Simple Summary
Cell developmental programs used in wound healing and development such as the epithelial-to-mesenchymal transition (EMT) are frequently coopted by solid tumors to increase motility, plasticity, and invasive characteristics which promote metastasis. Identifying and quantifying the presence and extent of these programs can help to aid in patient prognosis and dictate therapeutic decision making. Here, we review the methods and findings to detect and quantify these cellular transitions in both laboratory and clinical settings.
Abstract
The epithelial-to-mesenchymal transition (EMT) and its reversal, the mesenchymal-to-epithelial transition (MET) are critical components of the metastatic cascade in breast cancer and many other solid tumor types. Recent work has uncovered the presence of a variety of states encompassed within the EMT spectrum, each of which may play unique roles or work collectively to impact tumor progression. However, defining EMT status is not routinely carried out to determine patient prognosis or dictate therapeutic decision-making in the clinic. Identifying and quantifying the presence of various EMT states within a tumor is a critical first step to scoring patient tumors to aid in determining prognosis. Here, we review the major strides taken towards translating our understanding of EMT biology from bench to bedside. We review previously used approaches including basic immunofluorescence staining, flow cytometry, single-cell sequencing, and multiplexed tumor mapping. Future studies will benefit from the consideration of multiple methods and combinations of markers in designing a diagnostic tool for detecting and measuring EMT in patient tumors.
1. Introduction
During the progression of many solid tumors, cells at the primary tumor site undergo phenotypic changes in response to extracellular stimuli [1,2], one among these being an epithelial-to-mesenchymal transition (EMT). This embryonic developmental program increases invasive and migratory behavior that is advantageous to a metastasizing cancer cell [3], enabling them to disseminate to distant organs. Plasticity within this transition, including its reversal (mesenchymal-to-epithelial transition; MET) to regain epithelial and proliferative characteristics, has been demonstrated in metastatic colonization [4,5]. Indeed, patients whose tumors express high levels of EMT signatures have worse overall prognoses and increased rates of metastasis [6,7]. Previous works questioned the relevance of EMT in metastasis in breast [8] and pancreatic [9] cancers; however, these conclusions were based in either incomplete disruption of the intermediate EMT [10] or in lineage markers related to a highly mesenchymal state [11].
Critical to our understanding of epithelial–mesenchymal plasticity (EMP) and its underlying regulators is our ability to distinguish unique EMT states from one another for identification in vitro, in vivo, and in patient samples. EMP and heterogeneity have frequently been associated with poor patient outcomes [6,7]; however, no robust method for assessing either of these has been developed to complement histopathological assessment in the clinic setting. The presence and role of a variety of hybrid EMT states in disease progression and metastasis remains a lynchpin in EMT-based therapies. Current evidence suggests that rather than relying solely on an MET to revert from a fully mesenchymal state, metastasis may result from the high plasticity and adaptability of the intermediate or hybrid states [12,13], as observed by the presence of intermediate circulating tumor cell clusters (CTCs clusters) [14,15]. Regardless of a clear-cut mechanism, which is still currently in debate, the presence of an intermediate state appears to be critical to the formation of metastasis, either through plasticity or as a transitional state.
Here we review the various methods that have been utilized to identify the spectrum of E–M states within a sample, from flow cytometry to single-cell analysis of the intricate RNA and protein expression patterns found in mouse and human tumors. Ultimately, one or a combination of these methods could be applied to assess patient prognosis by providing rapid and comprehensive analysis of the EMT state and heterogeneity of tumors to inform disease aggression and treatment regimens.
2. Epithelial and Mesenchymal Markers
Several markers have been used over the years, firstly to determine the occurrence of EMT and, more recently, to distinguish various distinct states along the epithelial-to-mesenchymal spectrum. These markers are based on a range of properties, from those that inform stemness, to those that indicate morphological changes, and transcriptional regulators (EMT and MET TFs) of the transition. These markers have been used in various combinations, each with their own benefits and deficits, based on context, specificity, and ease of use (Table 1).
2.1. Morphological Markers of EMT
The first discovered and readily utilized markers for EMT relate directly to the morphological changes that cells undergo to enhance invasion and motility, such as loss of classical epithelial adherens tight junction proteins and gain of non-canonical alternative intermediate filaments. E-cadherin, a key component of adherens junctions, was first identified as lost in epithelial cells that gained invasive characteristics [16]. Other adherens and tight junctional components that are key indicators of the epithelial state include claudins, occludins, and catenins as well as desmosomal components, such as desmoglein and desmocollin [17]. On the other hand, markers, such as Vimentin [18,19], fibronectin, N-cadherin, and smooth muscle actin (SMA) [20], have all been used to identify mesenchymal-like cells as invasion and/or progression markers in multiple cancer types. Co-expression of one or more epithelial markers along with mesenchymal markers, e.g., E-cadherin and Vimentin, is frequently used to identify intermediate or hybrid EMT states [21]. Morphological markers serve as good tools for defining the EMT state because they reflect the morphology of the cells themselves. However, they are frequently expressed at varying degrees across the EMT spectrum and therefore make poor singular identifiers for any individual state.
2.2. Cell Surface Markers
While epithelial and mesenchymal markers, such as E-cadherin, vimentin, and fibronectin, serve to describe the internal cellular processes of EMT, they can be difficult to identify without permeabilization of the cell membrane, given their predominant intracellular localization. Consequently, cell surface markers and receptors have been adopted to identify and isolate E–M states while maintaining cell viability. EMT states were first stratified by CD44 and CD24 [22], and later by CD104 (ITGB4) [23], to identify tumorigenic populations of cells, linking the invasive and disease progressing nature of EMT to stem-like processes of cancer stem cells, particularly in breast cancer [24,25]. EpCAM, an epithelial cell adhesion molecule similar to E-cadherin, has been used in many ways to identify cells exhibiting an epithelial state, particularly CTCs [26], and as a marker for flow cytometry [27]. Notably, Pastushenko et al. [28] profiled a panel of cell surface markers to describe the transitions across an EMT, identifying a gating strategy using Epcam, CD106, CD51, and CD61 that accurately captured cells in an intermediate state. These will be discussed in more detail below. Cell surface markers are easy to access and utilize for cell sorting, but ultimately have proven inconsistent across tumor types and models for accurately defining EMT states.
2.3. Transcription Factors
In addition to morphological characteristics, gene regulators of EMT or MET, such as transcription factors, provide a finer metric for measuring the progression of a cell across epithelial and mesenchymal transitions, and have been reviewed extensively [29]. These markers provide specificity, particularly when paired with morphological features. Master EMT regulator TFs, such as Snail [30], Twist1/2 [31], and ZEB1 [32,33], were originally identified as repressors of E-cadherin and regulators of plasticity and EMT. PRRX1 has also been implicated in later EMT staged in colorectal [34], thyroid [35], and gastric [36] cancers. Conversely, OVOL1/2 are required for the suppression of EMT and induction of MET in breast [37] and skin [38] epithelial and cancer [39]. These markers, as well as others detailed in other reviews [29] have been used extensively in transcriptomics-based approaches to rank EMT [40] as well as image-based methods, described later in this review. Transcription factors provide detailed information on EMT state, especially when analyzed as a network. However, some TFs have tissue specific functions that can convolute a global EMT signature generated in this way.

Table 1.
A summary of key EMT marker proteins and their use and relevance in determining EMT state.
Table 1.
A summary of key EMT marker proteins and their use and relevance in determining EMT state.
| Marker | Method(s) | Importance | Sample Type | Source(s) |
|---|---|---|---|---|
| E-cadherin | IF, transcriptional EMT scores | One of the first epithelial markers. Used for many analysis methods | Human tissue, human cell lines, mouse tissue, mouse cell lines | Behrens 1989 [16] |
| EpCAM | Flow cytometry, circulating tumor cells | Epithelial marker used in flow cytometry and CTC detection. Frequently lost early in EMT | Human tissue, human cell lines, mouse tissue, mouse cell lines | Riethdorf 2007 [26] Schnell 2013 [27] |
| Vimentin | Intracellular flow cytometry, IF, transcriptional EMT scores | Mesenchymal marker used in many analysis methods | Human tissue, human cell lines, mouse tissue, mouse cell lines | Sommers 1991 [18] Thompson 1992 [19] |
| CD44/CD24 | Flow cytometry | Stemness markers first used to separate epithelial and mesenchymal states | Human tissue, human cell lines | Al-Hajj 2003 [22] |
| CD106/CD51/CD61 | Flow cytometry | Used to segregate multiple EMT states | Mouse tissue, mouse cell lines | Pastushenko 2018 [28] |
| CD104 (ITGβ4) | Flow cytometry | An improved marker to replace CD24 | Human tissue, human cell lines | Bierie 2017 [23] |
| Snail | IF, transcriptional methods, multiplexed image analysis | Transcriptional repressor of E-cadherin, responds to TGFβ signaling | Human tissue, human cell lines, mouse tissue, mouse cell lines | Cano 2000 [30] van Staalduinen 2018 [29] |
| Twist | IF, transcriptional methods | Hallmark EMT transcription factor | Human tissue, human cell lines, mouse tissue, mouse cell lines | Yang 2004 [31] van Staalduinen 2018 [29] |
| ZEB1 | IF, transcriptional methods, multiplexed image analysis | Hallmark EMT-driving transcription factor, repressor of E-cadherin | Human tissue, human cell lines, mouse tissue, mouse cell lines | Guaita 2002 [32] Eger 2005 [33] van Staalduinen 2018 [29] |
| PRRX1 | IF, transcriptional methods | EMT transcription factor prevalent in late EMT | Human tissue, human cell lines, mouse tissue, mouse cell lines | Takahashi 2013 [34] Hardin 2014 [35] Guo 2015 [36] |
| OVOL1/2 | Transcriptional methods | MET transcription factor responsible for maintaining and epithelial state | Human tissue, human cell lines, mouse tissue, mouse cell lines | Roca 2013 [39] Watanabe 2014 [37] Li 2014 [38] van Staalduinen 2018 [29] |
While several other cytoskeletal proteins, such as FSP1 [41] and α SMA [42], secreted proteins, including fibronectin [43] and MMPs [44], and epithelial junctional proteins, such as claudins and occludins [45], have been employed as EMT markers in different contexts, these have not specifically been used to identify intermediate/hybrid EMT states and could possibly highlight cells that reside in more extreme epithelial or mesenchymal states.
3. Model Systems Used to Study EMP
3.1. Cell Lines
Immortalized or cancer derived cell lines have been used for decades as models to understand cancer at a basic level. They are easy to work with, highly manipulable, and can provide a basis for testing novel drugs and therapies. Cell lines have been used to perfect many of the methods detailed below, including flow cytometry, immunofluorescence, and RNA-sequencing. Databases, such as the cancer cell line encyclopedia (CCLE) and ATCC, serve as repositories for data and frozen stocks of cell lines for research use. However, cell lines alone cannot recapitulate the complexities of an in vivo system, which can be achieved through orthotopic transplantation in mice, rats, and other model organisms. This model, therefore, serves as a necessary but simple steppingstone to understanding E–M heterogeneity and plasticity in patients.
3.2. Genetically Engineered Models
In efforts to recapitulate human tumor progression for laboratory study, many different non-human models have been generated that mimic aspects of patient disease to study the roles of EMT and MET in tumor development, progression, and metastasis. Most popular are genetically engineered mouse models (GEMMs), although zebrafish [46,47], drosophila [48,49], and sea urchin [50,51] models have been elegantly used to generate important insights in the field. These GEMMs provide an excellent framework for studying the metastatic cascade, allowing for spontaneous tumorigenesis in a specific tissue of choice, collection of organs and circulating tumor cells for basic research and diagnostic development, and testing novel drug targets in a complex living system. GEMMs have been used with great success to isolate and study EMP in vivo in breast [28], skin [28], pancreatic [52], and prostate [53] cancer. Additionally, immune compromised mouse models can host human-derived cell lines or patient-derived xenografts, which, when transplanted orthotopically into the tissue of origin, can recapitulate the nuanced disease of that individual for further study. Overall, mouse and other models of human cancer have been crucial in expanding on in vivo dynamics of the metastatic cascade where cell lines have fallen short.
3.3. Primary Human Tissue
Ultimately, the best tool for studying human disease is directly on human patient samples. This can be tumor or tissue biopsies either taken fresh or stored in a formalin fixative, as well as circulating tumor cells, cytological specimens, and secondary site biopsies, etc. However, acquiring patient samples and full patient data can be challenging and take years. Databases, such as The Cancer Genome Atlas (TCGA), contain complete sequenced genomes for thousands of primary patient samples and can be a helpful bioinformatic tool to begin transitioning from basic to translational research, such as validating cancer predictor genes or looking for large trends across many samples. EMP has been successfully identified, validated, and explored in patient cancers, including in circulating tumor cells [7,54,55], determining EMT gene signatures in primary tissue [56,57], and mapping EMT states at single-cell resolution [58,59], both substantiating its relevance in patient disease and opening new possibilities for diagnostic approaches.
3.4. Circulating Tumor Cells
Circulating tumor cells in the blood have served as a “window to cancer” for many years [60,61]. As a diagnostic tool, it is easy to implement on patients, requiring only a small blood sample, and can be used to screen tumor cells in a multitude of ways [62], including immunofluorescence [54], RNA in situ hybridization (ISH) [15], and RNA-sequencing [14]. CTC collection methods have also helped to validate the significance of an EMT in the metastatic cascade [15,54], as well as a possible requirement for the reversal, or MET, to colonize metastatic organs [63].
Traditionally, circulating tumor cells were harvested using the cell surface marker EpCAM [26], as most cancers of interest were epithelial in origin. However, EpCAM is lost during an EMT [64], leading to a misrepresentation of CTCs collected by this method. Indeed, even when captured with an EpCAM retrieval method, CTCs in breast and prostate cancer patients were found to co-express epithelial and mesenchymal markers in progressive disease [6], bringing up the question of how many EpCAM-negative mesenchymal cells were missed in the analysis. In response, other microfluidics-based methods of CTC capture have been adopted [65,66], although EpCAM-based methods still dominate patient diagnostics [67]. Non-specific capture methods have identified relatively equal populations of epithelial, intermediate, and mesenchymal CTCs, defined by EMT markers, such as E-cadherin and vimentin; however, multiple studies have found a correlation between high presence of mesenchymal CTCs and worsened patient prognosis [7,63]. Along with CTCs, microfluidics devices have also identified circulating tumor cell clusters (CTC clusters) which, although rare, are much more potent metastatic seeders than individual CTCs alone [14]. Under unbiased collection, identification and classification of CTCs and CTC clusters by microfluidics is a powerful diagnostic tool that can be combined with a multitude of other methods to understand EMP and its role in the metastatic cascade.
4. Methods
Flow Cytometry
Flow cytometry techniques can easily and readily detect cell populations expressing a combination of cell surface markers. Further, live cell populations can be sorted based on marker expression via fluorescence assisted cell sorting (FACS) for further study. This has made flow cytometry a very popular and easily applicable resource in many early studies defining E–M states. The invasive nature of cells undergoing an EMT elicited a natural link to cancer stem-cell like states, prompting these tumorigenic populations of cells to be initially isolated and described as CD44hi/CD24lo [22], ALDH+ [68] stem-like populations, and later linked to the EMT process [24,25]. Flow cytometry has provided a means of differentiating epithelial (EpCAM+/CD24hi/CD44-) and mesenchymal (EpCAM-/CD44hi/CD24-) in multiple cancer cell lines [69,70] as well as the breast [71] and prostate [53] mouse model to delineate differences between these states, understand the unique mechanisms that control EMT and MET, and determine their various roles in disease progression and the metastatic cascade. Further endeavors to increase the flow sorting sensitivity of E–M states has led to the discovery of novel EMT cell surface markers, such as CD104 (ITGB4) [23] as a supplement in addition to CD44/CD24 to define intermediate EMT states with cancer stem cells properties within human tumors, as well as combinations of EpCAM, CD51, CD61, and CD106 [28] to isolate multiple transitional intermediate/hybrid EMT states from mouse tumor models (Figure 1).
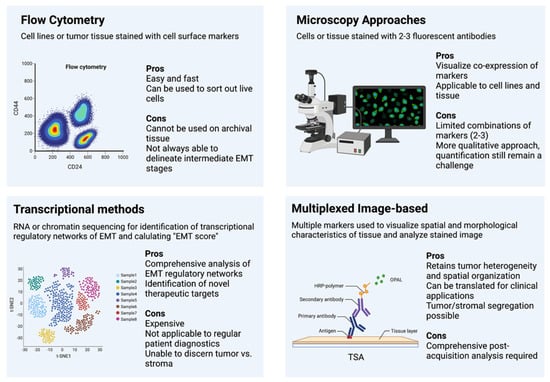
Figure 1.
Methods of Assessing EMT.
These works and the application of flow cytometry established a link between the hybrid or intermediate state and increased stemness and decreased patient prognosis using several cell surface markers; however, this technique is unable to consider the expression of intracellular markers, such as vimentin or ZEB1, which require cell permeabilization. In addition, this technique can only be used in live tissue, making it more challenging to study patient samples, which are often archival formalin-fixed paraffin embedded (FFPE) tissue. Additionally, evidence from our recent study on EMT states suggests that canonical cell surface markers (CD44, CD104, and EpCAM) are not sufficient to separate distinct intermediate states from one another [59]. Indeed, Pastushenko et al. [28] relied upon the co-staining of four markers and precise gating strategies to adequately separate these states. Thus, flow cytometry presents an excellent approach for basic biological analysis of E–M plasticity but has few applications for direct use on archival patient tissues.
5. Immunohistochemistry and Fluorescence
Immunohistochemistry is the most common form of immunostaining and has been used for decades to detect and label antigens in tissue sections [72]. Hematoxylin and eosin (H&E) staining for DNA and proteins, respectively, is the principal method for histological assessment of tumor grade and histological subtype. In addition to assessing tumor grade and histologic subtype, IHC staining for other biomarkers is routinely performed by pathologists for certain tumors, such as hormone receptors, HER2, and Ki67 in breast cancer, to provide prognostic and predicative information, and to stratify tumors into molecular intrinsic subtypes [73,74].
Immunofluorescence (IF) staining for EMT markers, particularly E-cadherin and Vimentin, has frequently been used alongside other methods for visualizing the co-expression of epithelial and mesenchymal markers as well as discerning the sub-cellular localization of proteins. However, it is rarely used as a comprehensive method for defining EMT states, owing, in part, to the limitation of fluorescence wavelengths that only allow visualization of a limited number of markers simultaneously. Efforts to combine changes in cell morphology with E-cadherin/Vimentin IF staining in a predictive EMT model have been partially successful in cell lines [75]; however, the application of this predictive model in vivo remains unclear. An immunofluorescence microscopy assay for cytoskeletal remodeling elements [76] has been successfully implemented as a readout for EMT disruption to screen a panel of transcription factor-targeting siRNAs to determine transcriptional nodes that control EMT [77], which can be useful for easy drug treatment and screening for future testable therapeutics targeting EMT. Moreover, recent work has combined fluorescent lineage tracing with intravital live microscopy to visualize and trace early and late EMT states in the primary tumor and metastatic sites, providing a much needed look inside the dynamic processes of tumor progression and EMT [13]. Combined immunofluorescence with other techniques, such as cell morphology or single-cell segmentation, has distinct advantages over bulk flow or sequencing by maintaining tumor architecture and spatial organization in the tissue. However, the limiting number of probes in classical IF presents the same drawbacks as flow cytometry and may be insufficient to describe the complexity of E–M states (Figure 1).
6. Transcription-Based Methods
RNA-based, and later chromatin-based, methods of assessing the EMT state have two main goals: to generate an EMT gene signature, or to characterize EMT gene networks across a spectrum of samples. This can be done in a variety of ways, although the goal is generally to further basic knowledge rather than apply directly to patients.
Bulk RNA-seq has been repeatedly used to generate EMT gene signatures or “EMT scores” to help standardize and define entrance into an EMT [56] or partial EMT states in many cancers [57] and correlate that gene signature with poor patient prognosis. This is useful in understanding the connection between EMT gene signatures and patient prognosis as well as defining EMT states for new model systems [59]. However, many groups have put forth their own signature or method for ranking EMT [40,56,57,78], calling into question a standardized signature for ubiquitous use. In a more exploratory approach, RNA-seq has been used to interrogate EMP using either isolated clonal states within an EMT [28,59], or an EMT induction and withdrawal (MET) time course. These experiments served to delineate the specific gene networks that are active during the transition from epithelial to mesenchymal and back [77,79] and to help distinguish targetable transcriptional networks in aggressive or metastatic cell states. Combinations of EMT induction and siRNA knockdown of EMT target transcription factors identified control nodes, such as TEAD2, FOSL2, SP1, and others that had not been previously associated with EMT [77].
Other approaches to visualize RNA expression, such as fluorescence in situ hybridization (FISH) probing for a panel of EMT markers, have had success in assessing EMP at a single-cell level in circulating tumor cells [15] before the more widely accepted single-cell sequencing approaches were robust enough to be used in this context.
Single-cell RNA sequencing has been particularly vital in assessing EMT in tumors or cells where the bulk RNA signature may not be sufficient to describe the heterogeneous populations within each sample. This can be applied to CTCs [55], tumor cells [80], or as part of induced time course [79,81] to delineate EMP in these samples as well as interrogate EMP and heterogeneity at multiple points during metastasis, including the primary tumor, CTCs, and metastatic sites, to see how EMT states may work individually or cooperatively to promote metastasis.
A leap in the field came from non-specific sequencing of accessible chromatin (ATAC-seq), which revealed large scale chromatin modification in response to progression through an EMT [28,82,83], indicating that large transcriptional shifts may be controlled through a combination of epigenetic and transcriptional regulation. Recently, multiple efforts have taken a multi-omics approach, combining RNA-seq and ATAC-seq to determine these combined epigenetic and transcriptional regulatory proteins, such as CTCF, the AP-1 complex, and the RUNX transcription factor family [59,84,85].
While transcription-based approaches have provided a wealth of data and greatly contribute to the understanding of EMT, MET, and the metastatic cascade, the cost of sequencing, the processing times, and the inability to segregate tumor from stroma still make it inapplicable to assess patient samples for routine diagnosis. However, these comprehensive analyses have pinpointed specific EMT indicators for further and more directed approaches, such as multiplexed staining (Figure 1).
7. Multiplexed Image-Based Methods
Akin to immunohistochemistry, image-based methods of assessing tumors have distinct advantages for patient diagnosis. They are relatively easy, can be done with high throughput, and most importantly, retain spatial organization and heterogeneity of the original tumors. However, immunohistochemistry or even immunofluorescence struggles to describe the complexity of patient tumor states that may affect disease progression and metastasis, particularly epithelial to mesenchymal plasticity. Previous efforts have combined immunohisto-fluoresence for E-cadherin and Vimentin with high content screening (HCS), introducing a method that combines cell segmentation, morphological evaluation, and marker expression to determine nuanced EMT states within a tumor sample at a single-cell level [75]. This has been implemented with various other probes and image analysis software to combine immunofluorescence and morphological features into a reliable patient diagnostic tool [86,87]. However, this method is limited by the number of markers that can be used. Others, in efforts to combat this issue, have relied on mass cytometry time courses to map changes and co-expression of E-cadherin, Vimentin, CD44, CD24, and others in individual cells undergoing a TGF-β-induced EMT in lung cancer [58]. Similar methods have been used for multiplexed identification and stratification of heterogeneity in breast cancer patient samples with 35 different markers [88]. Even newer technologies, such as Nanostring DSP, present exciting new platforms for high-plex spatial imaging of RNA and/or proteins. This platform is becoming increasingly useful to deconvolute tumor heterogeneity and the tumor microenvironment (TME) of specific tumor types [89]. These approaches rely on precise image analysis software that has only recently become sensitive enough to reliably segment individual cells and deconvolute multiplexed staining approaches. Considering these technological advances, image-based approaches to quantify EMT progression in tumors are becoming more popular and easily implementable; these technologies have been reviewed extensively elsewhere [90]. Recently, we have employed a multiplexed, multi-round tyramide signal amplification (TSA) staining method using six canonical EMT markers that was used with cell segmentation and morphological features to define an EMT heterogeneity score and overall tumor EMT score in a model system of EMT, and further validated in a cohort of breast cancer patient samples [59]. Notably, this method was the first to reliably segment out stromal tissue, such as fibroblasts, which can surround tumors and frequently express Vimentin and ZEB1, mesenchymal markers that would skew an analysis of tumor composition. When implemented with widely practiced immunohistochemical approaches to determining tumor grade and composition, this method can help to elucidate the complexity of patient tumor heterogeneity and EMT state in the clinic to better inform prognosis and treatment regimes (Figure 1).
8. Conclusions
Endeavors to characterize, quantify, and stratify epithelial–mesenchymal cell states in research models and patient samples has spanned decades. With each technological leap, the field gains more knowledge and insight into the markers and methods that can best and most simply stratify these phenotypic cell states. While no method is obsolete, some, such as immunohistochemistry, have made way for more complex and descriptive methods, such as multiplexed immunofluorescence. Ultimately, when patient diagnosis is the goal, approaches should be tailored for these specific needs, such as ease of use, number of samples at a time, cost, and accurate resolution of tumor tissue and individual cells. Flow cytometry and immunohistochemistry are simple and easy to implement, but lack the complexity and standardization required to reliably identify and score EMT states in patient samples from many different tumors. Transcriptional and chromatin-based methods provide this complexity and have pulled back the veil on the intricate transcriptional and chromatin regulatory networks that controlled epithelial–mesenchymal plasticity. However, they are difficult to implement on fresh samples and are quite cost prohibitive. For this reason, they remain a strong tool for use on test cohorts and in vitro or in vivo models of EMT but are unlikely to be adopted for routine diagnostics. Circulating tumor cells have been excellent diagnostic tools in patients for many years as they are easy to sample and provide a heterogeneous window into the tumor itself. CTCs also provide a background for testing many prognostic tools and have been tested out with many methods, past, present, and future. Image-based approaches have built upon what the field has learned about most descriptive and succinct E–M markers, as well as tissue and single-cell segmentation to create a robust tool to apply across many patient samples and in many contexts. In the future, such a method can be used to complement histopathological assessment in a clinical setting to provide a rapid and comprehensive analysis of E–M heterogeneity and the EMT tumor score to predict disease progression and inform treatment regimens.
Author Contributions
Conceptualization, M.S.B. and D.R.P.; writing—original draft preparation, M.S.B.; writing—review and editing, M.S.B., K.E.M. and D.R.P.; visualization, M.S.B.; supervision, D.R.P.; project administration, D.R.P.; funding acquisition, D.R.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (under grant number P20GM104416), a Prouty Pilot Grant from Friends of the Norris Cotton Cancer Center, funding from The Elmer R. Pfefferkorn & Allan U. Munck Education and Research Fund at the Geisel School of Medicine at Dartmouth, an NCI Cancer Center Support Grant (5P30CA023108-40) and funding from the NIH 5R00CA201574-05 (to D.R.P.) The APC was funded by discretionary funds from the Norris Cotton Cancer Center.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hay, E.D. An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs 1995, 154, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Donaher, J.L.; Murphy, D.A.; Chau, S.; Yang, J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 2012, 22, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef]
- Markiewicz, A.; Topa, J.; Nagel, A.; Skokowski, J.; Seroczynska, B.; Stokowy, T.; Welnicka-Jaskiewicz, M.; Zaczek, A.; Markiewicz, A.; Topa, J.; et al. Spectrum of Epithelial-Mesenchymal Transition Phenotypes in Circulating Tumour Cells from Early Breast Cancer Patients. Cancers 2019, 11, 59. [Google Scholar] [CrossRef]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Ye, X.; Brabletz, T.; Kang, Y.; Longmore, G.D.; Nieto, M.A.; Stanger, B.Z.; Yang, J.; Weinberg, R.A. Upholding a role for EMT in breast cancer metastasis. Nature 2017, 547, E1–E3. [Google Scholar] [CrossRef]
- Aiello, N.M.; Brabletz, T.; Kang, Y.; Nieto, M.A.; Weinberg, R.A.; Stanger, B.Z. Upholding a role for EMT in pancreatic cancer metastasis. Nature 2017, 547, E7–E8. [Google Scholar] [CrossRef] [PubMed]
- Ognjenovic, N.B.; Bagheri, M.; Mohamed, G.A.; Xu, K.; Chen, Y.; Mohamed Saleem, M.A.; Brown, M.S.; Nagaraj, S.H.; Muller, K.E.; Gerber, S.A.; et al. Limiting Self-Renewal of the Basal Compartment by PKA Activation Induces Differentiation and Alters the Evolution of Mammary Tumors. Dev. Cell 2020, 55, 544–557.e6. [Google Scholar] [CrossRef] [PubMed]
- Lüönd, F.; Sugiyama, N.; Bill, R.; Tiede, S.; Van Rheenen, J.; Correspondence, G.C. Distinct contributions of partial and full EMT to breast cancer malignancy. Dev. Cell 2021, 56, 3203–3221.e11. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Behrens, J.; Mareel, M.M.; Van Roy, F.M.; Birchmeier, W. Dissecting Tumor Cell Invasion: Epithelial Cells Acquire Invasive Properties after the Loss of Uvomorulin-mediated Cell-Cell Adhesion. J. Cell Biol. 1989, 108, 2435–2447. [Google Scholar] [CrossRef]
- Hendrix, M.J.C.; Seftor, E.A.; Seftor, R.E.B.; Trevor, K.T. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J. Pathol. 1997, 150, 483–495. [Google Scholar]
- Sommers, C.L.; Thompson, E.W.; Torri, J.A.; Kemler, R.; Gelmann, E.P.; Byers, S.W. Cell adhesion molecule uvomorulin expression in human breast cancer cell lines: Relationship to morphology and invasive capacities. Cell Growth Differ. 1991, 2, 365–372. [Google Scholar]
- Thompson, E.W.; Paik, S.; Brünner, N.; Sommers, C.L.; Zugmaier, G.; Clarke, R.; Shima, T.B.; Torri, J.; Donahue, S.; Lippman, M.E.; et al. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J. Cell. Physiol. 1992, 150, 534–544. [Google Scholar] [CrossRef]
- Boyer, B.; Thiery, J.P. Epithelium-mesenchyme interconversion as example of epithelial plasticity. Apmis 1993, 101, 257–268. [Google Scholar] [CrossRef]
- Park, S.-M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Pierce, S.E.; Kroeger, C.; Stover, D.G.; Pattabiraman, D.R.; Thiru, P.; Liu Donaher, J.; Reinhardt, F.; Chaffer, C.L.; Keckesova, Z.; et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2337–E2346. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Morel, A.-P.; Lièvre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of Breast Cancer Stem Cells through Epithelial-Mesenchymal Transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the CellSearch System. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef]
- Schnell, U.; Cirulli, V.; Giepmans, B.N.G. EpCAM: Structure and function in health and disease. Biochim. Biophys. Acta Biomembr. 2013, 1828, 1989–2001. [Google Scholar] [CrossRef]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- van Staalduinen, J.; Baker, D.; ten Dijke, P.; van Dam, H. Epithelial–mesenchymal-transition-inducing transcription factors: New targets for tackling chemoresistance in cancer? Oncogene 2018, 37, 6195–6211. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef]
- Guaita, S.; Puig, I.; Franci, C.; Garrido, M.; Dominguez, D.; Batlle, E.; Sancho, E.; Dedhar, S.; De Herreros, A.G.; Baulida, J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 2002, 277, 39209–39216. [Google Scholar] [CrossRef] [PubMed]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Sawada, G.; Kurashige, J.; Uchi, R.; Matsumura, T.; Ueo, H.; Takano, Y.; Akiyoshi, S.; Eguchi, H.; Sudo, T.; et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br. J. Cancer 2013, 109, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Hardin, H.; Guo, Z.; Shan, W.; Montemayor-Garcia, C.; Asioli, S.; Yu, X.-M.; Harrison, A.D.; Chen, H.; Lloyd, R.V. The roles of the epithelial-mesenchymal transition marker PRRX1 and miR-146b-5p in papillary thyroid carcinoma progression. Am. J. Pathol. 2014, 184, 2342–2354. [Google Scholar] [CrossRef]
- Guo, J.; Fu, Z.; Wei, J.; Lu, W.; Feng, J.; Zhang, S. PRRX1 promotes epithelial–mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer. Med. Oncol. 2015, 32, 393. [Google Scholar] [CrossRef]
- Watanabe, K.; Villarreal-Ponce, A.; Sun, P.; Salmans, M.L.; Fallahi, M.; Andersen, B.; Dai, X. Mammary Morphogenesis and Regeneration Require the Inhibition of EMT at Terminal End Buds by Ovol2 Transcriptional Repressor. Dev. Cell 2014, 29, 59–74. [Google Scholar] [CrossRef]
- Li, S.; Yang, J. Ovol Proteins: Guardians against EMT during Epithelial Differentiation. Dev. Cell 2014, 29, 1–2. [Google Scholar] [CrossRef][Green Version]
- Roca, H.; Hernandez, J.; Weidner, S.; McEachin, R.C.; Fuller, D.; Sud, S.; Schumann, T.; Wilkinson, J.E.; Zaslavsky, A.; Li, H.; et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE 2013, 8, e76773. [Google Scholar] [CrossRef]
- Chakraborty, P.; George, J.T.; Tripathi, S.; Levine, H.; Jolly, M.K. Comparative Study of Transcriptomics-Based Scoring Metrics for the Epithelial-Hybrid-Mesenchymal Spectrum. Front. Bioeng. Biotechnol. 2020, 8, 220. [Google Scholar] [CrossRef]
- Xue, C.; Plieth, D.; Venkov, C.; Xu, C.; Neilson, E.G. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003, 63, 3386–3394. [Google Scholar] [PubMed]
- Gabbiani, G.; Kapanci, Y.; Barazzone, P.; Franke, W.W. Immunochemical identification of intermediate-sized filaments in human neoplastic cells. A diagnostic aid for the surgical pathologist. Am. J. Pathol. 1981, 104, 206. [Google Scholar] [PubMed]
- Yang, Z.; Zhang, X.; Gang, H.; Li, X.; Li, Z.; Wang, T.; Han, J.; Luo, T.; Wen, F.; Wu, X. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem. Biophys. Res. Commun. 2007, 358, 925–930. [Google Scholar] [CrossRef]
- Gilles, C.; Newgreen, D.F.; Sato, H.; Thompson, E.W. Matrix Metalloproteases and Epithelial-to-Mesenchymal Transition: Implications for Carcinoma Metastasis. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Ikenouchi, J.; Matsuda, M.; Furuse, M.; Tsukita, S. Regulation of tight junctions during the epithelium-mesenchyme transition:direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 2003, 116, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Amack, J.D. Cellular dynamics of EMT: Lessons from live in vivo imaging of embryonic development. Cell Commun. Signal. 2021, 19, 79. [Google Scholar] [CrossRef]
- Chen, L.; Boleslaw Olszewski, M.; Kruithof-de Julio, M.; Snaar-Jagalska, B.E. Zebrafish Microenvironment Elevates EMT and CSC-Like Phenotype of Engrafted Prostate Cancer Cells. Cells 2020, 9, 797. [Google Scholar] [CrossRef]
- Campbell, K.; Lebreton, G.; Franch-Marro, X.; Casanova, J. Differential roles of the Drosophila EMT-inducing transcription factors Snail and Serpent in driving primary tumour growth. PLOS Genet. 2018, 14, e1007167. [Google Scholar] [CrossRef]
- Campbell, K.; Rossi, F.; Adams, J.; Pitsidianaki, I.; Barriga, F.M.; Garcia-Gerique, L.; Batlle, E.; Casanova, J.; Casali, A. Collective cell migration and metastases induced by an epithelial-to-mesenchymal transition in Drosophila intestinal tumors. Nat. Commun. 2019, 10, 2311. [Google Scholar] [CrossRef]
- Romancino, D.P.; Anello, L.; Lavanco, A.; Buffa, V.; Di Bernardo, M.; Bongiovanni, A. A sea urchin in vivo model to evaluate Epithelial-Mesenchymal Transition. Dev. Growth Differ. 2017, 59, 141–151. [Google Scholar] [CrossRef]
- Massri, A.J.; Schiebinger, G.R.; Berrio, A.; Wang, L.; Wray, G.A.; McClay, D.R. Methodologies for Following EMT In Vivo at Single Cell Resolution. Methods Mol. Biol. 2021, 2179, 303–314. [Google Scholar] [CrossRef]
- Krebs, A.M.; Mitschke, J.; Losada, M.L.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Ruscetti, M.; Quach, B.; Dadashian, E.L.; Mulholland, D.J.; Wu, H. Tracking and Functional Characterization of Epithelial-Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Res. 2015, 75, 2749–2759. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-M.; Xu, S.-C.; Li, J.; Han, K.-Q.; Pi, H.-F.; Zheng, L.; Zuo, G.-H.; Huang, X.-B.; Li, H.-Y.; Zhao, H.-Z.; et al. Epithelial–mesenchymal transition markers expressed in circulating tumor cells in hepatocellular carcinoma patients with different stages of disease. Cell Death Dis. 2013, 4, e831. [Google Scholar] [CrossRef]
- Lapin, M.; Tjensvoll, K.; Oltedal, S.; Javle, M.; Smaaland, R.; Gilje, B.; Nordgård, O. Single-cell mRNA profiling reveals transcriptional heterogeneity among pancreatic circulating tumour cells. BMC Cancer 2017, 17, 390. [Google Scholar] [CrossRef]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.-J.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- George, J.T.; Jolly, M.K.; Xu, S.; Somarelli, J.A.; Levine, H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017, 77, 6415–6428. [Google Scholar] [CrossRef] [PubMed]
- Karacosta, L.G.; Anchang, B.; Ignatiadis, N.; Kimmey, S.C.; Benson, J.A.; Shrager, J.B.; Tibshirani, R.; Bendall, S.C.; Plevritis, S.K. Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat. Commun. 2019, 10, 5587. [Google Scholar] [CrossRef]
- Brown, M.S.; Abdollahi, B.; Wilkins, O.M.; Chakraborty, P.; Ognjenovic, N.B.; Muller, K.E.; Kumar Jolly, M.; Hassanpour, S.; Pattabiraman, D.R. Dynamic plasticity within the EMT spectrum, rather than static mesenchymal traits, drives tumor heterogeneity and metastatic progression of breast cancers. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- Fabisiewicz, A.; Grzybowska, E. CTC clusters in cancer progression and metastasis. Med. Oncol. 2017, 34, 12. [Google Scholar] [CrossRef] [PubMed]
- Revenco, T.; Nicodème, A.; Pastushenko, I.; Sznurkowska, M.K.; Latil, M.; Sotiropoulou, P.A.; Dubois, C.; Moers, V.; Lemaire, S.; de Maertelaer, V.; et al. Context Dependency of Epithelial-to-Mesenchymal Transition for Metastasis. Cell Rep. 2019, 29, 1458–1468.e3. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, S.; Liu, Z.; Huang, J.; Pu, X.; Li, J.; Yang, D.; Deng, H.; Yang, N.; Xu, J. Classification of Circulating Tumor Cells by Epithelial-Mesenchymal Transition Markers. PLoS ONE 2015, 10, e0123976. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid biopsy: From discovery to clinical application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Eslami, S.Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Hérouël, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.-A.; Huang, S. Stemness of the hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sakane, K.; Tominaga, K.; Gotoh, N.; Niwa, T.; Kikuchi, Y.; Tada, K.; Goshima, N.; Semba, K.; Inoue, J.I. Intratumoral bidirectional transitions between epithelial and mesenchymal cells in triple-negative breast cancer. Cancer Sci. 2017, 108, 1210–1222. [Google Scholar] [CrossRef]
- Bhatia, S.; Monkman, J.; Toh, A.K.L.; Nagaraj, S.H.; Thompson, E.W. Targeting epithelial–mesenchymal plasticity in cancer: Clinical and preclinical advances in therapy and monitoring. Biochem. J. 2017, 474, 3269–3306. [Google Scholar] [CrossRef]
- de Matos, L.L.; Trufelli, D.C.; de Matos, M.G.L.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark. Insights 2010, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Inwald, E.C.; Koller, M.; Klinkhammer-Schalke, M.; Zeman, F.; Hofstädter, F.; Gerstenhauer, M.; Brockhoff, G.; Ortmann, O. 4-IHC classification of breast cancer subtypes in a large cohort of a clinical cancer registry: Use in clinical routine for therapeutic decisions and its effect on survival. Breast Cancer Res. Treat. 2015, 153, 647. [Google Scholar] [CrossRef] [PubMed]
- Leggett, S.E.; Sim, J.Y.; Rubins, J.E.; Neronha, Z.J.; Williams, E.K.; Wong, I.Y. Morphological single cell profiling of the epithelial-mesenchymal transition. Integr. Biol. 2016, 8, 1133–1144. [Google Scholar] [CrossRef]
- Lotz-Jenne, C.; Lüthi, U.; Ackerknecht, S.; Lehembre, F.; Fink, T.; Stritt, M.; Wirth, M.; Pavan, S.; Bill, R.; Regenass, U.; et al. A high-content EMT screen identifies multiple receptor tyrosine kinase inhibitors with activity on TGFβ receptor. Oncotarget 2016, 7, 25983–26002. [Google Scholar] [CrossRef]
- Meyer-Schaller, N.; Cardner, M.; Diepenbruck, M.; Saxena, M.; Tiede, S.; Lüönd, F.; Ivanek, R.; Beerenwinkel, N.; Christofori, G. A Hierarchical Regulatory Landscape during the Multiple Stages of EMT. Dev. Cell 2019, 48, 539–553.e6. [Google Scholar] [CrossRef]
- Cook, D.P.; Vanderhyden, B.C. Transcriptional census of epithelial-mesenchymal plasticity in cancer. Sci. Adv. 2022, 8, 7640. [Google Scholar] [CrossRef]
- Cook, D.P.; Vanderhyden, B.C. Context specificity of the EMT transcriptional response. Nat. Commun. 2020, 11, 2142. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef]
- Deshmukh, A.P.; Vasaikar, S.V.; Tomczak, K.; Tripathi, S.; den Hollander, P.; Arslan, E.; Chakraborty, P.; Soundararajan, R.; Jolly, M.K.; Rai, K.; et al. Identification of EMT signaling cross-talk and gene regulatory networks by single-cell RNA sequencing. Proc. Natl. Acad. Sci. USA 2021, 118, e2102050118. [Google Scholar] [CrossRef]
- Latil, M.; Nassar, D.; Beck, B.; Boumahdi, S.; Wang, L.; Brisebarre, A.; Dubois, C.; Nkusi, E.; Lenglez, S.; Checinska, A.; et al. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. Cell Stem Cell 2017, 20, 191–204.e5. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Li, G.; Long, Y.; Liang, W.; Cui, H.; Zhang, B.; Tan, Y.; Li, Y.; Shen, L.; Deng, D.; et al. Integrative multi-omics analysis of a colon cancer cell line with heterogeneous Wnt activity revealed RUNX2 as an epigenetic regulator of EMT. Oncogene 2020, 39, 5152–5164. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.S.; Hussein, S.; Song, S.; Chakraborty, P.; Jolly, M.K.; Toneff, M.J.; Lin, Y.C.; Taube, J.H. Gene expression and chromatin accessibility during progressive EMT and MET linked to dynamic CTCF engagement. bioRxiv 2020. [Google Scholar] [CrossRef]
- Godin, L.; Balsat, C.; Van Eycke, Y.R.; Allard, J.; Royer, C.; Remmelink, M.; Pastushenko, I.; D’Haene, N.; Blanpain, C.; Salmon, I.; et al. A novel approach for quantifying cancer cells showing hybrid epithelial/mesenchymal states in large series of tissue samples: Towards a new prognostic marker. Cancers 2020, 12, 906. [Google Scholar] [CrossRef] [PubMed]
- Navas, T.; Kinders, R.J.; Lawrence, S.M.; Ferry-Galow, K.V.; Borgel, S.; Hollingshead, M.G.; Srivastava, A.K.; Alcoser, S.Y.; Makhlouf, H.R.; Chuaqui, R.; et al. Clinical evolution of epithelial–mesenchymal transition in human carcinomas. Cancer Res. 2020, 80, 304–318. [Google Scholar] [CrossRef]
- Jackson, H.W.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef]
- Bergholtz, H.; Carter, J.M.; Cesano, A.; Cheang, M.C.U.; Church, S.E.; Divakar, P.; Fuhrman, C.A.; Goel, S.; Gong, J.; Guerriero, J.L.; et al. Best practices for spatial profiling for breast cancer research with the GeoMx® digital spatial profiler. Cancers 2021, 13, 4456. [Google Scholar] [CrossRef]
- Lewis, S.M.; Asselin-Labat, M.-L.; Nguyen, Q.; Berthelet, J.; Tan, X.; Wimmer, V.C.; Merino, D.; Rogers, K.L.; Naik, S.H. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 2021, 18, 997–1012. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).