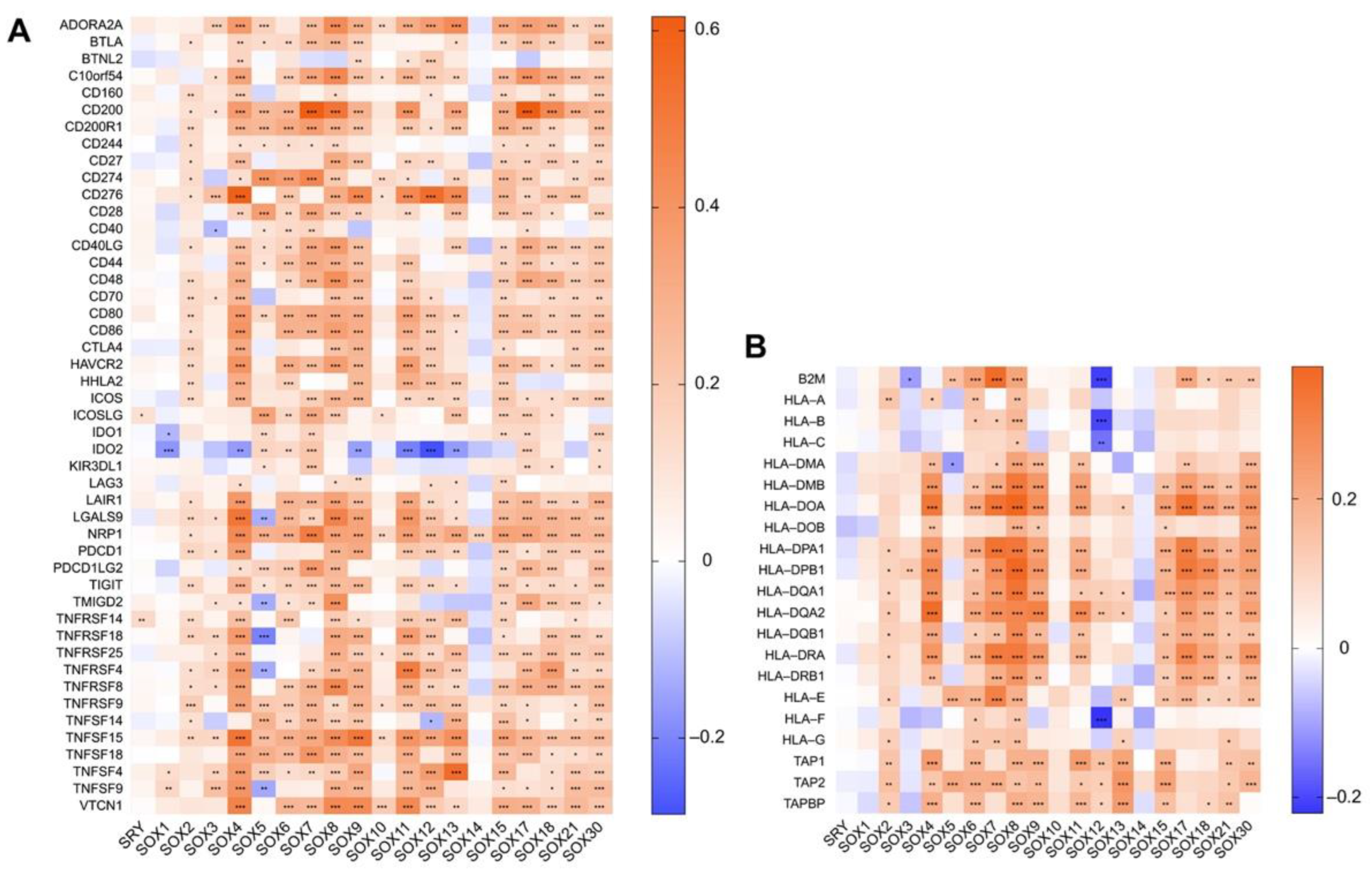
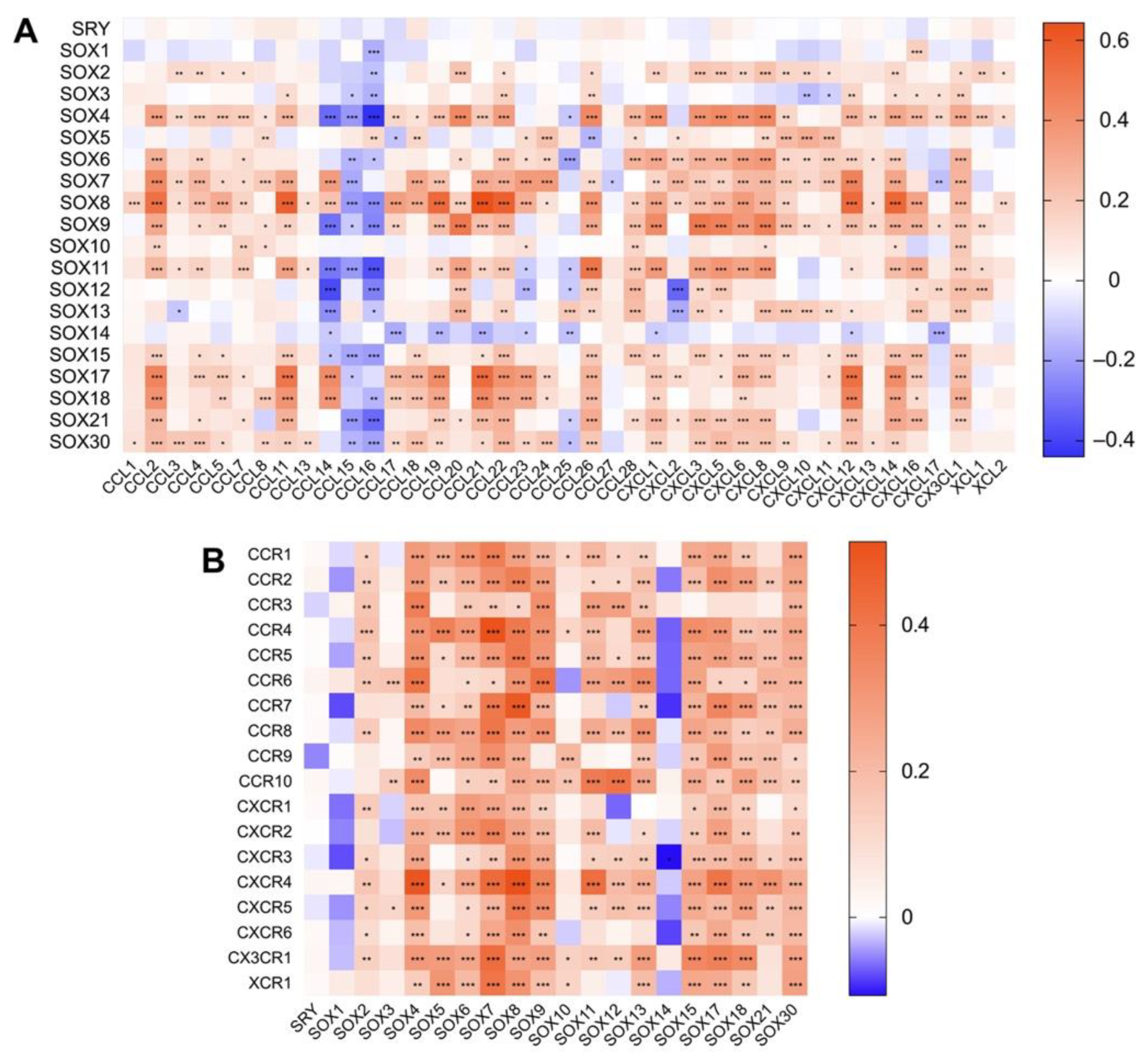
4.2. Association between the Expression of SOX and That of Immune Components in Hepatocellular Carcinoma
5. SOX Transcription Factors and Translational Potential
5.1. SOX Factors as Biomarkers for Patient Stratification Treatment
5.2. Targeting SOX Proteins Degradation
5.3. SOX Factors as Peptide Vaccine Boost Anti-Tumor Immune Response
5.4. Tumor-Targeted Delivery of siRNA to Silence SOX Expression
5.5. Targeting Endogenous SOX Expression by Artificial Transcription Factors-Based Technologies
6. Discussion and Outlook
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| TFs | Transcription factors |
| DBD | DNA-binding domain |
| TME | Tumor microenvironment |
| SRY | Sex-determining region Y |
| HMG | High-mobility group |
| SOX | Sex-determining region Y-related high-mobility group box |
| HCC | Hepatocellular carcinoma |
| TIME | Tumor immune microenvironment |
| CSCs | Cancer stem cells |
| TICs | Tumor-initiating cells |
| EMT | Epithelial-mesenchymal transition |
| miRNA | MicroRNA |
| HBV | Hepatitis B virus |
| SNP | Single-nucleotide polymorphism |
| 3′UTR | 3′ untranslated region |
| AFP | Alpha-fetoprotein |
| PVTT | Portal vein tumor thrombosis |
| HSCs | Hepatic stellate cells |
| MDSCs | Myeloid-derived suppressor cells |
| IFNI | Type I interferon |
| APCs | Antigen-presenting cells |
| DCs | Dendritic cells |
| STING | Stimulator of interferon genes |
| Treg | Regulatory T cell |
| ISG.RS | Interferon-stimulated genes resistance signature |
| ICIs | Immune checkpoint inhibitors |
| TANs | Tumor-associated neutrophils |
| TNBC | Triple-negative breast cancer |
| TAM | Tumor-associated macrophage |
| DCreg | Regulatory dendritic cell |
| Breg | Regulatory B cell |
| HLA | Human leukocyte antigen |
| SMO | Smoothened |
| PROTACs | Proteolysis-targeting chimeras |
| SAHA | Suberoylanilide hydroxamic acid |
| CTLs | Cytotoxic T lymphocytes |
| siRNAs | Small interfering RNAs |
| NPs | Nanoparticles |
| CL-siSOX2 | siSOX2 delivered by cationic lipoplex |
| RGDfC-SeNPs | RGDfC-modified functionalized selenium nanoparticles |
| ATFs | Artificial transcription factors |
| ZF | Zinc-finger |
| SRR1 | SOX2 regulatory region I |
| SKD | Kruppel Associated box |
References
- Ptashne, M.; Gann, A. Transcriptional activation by recruitment. Nature 1997, 386, 569–577. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef]
- Look, A.T. Oncogenic Transcription Factors in the Human Acute Leukemias. Science 1997, 278, 1059–1064. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2021, 19, 23–36. [Google Scholar] [CrossRef]
- Wilson, G.K.; Tennant, D.; McKeating, J.A. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: Current understanding and future directions. J. Hepatol. 2014, 61, 1397–1406. [Google Scholar] [CrossRef]
- Calissi, G.; Lam, E.W.-F.; Link, W. Therapeutic strategies targeting FOXO transcription factors. Nat. Rev. Drug Discov. 2020, 20, 21–38. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in 2020, 70, 313. [Google Scholar] [CrossRef]
- Gubbay, J.; Collignon, J.; Koopman, P.; Capel, B.; Economou, A.; Munsterberg, A.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990, 346, 245–250. [Google Scholar] [CrossRef]
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX Family of Developmental Transcription Factors Based on Sequence and Structural Indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef]
- Wegner, M. All purpose Sox: The many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010, 42, 381–390. [Google Scholar] [CrossRef]
- Liang, Z.; Xu, J.; Gu, C. Novel role of the SRY-related high-mobility-group box D gene in cancer. Semin. Cancer Biol. 2020, 67, 83–90. [Google Scholar] [CrossRef]
- Liu, C.-F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- Harley, V.; Lovell-Badge, R.; Goodfellow, P.N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994, 22, 1500–1501. [Google Scholar] [CrossRef]
- Williams, C.A.; Soufi, A.; Pollard, S.M. Post-translational modification of SOX family proteins: Key biochemical targets in cancer? Semin. Cancer Biol. 2019, 67, 30–38. [Google Scholar] [CrossRef]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef]
- Sarkar, A.; Hochedlinger, K. The Sox Family of Transcription Factors: Versatile Regulators of Stem and Progenitor Cell Fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Xu, J.; Watts, J.A.; Pope, S.D.; Gadue, P.; Kamps, M.; Plath, K.; Zaret, K.S.; Smale, S.T. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009, 23, 2824–2838. [Google Scholar] [CrossRef]
- Nishino, K.; Hattori, N.; Tanaka, S.; Shiota, K. DNA Methylation-mediated Control of Sry Gene Expression in Mouse Gonadal Development. J. Biol. Chem. 2004, 279, 22306–22313. [Google Scholar] [CrossRef]
- Katoh, H.; Ojima, H.; Kokubu, A.; Saito, S.; Kondo, T.; Kosuge, T.; Hosoda, F.; Imoto, I.; Inazawa, J.; Hirohashi, S.; et al. Genetically Distinct and Clinically Relevant Classification of Hepatocellular Carcinoma: Putative Therapeutic Targets. Gastroenterology 2007, 133, 1475–1486. [Google Scholar] [CrossRef]
- Xue, T.-C.; Zhang, L.; Ren, Z.-G.; Chen, R.-X.; Cui, J.-F.; Ge, N.-L.; Ye, S.-L. Sex-Determination Gene SRY Potentially Associates with Poor Prognosis but Not Sex Bias in Hepatocellular Carcinoma. Am. J. Dig. Dis. 2014, 60, 427–435. [Google Scholar] [CrossRef]
- Murakami, S.; Chishima, S.; Uemoto, H.; Sakamoto, E.; Sato, T.; Kurabe, N.; Kawasaki, Y.; Shibata, T.; Akiyama, H.; Tashiro, F. The male-specific factor Sry harbors an oncogenic function. Oncogene 2013, 33, 2978–2986. [Google Scholar] [CrossRef]
- Murakami, S.; Ninomiya, W.; Sakamoto, E.; Shibata, T.; Akiyama, H.; Tashiro, F. SRY and OCT4 Are Required for the Acquisition of Cancer Stem Cell-Like Properties and Are Potential Differentiation Therapy Targets. Stem Cells 2015, 33, 2652–2663. [Google Scholar] [CrossRef]
- Liu, C.; Ren, Y.-F.; Dong, J.; Ke, M.-Y.; Ma, F.; Monga, S.P.; Wu, R.; Lv, Y.; Zhang, X.-F. Activation of SRY accounts for male-specific hepatocarcinogenesis: Implication in gender disparity of hepatocellular carcinoma. Cancer Lett. 2017, 410, 20–31. [Google Scholar] [CrossRef]
- Pritchett, J.; Harvey, E.; Athwal, V.; Berry, A.; Rowe, C.; Oakley, F.; Moles, A.; Mann, D.A.; Bobola, N.; Sharrocks, A.D.; et al. Osteopontin is a novel downstream target of SOX9 with diagnostic implications for progression of liver fibrosis in humans. Hepatology 2012, 56, 1108–1116. [Google Scholar] [CrossRef]
- Liu, C.; Liu, L.; Chen, X.; Cheng, J.; Zhang, H.; Shen, J.; Shan, J.; Xu, Y.; Yang, Z.; Lai, M.; et al. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 2016, 64, 117–129. [Google Scholar] [CrossRef]
- Campbell, J.S.; Hughes, S.D.; Gilbertson, D.G.; Palmer, T.E.; Holdren, M.S.; Haran, A.C.; Odell, M.M.; Bauer, R.L.; Ren, H.-P.; Haugen, H.S.; et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc. Natl. Acad. Sci. 2005, 102, 3389–3394. [Google Scholar] [CrossRef]
- Uchikawa, M.; Kamachi, Y.; Kondoh, H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: Their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999, 84, 103–120. [Google Scholar] [CrossRef]
- Shih, Y.-L.; Hsieh, C.-B.; Yan, M.-D.; Tsao, C.-M.; Hsieh, T.-Y.; Liu, C.-H.; Lin, Y.-W. Frequent concomitant epigenetic silencing ofSOX1and secreted frizzled-related proteins (SFRPs) in human hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2012, 28, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.-M.; Yan, M.-D.; Shih, Y.-L.; Yu, P.-N.; Kuo, C.-C.; Lin, W.-C.; Li, H.-J.; Lin, Y.-W. SOX1 functions as a tumor suppressor by antagonizing the WNT/β-catenin signaling pathway in hepatocellular carcinoma. Hepatology 2012, 56, 2277–2287. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.K.; Guan, X.Y.; Ma, S. Cancer stem cells in hepatocellular carcinoma—from origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 19, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Sun, C.; Sun, L.; Li, Y.; Kang, X.; Zhang, S.; Liu, Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med. Oncol. 2013, 30, 503. [Google Scholar] [CrossRef]
- Pu, J.; Wu, X.; Wu, Y.; Shao, Z.; Luo, C.; Tang, Q.; Wang, J.; Wei, H.; Lu, Y. Anti-oncogenic effects of SOX2 silencing on hepatocellular carcinoma achieved by upregulating miR-222-5p-dependent CYLD via the long noncoding RNA CCAT1. Aging 2021, 13, 12207–12223. [Google Scholar] [CrossRef]
- Wen, W.; Han, T.; Chen, C.; Huang, L.; Sun, W.; Wang, X.; Chen, S.-Z.; Xiang, D.-M.; Tang, L.; Cao, D.; et al. Cyclin G1 Expands Liver Tumor-Initiating Cells by Sox2 Induction via Akt/mTOR Signaling. Mol. Cancer Ther. 2013, 12, 1796–1804. [Google Scholar] [CrossRef]
- Gnoni, A.; Licchetta, A.; Memeo, R.; Argentiero, A.; Solimando, A.G.; Longo, V.; Delcuratolo, S.; Brunetti, O. Role of BRAF in Hepatocellular Carcinoma: A Rationale for Future Targeted Cancer Therapies. Medicina 2019, 55, 754. [Google Scholar] [CrossRef]
- Krstic, J.; Reinisch, I.; Schindlmaier, K.; Galhuber, M.; Riahi, Z.; Berger, N.; Kupper, N.; Moyschewitz, E.; Auer, M.; Michenthaler, H.; et al. Fasting improves therapeutic response in hepatocellular carcinoma through p53-dependent metabolic synergism. Sci. Adv. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.-J.; Li, T.-W.; Zhou, L.; Wong, T.-L.; Liu, X.; Ma, V.W.; Lo, C.-M.; Man, K.; Lee, T.K.; Ning, W.; et al. FSTL1 Secreted by Activated Fibroblasts Promotes Hepatocellular Carcinoma Metastasis and Stemness. Cancer Res. 2021, 81, 5692–5705. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Zhang, Q.; Shen, J.; Zhang, H.; Shan, J.; Duan, G.; Guo, D.; Chen, X.; Cheng, J.; et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology 2016, 64, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, C.; Ungerleider, N.; Chen, W.; Song, K.; Wang, Y.; Kwon, H.; Ma, W.; Wu, T. A Transforming Growth Factor-β and H19 Signaling Axis in Tumor-Initiating Hepatocytes That Regulates Hepatic Carcinogenesis. Hepatology 2018, 69, 1549–1563. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, T.S.R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, D.; Jiang, J.; Dong, L. Loss of miR-638 promotes invasion and epithelial-mesenchymal transition by targeting SOX2 in hepatocellular carcinoma. Oncol. Rep. 2016, 37, 323–332. [Google Scholar] [CrossRef]
- Sakurai, T.; Kudo, M.; Umemura, A.; He, G.; Elsharkawy, A.M.; Seki, E.; Karin, M. p38α Inhibits Liver Fibrogenesis and Consequent Hepatocarcinogenesis by Curtailing Accumulation of Reactive Oxygen Species. Cancer Res. 2013, 73, 215–224. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, F.; Yang, N.; Zhu, N.; Fu, Y.; Zhang, H.-B.; Yang, G.-S. Overexpression of Sox3 is associated with promoted tumor progression and poor prognosis in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 7873–7881. [Google Scholar]
- Vicentic, J.M.; Drakulic, D.; Garcia, I.; Vukovic, V.; Aldaz, P.; Puskas, N.; Nikolic, I.; Tasic, G.; Raicevic, S.; Garros-Regulez, L.; et al. SOX3 can promote the malignant behavior of glioblastoma cells. Cell. Oncol. 2018, 42, 41–54. [Google Scholar] [CrossRef]
- Qiu, M.; Chen, D.; Shen, C.; Shen, J.; Zhao, H.; He, Y. Sex-determining region Y-box protein 3 induces epithelial-mesenchymal transition in osteosarcoma cells via transcriptional activation of Snail1. J. Exp. Clin. Cancer Res. 2017, 36, 46. [Google Scholar] [CrossRef]
- Tiwari, N.; Tiwari, V.K.; Waldmeier, L.; Balwierz, P.J.; Arnold, P.; Pachkov, M.; Meyer-Schaller, N.; Schübeler, D.; van Nimwegen, E.; Christofori, G. Sox4 Is a Master Regulator of Epithelial-Mesenchymal Transition by Controlling Ezh2 Expression and Epigenetic Reprogramming. Cancer Cell 2013, 23, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, S.J.; van Boxtel, R.; Coffer, P.J. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: Friend or foe? Oncogene 2012, 32, 3397–3409. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-L.; Sun, Y.-M.; Chau, G.-Y.; Chau, Y.-P.; Lai, T.-C.; Wang, J.-L.; Horng, J.-T.; Hsiao, M.; Tsou, A.-P. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 2008, 27, 5578–5589. [Google Scholar] [CrossRef] [PubMed]
- Hur, W.; Rhim, H.; Jung, C.K.; Kim, J.D.; Bae, S.H.; Jang, J.W.; Yang, J.M.; Oh, S.-T.; Kim, D.G.; Wang, H.J.; et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: Clinical implication and functional analysis in vitro. Carcinogenesis 2010, 31, 1298–1307. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Hao, M.; Zhang, X.; Zhang, J.; Xie, Q.; Wang, Y.; Guo, M.; Zhuang, H.; Lu, F. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2012, 33, 476–486. [Google Scholar] [CrossRef]
- Chen, Z.-z.; Huang, L.; Wu, Y.-h.; Zhai, W.-j.; Zhu, P.-p.; Gao, Y.-f. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat. Commun. 2016, 7, 12598. [Google Scholar] [CrossRef]
- Sandbothe, M.; Buurman, R.; Reich, N.; Greiwe, L.; Vajen, B.; Gürlevik, E.; Schäffer, V.; Eilers, M.; Kühnel, F.; Vaquero, A.; et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J. Hepatol. 2017, 66, 1012–1021. [Google Scholar] [CrossRef]
- Tsai, C.-N.; Yu, S.-C.; Lee, C.-W.; Pang, J.-H.S.; Wu, C.-H.; Lin, S.-E.; Chung, Y.-H.; Tsai, C.-L.; Hsieh, S.-Y.; Yu, M.-C. SOX4 activates CXCL12 in hepatocellular carcinoma cells to modulate endothelial cell migration and angiogenesis in vivo. Oncogene 2020, 39, 4695–4710. [Google Scholar] [CrossRef]
- Huang, J.-L.; Wang, X.-K.; Liao, X.-W.; Han, C.-Y.; Yu, T.-D.; Huang, K.-T.; Yang, C.-K.; Liu, X.-G.; Yu, L.; Zhu, G.-Z.; et al. SOX4 as biomarker in hepatitis B virus-associated hepatocellular carcinoma. J. Cancer 2021, 12, 3486–3500. [Google Scholar] [CrossRef]
- Wang, H.; Huo, X.; Yang, X.-R.; He, J.; Cheng, L.; Wang, N.; Deng, X.; Jingyuan, F.; Wang, N.; Wang, C.; et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 2017, 16, 1–15. [Google Scholar] [CrossRef]
- Wang, J.; Tang, Q.; Lu, L.; Luo, Z.; Li, W.; Lu, Y.; Pu, J. LncRNA OIP5-AS1 interacts with miR-363-3p to contribute to hepatocellular carcinoma progression through up-regulation of SOX4. Gene Ther. 2020, 27, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Fei, X.; Ren, X.; Xiao, J.X.; Chen, Y.; Wang, J. Pseudogene AKR1B10P1 enhances tumorigenicity and regulates epithelial-mesenchymal transition in hepatocellular carcinoma via stabilizing SOX4. J. Cell. Mol. Med. 2020, 24, 11779–11790. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.F.; Ahmed, M.B.; Alam, N.; Alemayohu, M.A.; Allen, C.; Alraddadi, R.; Alvisguzman, N.; Amoako, Y.A.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Shang, J.; Zheng, Y.; Guo, X.; Mo, J.; Xie, X.; Xiong, Y.; Liu, Y.; Wu, K.; Wu, J. Hepatitis B virus replication and sex-determining region Y box 4 production are tightly controlled by a novel positive feedback mechanism. Sci. Rep. 2015, 5, 10066. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, M.; Xi, J.; Liu, H.; Guan, G.; Shen, C.; Guo, Z.; Zhang, T.; Xu, Q.; Kudereti, D.; et al. Sex-determining region Y box 4 (SOX4) suppresses Hepatitis B virus replication by inhibiting hepatocyte nuclear factor 4α expression. Antivir. Res. 2020, 176, 104745. [Google Scholar] [CrossRef]
- Jiao, F.; Long, L.; Ding, S.; Xie, X.; Jia, L.; Lu, F. Complete genome sequencing and clinical analysis of intrahepatic hepatitis B virus cccDNA from HCC. Microb. Pathog. 2017, 109, 49–55. [Google Scholar] [CrossRef]
- Kang, J.; Wang, J.; Cheng, J.; Cao, Z.; Chen, R.; Li, H.; Liu, S.; Chen, X.; Sui, J.; Lu, F. Down-regulation of NTCP expression by cyclin D1 in hepatitis B virus-related hepatocellular carcinoma has clinical significance. Oncotarget 2016, 8, 56041–56050. [Google Scholar] [CrossRef]
- Ek, S.; Dictor, M.; Jerkeman, M.; Jirström, K.; Borrebaeck, C.A.K. Nuclear expression of the non–B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood 2008, 111, 800–805. [Google Scholar] [CrossRef]
- Weigle, B.; Ebner, R.; Temme, A.; Schwind, S.; Schmitz, M.; Kiessling, A.; Rieger, M.; Schackert, G.; Schackert, H.; Rieber, E. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol. Rep. 2005, 13. [Google Scholar] [CrossRef]
- De Bont, J.M.; Kros, J.; Passier, M.M.C.J.; Reddingius, R.E.; Smitt, P.A.E.S.; Luider, T.M.; den Boer, M.L.; Pieters, R. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro-Oncol. 2008, 10, 648–660. [Google Scholar] [CrossRef]
- Brennan, D.J.; Ek, S.; Doyle, E.; Drew, T.; Foley, M.; Flannelly, G.; O’Connor, D.; Gallagher, W.; Kilpinen, S.; Kallioniemi, O.; et al. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur. J. Cancer 2009, 45, 1510–1517. [Google Scholar] [CrossRef]
- Zvelebil, M.; Oliemuller, E.; Gao, Q.; Wansbury, O.; Mackay, A.; Kendrick, H.; Smalley, M.J.; Reis-Filho, J.S.; A Howard, B. Embryonic mammary signature subsets are activated in Brca1−/− and basal-like breast cancers. Breast Cancer Res. 2013, 15, R25. [Google Scholar] [CrossRef] [PubMed]
- Beekman, R.; Amador, V.; Campo, E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr. Opin. Hematol. 2018, 25, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Oliemuller, E.; Kogata, N.; Bland, P.; Kriplani, D.; Daley, F.; Haider, S.; Shah, V.; Sawyer, E.; A Howard, B. SOX11 promotes invasive growth and ductal carcinomain situprogression. J. Pathol. 2017, 243, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, J.Y.; Zhong, Y.; Xie, L.; Li, J.S. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz. J. Med Biol. Res. 2019, 52, e8631. [Google Scholar] [CrossRef]
- Liu, Z.; Zhong, Y.; Chen, Y.J.; Chen, H. SOX11 regulates apoptosis and cell cycle in hepatocellular carcinoma via Wnt/β-catenin signaling pathway. Biotechnol. Appl. Biochem. 2018, 66, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Wang, C.; Liu, J.; Wang, Q.; Zhang, D.; Zhu, S.; Xu, S.; Kang, M.; He, S. Sox12 Is a Cancer Stem-Like Cell Marker in Hepatocellular Carcinoma. Mol. Cells 2017, 40, 847–854. [Google Scholar] [CrossRef]
- Yuan, P.; Meng, L.; Wang, N. SOX12 upregulation is associated with metastasis of hepatocellular carcinoma and increases CDK4 and IGF2BP1 expression. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3821–3826. [Google Scholar]
- Huang, W.; Chen, Z.; Shang, X.; Tian, D.; Wang, D.; Wu, K.; Fan, D.; Xia, L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology 2015, 61, 1920–1933. [Google Scholar] [CrossRef]
- Jiang, T.; Guan, L.-Y.; Ye, Y.-S.; Liu, H.-Y.; Li, R. MiR-874 inhibits metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting SOX12. Am. J. Cancer Res. 2017, 7, 1310–1321. [Google Scholar]
- Zhang, W.; Liu, K.; Liu, S.; Ji, B.; Liu, Y. MicroRNA-744 inhibits migration and invasion of hepatocellular carcinoma cells by targeting SOX12. Oncol. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Meng, Y.; Luo, C.; He, S.; Qin, F.; Yin, Y.; Huang, J.; Zhao, H.; Hu, J.; Deng, Z.; et al. lncRNA PRR34-AS1 promotes HCC development via modulating Wnt/β-catenin pathway by absorbing miR-296-5p and upregulating E2F2 and SOX12. Mol. Ther.-Nucleic Acids 2021, 25, 37–52. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, X.; Lu, F.; Zhang, T.; Hao, M.; Wang, Y.; Zhao, J.; McCrae, M.A.; Zhuang, H. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer 2011, 118, 2431–2442. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y. miR-96 targets SOX6 and promotes proliferation, migration, and invasion of hepatocellular carcinoma. Biochem. Cell Biol. 2018, 96, 365–371. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, J.; Apaer, S.; Yao, G.; Li, T. microRNA-19a-3p and microRNA-376c-3p Promote Hepatocellular Carcinoma Progression Through SOX6-Mediated Wnt/β-Catenin Signaling Pathway. Int. J. Gen. Med. 2021, ume 14, 89–102. [Google Scholar] [CrossRef]
- Xiong, G.; Wang, Y.; Ding, Q.; Yang, L. Hsa-mir-1269 genetic variant contributes to hepatocellular carcinoma susceptibility through affecting SOX6. Am. J. Transl. Res. 2015, 7, 2091–2098. [Google Scholar]
- Guo, X.; Yang, M.; Gu, H.; Zhao, J.; Zou, L. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol. 2013, 37, 732–736. [Google Scholar] [CrossRef]
- Wang, N.; Han, S.; Wang, X.; Peng, R.; Li, X. SOX5 promotes epithelial–mesenchymal transition and cell invasion via regulation of Twist1 in hepatocellular carcinoma. Med. Oncol. 2015, 32, 461. [Google Scholar] [CrossRef]
- Feng, M.; Fang, F.; Fang, T.; Jiao, H.; You, S.; Wang, X.; Zhao, W. Sox13 promotes hepatocellular carcinoma metastasis by transcriptionally activating Twist1. Lab. Investig. 2020, 100, 1400–1410. [Google Scholar] [CrossRef]
- Kamachi, Y.; Uchikawa, M.; Kondoh, H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 2000, 16, 182–187. [Google Scholar] [CrossRef]
- Wilson, M.; Koopman, P. Matching SOX: Partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002, 12, 441–446. [Google Scholar] [CrossRef]
- Lefebvre, V.; Li, P.; De Crombrugghe, B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998, 17, 5718–5733. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Fang, F.; Fang, T.; You, Y.; Feng, M.; Wang, X.; Yin, Z.; Zhao, W. SOX13 regulates cancer stem-like properties and tumorigenicity in hepatocellular carcinoma cells. Am. J. Cancer Res. 2021, 11, 760–772. [Google Scholar] [PubMed]
- Furuyama, K.; Kawaguchi, Y.; Akiyama, H.; Horiguchi, M.; Kodama, S.; Kuhara, T.; Hosokawa, S.; Elbahrawy, A.; Soeda, T.; Koizumi, M.; et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2010, 43, 34–41. [Google Scholar] [CrossRef]
- Leung, C.O.-N.; Mak, W.-N.; Kai, A.K.-L.; Chan, K.-S.; Lee, K.W.; Oi-Ning, L.C.; Lo, R.C.-L. Sox9 confers stemness properties in hepatocellular carcinoma through Frizzled-7 mediated Wnt/β-catenin signaling. Oncotarget 2016, 7, 29371–29386. [Google Scholar] [CrossRef]
- Xiao, Y.; Sun, Y.; Liu, G.; Zhao, J.; Gao, Y.; Yeh, S.; Gong, L.; Chang, C. Androgen receptor (AR)/miR-520f-3p/SOX9 signaling is involved in altering hepatocellular carcinoma (HCC) cell sensitivity to the Sorafenib therapy under hypoxia via increasing cancer stem cells phenotype. Cancer Lett. 2018, 444, 175–187. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Zhi, X.; Ding, W.; Xiong, J.; Tao, T.; Yang, Y.; Zhang, H.; Zi, X.; Zhou, W.; et al. SOX9 enhances sorafenib resistance through upregulating ABCG2 expression in hepatocellular carcinoma. Biomed. Pharmacother. 2020, 129, 110315. [Google Scholar] [CrossRef]
- Morrison, S.J.; Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef]
- Sugiarto, S.; Persson, A.I.; Munoz, E.G.; Waldhuber, M.; Lamagna, C.; Andor, N.; Hanecker, P.; Ayers-Ringler, J.; Phillips, J.; Siu, J.; et al. Asymmetry-Defective Oligodendrocyte Progenitors Are Glioma Precursors. Cancer Cell 2011, 20, 328–340. [Google Scholar] [CrossRef]
- Pine, S.R.; Ryan, B.M.; Varticovski, L.; Robles, A.I.; Harris, C.C. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2195–2200. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Miyauchi, Y.; Kojima, H.; Yamaoka, R.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; et al. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci. Rep. 2016, 6, 30489. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Y.; Li, X.; Liu, B.; Han, S.; Li, X.; Zhang, B.; Li, J.; Sun, S. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed. Pharmacother. 2019, 121, 109517. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.-R.; Wong, C.C.-L. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells 2021, 10, 1715. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Lu, Q.; Liu, Y.; Zhao, J.; Zhang, Q.; Hu, L.; Shi, Z.; Tu, Y.; Xiao, Z.; Xu, Q.; et al. Effect of the Hypoxia Inducible Factor on Sorafenib Resistance of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 641522. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xiong, L.; Sun, T.; Peng, R.; Zou, L.; Zhu, H.; Zhang, J.; Li, H.; Zhao, J. Expression features of SOX9 associate with tumor progression and poor prognosis of hepatocellular carcinoma. Diagn. Pathol. 2012, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, Y.; Hou, B.; Wang, Y.; Deng, D.; Fu, Z.; Xu, Z. A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives tumor growth and metastasis in hepatocellular carcinoma through the Wnt/β-catenin pathway. Mol. Oncol. 2019, 13, 2194–2210. [Google Scholar] [CrossRef]
- Chen, Y.; Qian, B.; Sun, X.; Kang, Z.; Huang, Z.; Ding, Z.; Dong, L.; Chen, J.; Zhang, J.; Zang, Y. Sox9/INHBB axis-mediated crosstalk between the hepatoma and hepatic stellate cells promotes the metastasis of hepatocellular carcinoma. Cancer Lett. 2020, 499, 243–254. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Xiong, L.; Kong, X.; Xu, Y.; Liu, C.; Zou, L.; Li, Z.; Zhao, J.; Lin, N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012, 586, 4362–4370. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, W.; Liu, K.; Liu, S.; Ji, B.; Wang, Y. miR-138 suppresses cell proliferation and invasion by inhibiting SOX9 in hepatocellular carcinoma. Am. J. Transl. Res. 2016, 8, 2159–2168. [Google Scholar]
- Zhang, H.; Zhang, Z.; Gao, L.; Qiao, Z.; Yu, M.; Yu, B.; Yang, T. miR-1-3p suppresses proliferation of hepatocellular carcinoma through targeting SOX9. OncoTargets Ther. 2019, ume 12, 2149–2157. [Google Scholar] [CrossRef]
- Li, B.; Liu, D.; Yang, P.; Li, H.-Y.; Wang, D. miR-613 inhibits liver cancer stem cell expansion by regulating SOX9 pathway. Gene 2019, 707, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.-X.; Zhan, G.-F.; Wu, J.-C.; Fang, H.; Yang, S.-L. LncRNA SNHG14 Sponges miR-206 to Affect Proliferation, Apoptosis, and Metastasis of Hepatocellular Carcinoma Cells by Regulating SOX9. Am. J. Dig. Dis. 2021. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, W.-L.; Zhang, Y.; Guo, S.-P.; Zhang, J.; Li, Q.-L. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol. Res. 2007, 37, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Sze, K.M.-F.; Wong, K.L.-T.; Chu, G.K.-Y.; Lee, J.M.-F.; Yau, T.-O.; Ng, I.O.-L. Loss of phosphatase and tensin homolog enhances cell invasion and migration through aKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology 2011, 53, 1558–1569. [Google Scholar] [CrossRef]
- Yothaisong, S.; Dokduang, H.; Techasen, A.; Namwat, N.; Yongvanit, P.; Bhudhisawasdi, V.; Puapairoj, A.; Riggins, G.J.; Loilome, W. Increased activation of PI3K/AKT signaling pathway is associated with cholangiocarcinoma metastasis and PI3K/mTOR inhibition presents a possible therapeutic strategy. Tumor Biol. 2013, 34, 3637–3648. [Google Scholar] [CrossRef]
- Chen, J.; Debebe, A.; Zeng, N.; Kopp, J.; He, L.; Sander, M.; Stiles, B.L. Transformation of SOX9+ cells by Pten deletion synergizes with steatotic liver injury to drive development of hepatocellular and cholangiocarcinoma. Sci. Rep. 2021, 11, 11823. [Google Scholar] [CrossRef]
- Ma, X.-L.; Hu, B.; Tang, W.-G.; Xie, S.-H.; Ren, N.; Guo, L.; Lu, R.-Q. CD73 sustained cancer-stem-cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 11–16. [Google Scholar] [CrossRef]
- Zeng, R.; Wang, C.; Wang, W.; Wang, S. Long non-coding RNA DUXAP9 promotes hepatocellular carcinoma cell stemness via directly interacting with sox9. Environ. Toxicol. 2021, 36, 1793–1801. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef]
- Coulouarn, C.; Corlu, A.; Glaise, D.; Guénon, I.; Thorgeirsson, S.S.; Clément, B. Hepatocyte–Stellate Cell Cross-Talk in the Liver Engenders a Permissive Inflammatory Microenvironment That Drives Progression in Hepatocellular Carcinoma. Cancer Res. 2012, 72, 2533–2542. [Google Scholar] [CrossRef]
- Ji, J.; Eggert, T.; Budhu, A.; Forgues, M.; Takai, A.; Dang, H.; Ye, Q.; Lee, J.-S.; Kim, J.H.; Greten, T.F.; et al. Hepatic stellate cell and monocyte interaction contributes to poor prognosis in hepatocellular carcinoma. Hepatology 2015, 62, 481–495. [Google Scholar] [CrossRef]
- Kang, N.; Gores, G.J.; Shah, V.H. Hepatic stellate cells: Partners in crime for liver metastases? Hepatology 2011, 54, 707–713. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, C.; Zhu, L.; Liu, H.; Liu, S.; Zhao, N.; Wu, J.; Huang, X.; Zhang, Y.; Jin, J.; et al. Oncogenicity of the transcription factor SOX8 in hepatocellular carcinoma. Med. Oncol. 2014, 31, 918. [Google Scholar] [CrossRef]
- Higashijima, Y.; Kanki, Y. Molecular mechanistic insights: The emerging role of SOXF transcription factors in tumorigenesis and development. Semin. Cancer Biol. 2019, 67, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, Y.; Wang, J.; Min, Z. The Suppressive Role of SOX7 in Hepatocarcinogenesis. PLoS ONE 2014, 9, e97433. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, S.; Wu, J.; Lu, Z.; Yang, J.; Wu, H.; Chen, H.; Lin, B.; Cao, T. Clinical significance and prognostic value of SOX7 expression in liver and pancreatic carcinoma. Mol. Med. Rep. 2017, 16, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Liu, J.; Yang, Z.; Wu, L.; Xie, H.; Jiang, C.; Lin, B.; Chen, T.; Xing, C.; Liu, Z.; et al. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/β-catenin signaling pathway. Oncotarget 2016, 7, 28000–28012. [Google Scholar] [CrossRef]
- Wu, G.-G.; Li, W.-H.; He, W.-G.; Jiang, N.; Zhang, G.-X.; Chen, W.; Yang, H.-F.; Liu, Q.-L.; Huang, Y.-N.; Zhang, L.; et al. Mir-184 Post-Transcriptionally Regulates SOX7 Expression and Promotes Cell Proliferation in Human Hepatocellular Carcinoma. PLoS ONE 2014, 9, e88796. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Yu, Z.; Li, J.; Sun, R.; Kan, Q. miR-935 Promotes Liver Cancer Cell Proliferation and Migration by Targeting SOX7. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 427–435. [Google Scholar] [CrossRef]
- Zhang, W.; Glöckner, S.C.; Guo, M.; Machida, E.O.; Wang, D.H.; Easwaran, H.; Van Neste, L.; Herman, J.G.; Schuebel, K.E.; Watkins, D.N.; et al. Epigenetic Inactivation of the Canonical Wnt Antagonist SRY-Box Containing Gene 17 in Colorectal Cancer. Cancer Res. 2008, 68, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, Y.; Liu, S.; Herman, J.G.; Lu, F.; Guo, M. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics 2010, 5, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wei, Z.; Jia, H.; Zhao, W.; Yang, G.; Zhao, H. Knockdown of SOX18 inhibits the proliferation, migration and invasion of hepatocellular carcinoma cells. Oncol. Rep. 2015, 34, 1121–1128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.; Du, F.; Dang, Y.; Li, X.; Qian, M.; Feng, W.; Qiao, C.; Fan, D.; Nie, Y.; Wu, K.; et al. Fibroblast Growth Factor 19–Mediated Up-regulation of SYR-Related High-Mobility Group Box 18 Promotes Hepatocellular Carcinoma Metastasis by Transactivating Fibroblast Growth Factor Receptor 4 and Fms-Related Tyrosine Kinase 4. Hepatology 2019, 71, 1712–1731. [Google Scholar] [CrossRef]
- Sun, Y.; Lei, B.; Huang, Q. SOX18 Affects Cell Viability, Migration, Invasiveness, and Apoptosis in Hepatocellular Carcinoma (HCC) Cells by Participating in Epithelial-to-Mesenchymal Transition (EMT) Progression and Adenosine Monophosphate Activated Protein Kinase (AMPK)/Mammalian Target of Rapamycin (mTOR). Med. Sci. Monit. 2019, 25, 6244–6254. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Zhong, J.; Wu, C.; Yang, G.; Zhong, Y.; Zhang, J.; Tang, A. Decreased expression of SRY-box containing gene 30 is related to malignant phenotypes of human bladder cancer and correlates with poor prognosis. BMC Cancer 2018, 18, 642. [Google Scholar] [CrossRef]
- Han, F.; Liu, W.-B.; Shi, X.-Y.; Yang, J.-T.; Zhang, X.; Li, Z.-M.; Jiang, X.; Yin, L.; Li, J.-J.; Huang, C.; et al. SOX30 Inhibits Tumor Metastasis through Attenuating Wnt-Signaling via Transcriptional and Posttranslational Regulation of β-Catenin in Lung Cancer. eBioMedicine 2018, 31, 253–266. [Google Scholar] [CrossRef]
- Tao, J.; Liu, Z.; Wang, Y.; Wang, L.; Yin, G.; Yang, W.; Tu, K.; Liu, Q. MicroRNA-645 represses hepatocellular carcinoma progression by inhibiting SOX30-mediated p53 transcriptional activation. Int. J. Biol. Macromol. 2018, 121, 214–222. [Google Scholar] [CrossRef]
- Lei, X.; Lei, Y.; Li, J.-K.; Du, W.-X.; Li, R.-G.; Yang, J.; Li, J.; Li, F.; Tan, H.-B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2019, 470, 126–133. [Google Scholar] [CrossRef]
- Pitt, J.M.; Vétizou, M.; Daillère, R.; Roberti, M.P.; Yamazaki, T.; Routy, B.; Lepage, P.; Boneca, I.G.; Chamaillard, M.; Kroemer, G.; et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016, 44, 1255–1269. [Google Scholar] [CrossRef]
- Phuengkham, H.; Ren, L.; Shin, I.W.; Lim, Y.T. Nanoengineered Immune Niches for Reprogramming the Immunosuppressive Tumor Microenvironment and Enhancing Cancer Immunotherapy. Adv. Mater. 2019, 31, e1803322. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Xie, Y.; Tan, Y.S.; Prince, M.E.; Moyer, J.S.; Nör, J.; Wolf, G.T. Telltale tumor infiltrating lymphocytes (TIL) in oral, head & neck cancer. Oral Oncol. 2016, 61, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.S.; Sansanaphongpricha, K.; Xie, Y.; Donnelly, C.; Luo, X.; Heath, B.R.; Zhao, X.; Bellile, E.L.; Hu, H.; Chen, H.; et al. Mitigating SOX2-potentiated Immune Escape of Head and Neck Squamous Cell Carcinoma with a STING-inducing Nanosatellite Vaccine. Clin. Cancer Res. 2018, 24, 4242–4255. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, C.; Li, Z.; Xiao, J.; Li, C.; Wang, X.; Kong, P.; Cao, J.; Huang, F.; Huang, Y.; et al. SOX2 promotes resistance of melanoma with PD-L1 high expression to T-cell-mediated cytotoxicity that can be reversed by SAHA. J. Immunother. Cancer 2020, 8, e001037. [Google Scholar] [CrossRef]
- Benci, J.L.; Johnson, L.R.; Choa, R.; Xu, Y.; Qiu, J.; Zhou, Z.; Xu, B.; Ye, D.; Nathanson, K.L.; June, C.H.; et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 2019, 178, 933–948.e14. [Google Scholar] [CrossRef]
- Benci, J.L.; Xu, B.; Qiu, Y.; Wu, T.J.; Dada, H.; Twyman-Saint Victor, C.; Cucolo, L.; Lee, D.S.M.; Pauken, K.E.; Huang, A.C.; et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016, 167, 1540–1554.e12. [Google Scholar] [CrossRef] [PubMed]
- Mollaoglu, G.; Jones, A.; Wait, S.J.; Mukhopadhyay, A.; Jeong, S.; Arya, R.; Camolotto, S.A.; Mosbruger, T.L.; Stubben, C.J.; Conley, C.J.; et al. The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment. Immunity 2018, 49, 764–779.e9. [Google Scholar] [CrossRef]
- Ma, T.; Hu, C.; Lal, B.; Zhou, W.; Ma, Y.; Ying, M.; Prinos, P.; Quiñones-Hinojosa, A.; Lim, M.; Laterra, J.; et al. Reprogramming Transcription Factors Oct4 and Sox2 Induce a BRD-Dependent Immunosuppressive Transcriptome in GBM-Propagating Cells. Cancer Res. 2021, 81, 2457–2469. [Google Scholar] [CrossRef]
- Kuo, M.-H.; Chen, P.-Y.; Yang, Y.-P.; Zheng, M.-Y.; Miao, C.-C.; Wen, K.-C.; Chang, K.-M.; Chou, S.-J.; Wang, M.-L.; Chiou, S.-H.; et al. Cytokine and Epigenetic Regulation of Programmed Death-Ligand 1 in Stem Cell Differentiation and Cancer Cell Plasticity. Stem Cells 2021, 39, 1298–1309. [Google Scholar] [CrossRef]
- Bagati, A.; Kumar, S.; Jiang, P.; Pyrdol, J.; Zou, A.E.; Godicelj, A.; Mathewson, N.D.; Cartwright, A.N.; Cejas, P.; Brown, M.; et al. Integrin αvβ6–TGFβ–SOX4 Pathway Drives Immune Evasion in Triple-Negative Breast Cancer. Cancer Cell 2020, 39, 54–67.e9. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Liu, G.; Peng, H.; Chen, A.; Zha, L.; Wang, Z. SOX4 contributes to TGF-β-induced epithelial–mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis. 2018, 5, 49–61. [Google Scholar] [CrossRef]
- Vervoort, S.J.; Lourenço, A.R.; van Boxtel, R.; Coffer, P.J. SOX4 Mediates TGF-β-Induced Expression of Mesenchymal Markers during Mammary Cell Epithelial to Mesenchymal Transition. PLoS ONE 2013, 8, e53238. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Mann, M.; Zhao, J.L.; Marinov, G.K.; Majumdar, D.; Garcia-Flores, Y.; Du, X.; Erikci, E.; Chowdhury, K.; Baltimore, D. The microRNA-212/132 cluster regulates B cell development by targeting Sox4. J. Exp. Med. 2015, 212, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Bezzi, M.; Seitzer, N.; Ishikawa, T.; Reschke, M.; Chen, M.; Wang, G.; Mitchell, C.; Ng, C.; Katon, J.; Lunardi, A.; et al. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat. Med. 2018, 24, 165–175. [Google Scholar] [CrossRef]
- Chen, Y.; Kim, J.; Yang, S.; Wang, H.; Wu, C.-J.; Sugimoto, H.; LeBleu, V.S.; Kalluri, R. Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell 2021, 39, 548–565.e6. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef]
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Xu, D.; Liu, X.; Wang, Y.; Zhou, K.; Wu, J.; Chen, J.C.; Chen, C.; Chen, L.; Zheng, J. Identification of immune subtypes and prognosis of hepatocellular carcinoma based on immune checkpoint gene expression profile. Biomed. Pharmacother. 2020, 126, 109903. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Gari, A.; Qu, C.; Chen, J. Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures. Front. Immunol. 2021, 12, 721030. [Google Scholar] [CrossRef] [PubMed]
- Pietrobono, S.; Gaudio, E.; Gagliardi, S.; Zitani, M.; Carrassa, L.; Migliorini, F.; Petricci, E.; Manetti, F.; Makukhin, N.; Bond, A.G.; et al. Targeting non-canonical activation of GLI1 by the SOX2-BRD4 transcriptional complex improves the efficacy of HEDGEHOG pathway inhibition in melanoma. Oncogene 2021, 40, 3799–3814. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Ye, H.; He, H.H.; Gerrin, S.J.; Chen, S.; Tanenbaum, B.A.; Cai, C.; Sowalsky, A.G.; He, L.; Wang, H.; et al. SOX9 drives WNT pathway activation in prostate cancer. J. Clin. Investig. 2016, 126, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Z.; Yu, X.; Li, Q.; Wang, Q.; Chang, A.; Huang, X.; Han, X.; Song, Y.; Hu, J.; et al. SOX9/miR-203a axis drives PI3K/AKT signaling to promote esophageal cancer progression. Cancer Lett. 2019, 468, 14–26. [Google Scholar] [CrossRef]
- Hua, X.; Huang, M.; Deng, X.; Xu, J.; Luo, Y.; Xie, Q.; Xu, J.; Tian, Z.; Li, J.; Zhu, J.; et al. The inhibitory effect of compound ChlA-F on human bladder cancer cell invasion can be attributed to its blockage of SOX2 protein. Cell Death Differ. 2019, 27, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Temme, A.; Senner, V.; Ebner, R.; Schwind, S.; Stevanovic, S.; Wehner, R.; Schackert, G.; Schackert, H.K.; Fussel, M.; et al. Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. Br. J. Cancer 2007, 96, 1293–1301. [Google Scholar] [CrossRef]
- Friedman, R.S.; Bangur, C.S.; Zasloff, E.J.; Fan, L.; Wang, T.; Watanabe, Y.; Kalos, M. Molecular and immunological evaluation of the transcription factor SOX-4 as a lung tumor vaccine antigen. J. Immunol. 2004, 172, 3319–3327. [Google Scholar] [CrossRef]
- Ueda, R.; Kinoshita, E.; Ito, R.; Kawase, T.; Kawakami, Y.; Toda, M. Induction of protective and therapeutic antitumor immunity by a DNA vaccine with a glioma antigen, SOX6. Int. J. Cancer 2008, 122, 2274–2279. [Google Scholar] [CrossRef]
- Ueda, R.; Ohkusu-Tsukada, K.; Fusaki, N.; Soeda, A.; Kawase, T.; Kawakami, Y.; Toda, M. Identification of HLA-A2- and A24-restricted T-cell epitopes derived from SOX6 expressed in glioma stem cells for immunotherapy. Int. J. Cancer 2009, 126, 919–929. [Google Scholar] [CrossRef]
- Schmitz, M.; Wehner, R.; Stevanovic, S.; Kiessling, A.; Rieger, M.A.; Temme, A.; Bachmann, M.; Rieber, E.P.; Weigle, B. Identification of a naturally processed T cell epitope derived from the glioma-associated protein SOX11. Cancer Lett. 2007, 245, 331–336. [Google Scholar] [CrossRef]
- Andey, T.; Bora-Singhal, N.; Chellappan, S.P.; Singh, M. Cationic lipoplexes for treatment of cancer stem cell-derived murine lung tumors. Nanomed. Nanotechnology, Biol. Med. 2019, 18, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Tang, G.; Chen, Y.; Wang, C.; Guo, M.; Xu, T.; Zhao, M.; Zhou, Y. Tumor-targeted delivery of siRNA to silence Sox2 gene expression enhances therapeutic response in hepatocellular carcinoma. Bioact. Mater. 2020, 6, 1330–1340. [Google Scholar] [CrossRef] [PubMed]
- Stolzenburg, S.; Rots, M.G.; Beltran, A.S.; Rivenbark, A.G.; Yuan, X.; Qian, H.; Strahl, B.D.; Blancafort, P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012, 40, 6725–6740. [Google Scholar] [CrossRef]
- Yokota, E.; Yamatsuji, T.; Takaoka, M.; Haisa, M.; Takigawa, N.; Miyake, N.; Ikeda, T.; Mori, T.; Ohno, S.; Sera, T.; et al. Targeted silencing of SOX2 by an artificial transcription factor showed antitumor effect in lung and esophageal squamous cell carcinoma. Oncotarget 2017, 8, 103063–103076. [Google Scholar] [CrossRef]
- A Marks, P. Discovery and development of SAHA as an anticancer agent. Oncogene 2007, 26, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Revia, R.A.; Stephen, Z.R.; Zhang, M. Theranostic Nanoparticles for RNA-Based Cancer Treatment. Accounts Chem. Res. 2019, 52, 1496–1506. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lin, Z.; Li, Y.; Zhao, M.; Wang, C.; Guo, M.; Zhang, B.; Zhu, B. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J. Mater. Chem. B 2017, 5, 6941–6952. [Google Scholar] [CrossRef]
- Sera, T. Zinc-finger-based artificial transcription factors and their applications. Adv. Drug Deliv. Rev. 2009, 61, 513–526. [Google Scholar] [CrossRef]
- Margolin, J.F.; Friedman, J.; Meyer, W.K.; Vissing, H.; Thiesen, H.J.; Rauscher, F.J. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 1994, 91, 4509–4513. [Google Scholar] [CrossRef]
- Groner, A.C.; Meylan, S.; Ciuffi, A.; Zangger, N.; Ambrosini, G.; Dénervaud, N.; Bucher, P.; Trono, D. KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading. PLoS Genet. 2010, 6, e1000869. [Google Scholar] [CrossRef]
- Kumar, P.; Mistri, T.K. Transcription factors in SOX family: Potent regulators for cancer initiation and development in the human body. Semin. Cancer Biol. 2019, 67, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Radoux, C.J.; Hercules, A.; Ochoa, D.; Dunham, I.; Zalmas, L.-P.; Hessler, G.; Ruf, S.; Shanmugasundaram, V.; Hann, M.M.; et al. The PROTACtable genome. Nat. Rev. Drug Discov. 2021, 20, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Henley, M.J.; Koehler, A.N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug Discov. 2021, 20, 669–688. [Google Scholar] [CrossRef] [PubMed]

| Strategy | Target | Biomarker | Cancer | Mechanism of Action | Clinical Trial |
|---|---|---|---|---|---|
| SOX factors as biomarkers for patient stratification treatment | |||||
| MRT-92 plus MZ1 [162] | SMO+ BRD4 | SOX2 | Melanoma | SMO and SOX2-BRD4 complex activate hedgehog/GLI signaling in canonical and noncanonical manners, respectively | / |
| Integrin αvβ6/8 mAb [151] | Integrin αvβ6/8 | SOX4 | TNBC | Integrin αv up-regulates SOX4 by activating TGFβ from a latent precursor | Phase II NCT03688230 |
| LGK974 [163] | WNT | SOX9 | Prostate cancer | SOX9 reactivates the WNT/β-catenin signaling | Phase I NCT01351103 |
| Rapamycin [164] | mTOR | SOX9 | Esophageal cancer | SOX9 inhibits miR-203a to activate the PI3K/AKT/mTOR signaling | Phase III NCT04736589 |
| BLU9931 [134] | FGFR4 | SOX18 | HCC | SOX18 transactivates FGFR4 and FLT4, and FGFR4-FGF19 in turn up-regulates SOX18 | / |
| Targeting SOX proteins degradation | |||||
| SAHA [145] | SOX2 | SOX2 | Melanoma | Promoting SOX2 acetylation and proteasome-dependent degradation | Phase I/II NCT02638090 |
| ChlA-F [165] | SOX2 | SOX2 | Bladder cancer | promoting SOX2 ubiquitination and protein degradation by enhancing the mRNA stability of USP8 and inhibiting SOX2 protein translation by activating c-Jun-miR-200c | / |
| SOX factors as peptide vaccine boost anti-tumor immune response | |||||
| SOX2-derived peptide [166] | SOX2 | SOX2 | Glioblastoma | As a tumor-specific vaccine antigen to augment CTLs response | Phase I NCT02157051 |
| SOX4-derived peptide [167] | SOX4 | SOX4 | Lung cancer | As a tumor-specific vaccine antigen to augment CTLs response | / |
| SOX6-derived peptide [168,169] | SOX6 | SOX6 | Glioblastoma | As a tumor-specific vaccine antigen to augment CTLs response | / |
| SOX11-derived peptide [170] | SOX11 | SOX11 | Glioblastoma | As a tumor-specific vaccine antigen to augment CTLs response | / |
| Tumor-targeted delivery of siRNA to silence SOX expression | |||||
| CL-siSOX2 [171] | SOX2 | SOX2 | Lung cancer | Deliver therapeutic siRNA targeting SOX2 to tumor in vivo through a lipoplex nanoparticle | / |
| RGDfC-SeNPs-siSOX2 [172] | SOX2 | SOX2 | HCC | Deliver therapeutic siRNA targeting SOX2 to tumor in vivo through a RGDfC-SeNP system | / |
| Targeting endogenous SOX expression by artificial transcription factors-based technologies | |||||
| ZF-552SKD [173] | SOX2 promoter | SOX2 | Breast cancer | ZF-based ATF inhibits SOX2 expression through recruiting transcriptional repressor SKD domain to the SOX2 promoter and recruiting co-repressor KAP1 to facilitate chromatin condensation | / |
| ZF-598SKD [173] | SOX2 promoter | SOX2 | Breast cancer | ZF-based ATF inhibits SOX2 expression through recruiting transcriptional repressor SKD domain to the SOX2 promoter and recruiting co-repressor KAP1 to facilitate chromatin condensation | / |
| ZF-619SKD [173] | SOX2 promoter | SOX2 | Breast cancer | ZF-based ATF inhibits SOX2 expression through recruiting transcriptional repressor SKD domain to the SOX2 promoter and recruiting co-repressor KAP1 to facilitate chromatin condensation | / |
| ZF-4203SKD [173] | SOX2 regulatory region I | SOX2 | Breast cancer | ZF-based ATF inhibits SOX2 expression through recruiting transcriptional repressor SKD domain to the SOX2 promoter and recruiting co-repressor KAP1 to facilitate chromatin condensation | / |
| ATF/SOX2 [174] | SOX2 promoter | SOX2 | Lung and esophageal SCC | ZF-based ATF inhibits SOX2 expression through targeting the SOX2 distal and proximal promoter region and a KOX transcriptional repressor domain | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Ji, X.; Xie, M.; Zhang, T.; Wang, Y.; Sun, M.; Huang, W.; Xia, L. Advance of SOX Transcription Factors in Hepatocellular Carcinoma: From Role, Tumor Immune Relevance to Targeted Therapy. Cancers 2022, 14, 1165. https://doi.org/10.3390/cancers14051165
Luo X, Ji X, Xie M, Zhang T, Wang Y, Sun M, Huang W, Xia L. Advance of SOX Transcription Factors in Hepatocellular Carcinoma: From Role, Tumor Immune Relevance to Targeted Therapy. Cancers. 2022; 14(5):1165. https://doi.org/10.3390/cancers14051165
Chicago/Turabian StyleLuo, Xiangyuan, Xiaoyu Ji, Meng Xie, Tongyue Zhang, Yijun Wang, Mengyu Sun, Wenjie Huang, and Limin Xia. 2022. "Advance of SOX Transcription Factors in Hepatocellular Carcinoma: From Role, Tumor Immune Relevance to Targeted Therapy" Cancers 14, no. 5: 1165. https://doi.org/10.3390/cancers14051165
APA StyleLuo, X., Ji, X., Xie, M., Zhang, T., Wang, Y., Sun, M., Huang, W., & Xia, L. (2022). Advance of SOX Transcription Factors in Hepatocellular Carcinoma: From Role, Tumor Immune Relevance to Targeted Therapy. Cancers, 14(5), 1165. https://doi.org/10.3390/cancers14051165






