Circular RNAs and Drug Resistance in Genitourinary Cancers: A Literature Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. CircRNAs and Drug Resistance in RCC
2.1. Gemcitabine Resistance in RCC
2.2. Sunitinib Resistance in RCC
3. CircRNAs and Drug Resistance in BCa
3.1. Gemcitabine and Doxorubicin Resistance in BCa
3.2. Cisplatin Resistance in BCa
4. CircRNAs and Drug Resistance in PCa
4.1. ADT Resistance in PCa
4.2. Docetaxel Resistance in PCa
4.3. ARSi Resistance in PCa
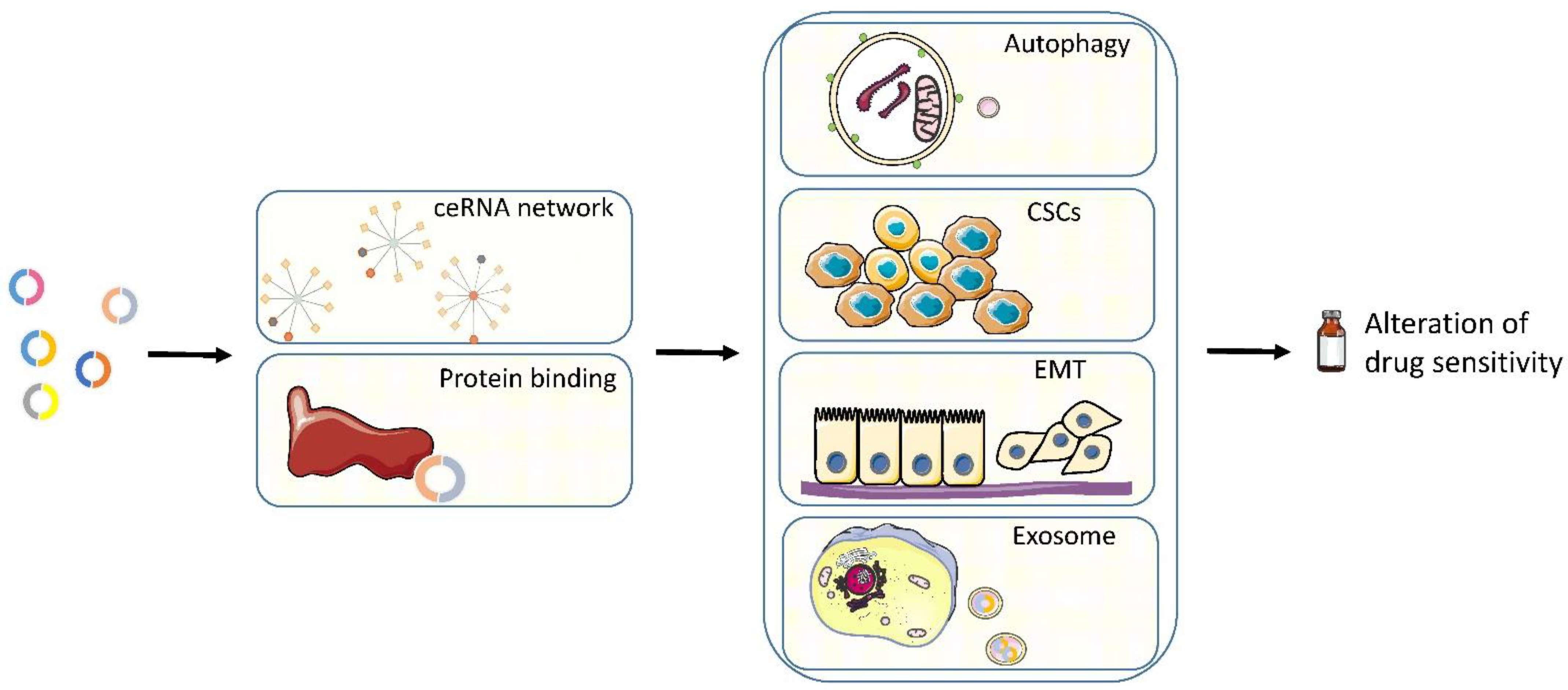
5. Mechanisms
5.1. CeRNA Network
5.2. Binding Proteins
5.3. Autophagy
5.4. Cancer Stem Cell (CSC)
5.5. EMT
5.6. Exosome
6. Limitations and Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef]
- Babjuk, M.; Burger, M.; Compérat, E.M.; Gontero, P.; Mostafid, A.H.; Palou, J.; van Rhijn, B.W.G.; Rouprêt, M.; Shariat, S.F.; Sylvester, R.; et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ)—2019 Update. Eur. Urol. 2019, 76, 639–657. [Google Scholar] [CrossRef]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Linares Espinós, E.; Lorch, A.; Neuzillet, Y.; et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2021, 79, 82–104. [Google Scholar] [CrossRef]
- Lam, T.B.L.; MacLennan, S.; Willemse, P.M.; Mason, M.D.; Plass, K.; Shepherd, R.; Baanders, R.; Bangma, C.H.; Bjartell, A.; Bossi, A.; et al. EAU-EANM-ESTRO-ESUR-SIOG Prostate Cancer Guideline Panel Consensus Statements for Deferred Treatment with Curative Intent for Localised Prostate Cancer from an International Collaborative Study (DETECTIVE Study). Eur. Urol. 2019, 76, 790–813. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Gastaldo, A.; Kempf, E.; González Del Alba, A.; Duran, I. Systemic treatment of renal cell cancer: A comprehensive review. Cancer Treat Rev. 2017, 60, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Teo, M.Y.; Rathkopf, D.E.; Kantoff, P. Treatment of Advanced Prostate Cancer. Annu. Rev. Med. 2019, 70, 479–499. [Google Scholar] [CrossRef]
- Patel, V.G.; Oh, W.K.; Galsky, M.D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J. Clin. 2020, 70, 404–423. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Li, R.; Jiang, J.; Shi, H.; Qian, H.; Zhang, X.; Xu, W. CircRNA: A rising star in gastric cancer. Cell Mol. Life Sci. 2020, 77, 1661–1680. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, T.; Chen, Y.; Xiao, T.; Li, W.; Cato, L.; Zhang, P.; Cotter, M.B.; Bowden, M.; Lis, R.T.; Zhao, S.G.; et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, E5207–E5215. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Feng, J.; Chen, K.; Ma, Y.; Gong, J.; Cai, F.; Jin, Y.; Gao, Y.; Xia, L.; Chang, H.; et al. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 2018, 46, D925–D929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2019, 20, 1420–1433. [Google Scholar] [CrossRef]
- Panda, A.C.; Gorospe, M. Detection and Analysis of Circular RNAs by RT-PCR. Bio Protoc. 2018, 8, e2775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611. [Google Scholar] [CrossRef] [PubMed]
- Stoll, L.; Sobel, J.; Rodriguez-Trejo, A.; Guay, C.; Lee, K.; Venø, M.T.; Kjems, J.; Laybutt, D.R.; Regazzi, R. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol. Metab. 2018, 9, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.L.; Su, M.; Wu, D.P. Functional roles of circular RNAs in Alzheimer’s disease. Ageing Res. Rev. 2020, 60, 101058. [Google Scholar] [CrossRef]
- Yang, X.; Ye, T.; Liu, H.; Lv, P.; Duan, C.; Wu, X.; Jiang, K.; Lu, H.; Xia, D.; Peng, E.; et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 2021, 20, 4. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Q.; Li, W.; Kang, Y.; Zhou, F.; Chang, D. Roles of circRNAs in prostate cancer: Expression, mechanism, application and potential. Int. J. Biochem. Cell Biol. 2021, 134, 105968. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Wang, P.; Fu, X.; Lin, W. Circular RNAs in renal cell carcinoma: Implications for tumorigenesis, diagnosis, and therapy. Mol. Cancer 2020, 19, 149. [Google Scholar] [CrossRef]
- Barata, P.C.; Rini, B.I. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef] [Green Version]
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.G.; Kolenko, V.M. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posadas, E.M.; Limvorasak, S.; Figlin, R.A. Targeted therapies for renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Bakouny, Z.; Hirsch, L.; Flippot, R.; Van Allen, E.M.; Wu, C.J.; Choueiri, T.K. Beyond conventional immune-checkpoint inhibition—Novel immunotherapies for renal cell carcinoma. Nat. Rev. Clin. Oncol. 2021, 18, 199–214. [Google Scholar] [CrossRef]
- Deleuze, A.; Saout, J.; Dugay, F.; Peyronnet, B.; Mathieu, R.; Verhoest, G.; Bensalah, K.; Crouzet, L.; Laguerre, B.; Belaud-Rotureau, M.A.; et al. Immunotherapy in Renal Cell Carcinoma: The Future Is Now. Int. J. Mol. Sci. 2020, 21, 2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, M.; Vale, N. Two Possible Strategies for Drug Modification of Gemcitabine and Future Contributions to Personalized Medicine. Molecules 2022, 27, 291. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Funt, S.A.; Rosenberg, J.E. Systemic, perioperative management of muscle-invasive bladder cancer and future horizons. Nat. Rev. Clin. Oncol. 2017, 14, 221–234. [Google Scholar] [CrossRef]
- Corrales, L.; Nogueira, A.; Passiglia, F.; Listi, A.; Caglevic, C.; Giallombardo, M.; Raez, L.; Santos, E.; Rolfo, C. Second-Line Treatment of Non-Small Cell Lung Cancer: Clinical, Pathological, and Molecular Aspects of Nintedanib. Front. Med. 2017, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Tella, S.H.; Kommalapati, A.; Borad, M.J.; Mahipal, A. Second-line therapies in advanced biliary tract cancers. Lancet Oncol. 2020, 21, e29–e41. [Google Scholar] [CrossRef]
- Boussios, S.; Seraj, E.; Zarkavelis, G.; Petrakis, D.; Kollas, A.; Kafantari, A.; Assi, A.; Tatsi, K.; Pavlidis, N.; Pentheroudakis, G. Management of patients with recurrent/advanced cervical cancer beyond first line platinum regimens: Where do we stand? A literature review. Crit. Rev. Oncol. Hematol. 2016, 108, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Casali, M.; Marcellini, M.; Casali, A.; Giuntini, T.; Galante, E.; Ferrone, C. Gemcitabine in pre-treated advanced renal carcinoma: A feasibility study. J. Exp. Clin. Cancer Res. 2001, 20, 195–198. [Google Scholar] [PubMed]
- Michaelson, M.D.; McKay, R.R.; Werner, L.; Atkins, M.B.; Van Allen, E.M.; Olivier, K.M.; Song, J.; Signoretti, S.; McDermott, D.F.; Choueiri, T.K. Phase 2 trial of sunitinib and gemcitabine in patients with sarcomatoid and/or poor-risk metastatic renal cell carcinoma. Cancer 2015, 121, 3435–3443. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Na’ara, S.; Gil, Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updat 2015, 23, 55–68. [Google Scholar] [CrossRef]
- Hu, J.; Guan, W.; Liu, P.; Dai, J.; Tang, K.; Xiao, H.; Qian, Y.; Sharrow, A.C.; Ye, Z.; Wu, L.; et al. Endoglin Is Essential for the Maintenance of Self-Renewal and Chemoresistance in Renal Cancer Stem Cells. Stem. Cell Rep. 2017, 9, 464–477. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Liu, G.; Cao, H.; Zhang, H.; Shao, F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem. Biophys. Res. Commun. 2019, 519, 172–178. [Google Scholar] [CrossRef]
- Cai, F.; Li, J.; Zhang, J.; Huang, S. Knockdown of Circ_CCNB2 Sensitizes Prostate Cancer to Radiation Through Repressing Autophagy by the miR-30b-5p/KIF18A Axis. Cancer Biother Radiopharm. 2020; Ahead of Print. [Google Scholar] [CrossRef]
- Bæk Møller, N.; Budolfsen, C.; Grimm, D.; Krüger, M.; Infanger, M.; Wehland, M.; Magnusson, N.E. Drug-Induced Hypertension Caused by Multikinase Inhibitors (Sorafenib, Sunitinib, Lenvatinib and Axitinib) in Renal Cell Carcinoma Treatment. Int. J. Mol. Sci. 2019, 20, 4712. [Google Scholar] [CrossRef] [Green Version]
- Escudier, B.; Szczylik, C.; Porta, C.; Gore, M. Treatment selection in metastatic renal cell carcinoma: Expert consensus. Nat. Rev. Clin. Oncol. 2012, 9, 327–337. [Google Scholar] [CrossRef]
- Nassif, E.; Thibault, C.; Vano, Y.; Fournier, L.; Mauge, L.; Verkarre, V.; Timsit, M.O.; Mejean, A.; Tartour, E.; Oudard, S. Sunitinib in kidney cancer: 10 years of experience and development. Expert Rev. Anticancer Ther. 2017, 17, 129–142. [Google Scholar] [CrossRef]
- Huang, K.B.; Pan, Y.H.; Shu, G.N.; Yao, H.H.; Liu, X.; Zhou, M.; Wei, J.H.; Chen, Z.H.; Lu, J.; Feng, Z.H.; et al. Circular RNA circSNX6 promotes sunitinib resistance in renal cell carcinoma through the miR-1184/GPCPD1/lysophosphatidic acid axis. Cancer Lett. 2021, 523, 121–134. [Google Scholar] [CrossRef]
- Zeng, X.; Tan, C.; Mo, M.; Qin, X.; Ma, X.; Huang, K.; Wang, X.; Liang, W.; Yang, L. CircRNA profiling identifies circRNF180 as a tumor suppressor in hepatocellular carcinoma. Epigenomics 2021, 13, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Cumberbatch, M.G.K.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T.; et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef] [Green Version]
- DeGeorge, K.C.; Holt, H.R.; Hodges, S.C. Bladder Cancer: Diagnosis and Treatment. Am. Fam. Physician. 2017, 96, 507–514. [Google Scholar]
- Lobo, N.; Mount, C.; Omar, K.; Nair, R.; Thurairaja, R.; Khan, M.S. Landmarks in the treatment of muscle-invasive bladder cancer. Nat. Rev. Urol. 2017, 14, 565–574. [Google Scholar] [CrossRef]
- Peyton, C.C.; Tang, D.; Reich, R.R.; Azizi, M.; Chipollini, J.; Pow-Sang, J.M.; Manley, B.; Spiess, P.E.; Poch, M.A.; Sexton, W.J.; et al. Downstaging and Survival Outcomes Associated With Neoadjuvant Chemotherapy Regimens Among Patients Treated With Cystectomy for Muscle-Invasive Bladder Cancer. JAMA Oncol. 2018, 4, 1535–1542. [Google Scholar] [CrossRef]
- Peyton, C.C.; Chipollini, J.; Azizi, M.; Kamat, A.M.; Gilbert, S.M.; Spiess, P.E. Updates on the use of intravesical therapies for non-muscle invasive bladder cancer: How, when and what. World J. Urol. 2019, 37, 2017–2029. [Google Scholar] [CrossRef]
- Huang, W.; Lu, Y.; Wang, F.; Huang, X.; Yu, Z. Circular RNA circRNA_103809 Accelerates Bladder Cancer Progression and Enhances Chemo-Resistance by Activation of miR-516a-5p/FBXL18 Axis. Cancer Manag. Res. 2020, 12, 7561–7568. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.; Guo, W.X.; Ren, M.S. Circular RNA hsa_circ_103809 promotes cell migration and invasion of gastric cancer cells by binding to microRNA-101-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6064–6071. [Google Scholar] [PubMed]
- Qiu, X.; Wang, Q.; Song, H.; Shao, D.; Xue, J. circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol. Lett. 2020, 19, 3725–3730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Shen, M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 555–566. [Google Scholar] [PubMed]
- Bian, L.; Zhi, X.; Ma, L.; Zhang, J.; Chen, P.; Sun, S.; Li, J.; Sun, Y.; Qin, J. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem. Biophys. Res. Commun. 2018, 505, 346–352. [Google Scholar] [CrossRef]
- Xie, F.; Zhao, N.; Zhang, H.; Xie, D. Circular RNA CircHIPK3 Promotes Gemcitabine Sensitivity in Bladder Cancer. J. Cancer 2020, 11, 1907–1912. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xia, L.; Dong, L.; Wang, J.; Xiao, Q.; Yu, X.; Zhu, H. CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Sponging miR-330-5p and Targets RASSF1. Cancer Manag. Res. 2020, 12, 921–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Xi, X.; Xiao, S.; Xiao, X. Silencing of circHIPK3 Sensitizes Paclitaxel-Resistant Breast Cancer Cells to Chemotherapy by Regulating HK2 Through Targeting miR-1286. Cancer Manag. Res. 2021, 13, 5573–5585. [Google Scholar] [CrossRef]
- Han, C.; Wang, S.; Wang, H.; Zhang, J. Exosomal circ-HIPK3 Facilitates Tumor Progression and Temozolomide Resistance by Regulating miR-421/ZIC5 Axis in Glioma. Cancer Biother. Radiopharm. 2021, 36, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, C.; Liu, X.; Wang, Y.; Zhao, R.; Yang, Y.; Zheng, X.; Zhang, Y.; Zhang, X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine 2019, 48, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Zhang, F. Circular RNA VANGL1 knockdown suppressed viability, promoted apoptosis, and increased doxorubicin sensitivity through targeting miR-145-5p to regulate SOX4 in bladder cancer cells. Open Med. 2021, 16, 1010–1021. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhou, W.; Duan, L.; Zhang, J.; Lu, X.; Jin, L.; Yu, Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J. Cell Physiol. 2019, 234, 3887–3896. [Google Scholar] [CrossRef]
- Yang, L.; Zeng, Z.; Kang, N.; Yang, J.C.; Wei, X.; Hai, Y. Circ-VANGL1 promotes the progression of osteoporosis by absorbing miRNA-217 to regulate RUNX2 expression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 949–957. [Google Scholar]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Herr, H.; Soloway, M. Cisplatin, Neoadjuvant Chemotherapy and Bladder Cancer. Urology 2022, 159, 2–5. [Google Scholar] [CrossRef]
- Iacovino, M.L.; Miceli, C.C.; De Felice, M.; Barone, B.; Pompella, L.; Chiancone, F.; Di Zazzo, E.; Tirino, G.; Della Corte, C.M.; Imbimbo, C.; et al. Novel Therapeutic Opportunities in Neoadjuvant Setting in Urothelial Cancers: A New Horizon Opened by Molecular Classification and Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2022, 23, 1133. [Google Scholar] [CrossRef]
- Leow, J.J.; Martin-Doyle, W.; Rajagopal, P.S.; Patel, C.G.; Anderson, E.M.; Rothman, A.T.; Cote, R.J.; Urun, Y.; Chang, S.L.; Choueiri, T.K.; et al. Adjuvant chemotherapy for invasive bladder cancer: A 2013 updated systematic review and meta-analysis of randomized trials. Eur. Urol. 2014, 66, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Pfister, C.; Gravis, G.; Flechon, A.; Chevreau, C.; Mahammedi, H.; Laguerre, B.; Guillot, A.; Joly, F.; Soulie, M.; Allory, Y. 652O Dose-dense methotrexate, vinblastine, doxorubicin and cisplatin (dd-MVAC) or gemcitabine and cisplatin (GC) as perioperative chemotherapy for patients with muscle-invasive bladder cancer (MIBC): Results of the GETUG/AFU VESPER V05 phase III trial. Ann. Oncol. 2021, 32, S678. [Google Scholar] [CrossRef]
- Wei, W.; Sun, J.; Zhang, H.; Xiao, X.; Huang, C.; Wang, L.; Zhong, H.; Jiang, Y.; Zhang, X.; Jiang, G. Circ0008399 interaction with WTAP promotes assembly and activity of the m6A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 2021, 81, 6142–6156. [Google Scholar] [CrossRef]
- Priem, D.; Devos, M.; Druwé, S.; Martens, A.; Slowicka, K.; Ting, A.T.; Pasparakis, M.; Declercq, W.; Vandenabeele, P.; van Loo, G.; et al. A20 protects cells from TNF-induced apoptosis through linear ubiquitin-dependent and -independent mechanisms. Cell Death Dis. 2019, 10, 692. [Google Scholar] [CrossRef]
- Su, Y.; Yang, W.; Jiang, N.; Shi, J.; Chen, L.; Zhong, G.; Bi, J.; Dong, W.; Wang, Q.; Wang, C.; et al. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int. J. Biol. Sci. 2019, 15, 441–452. [Google Scholar] [CrossRef]
- Gong, P.; Xu, R.; Zhuang, Q.; He, X. A novel circular RNA (hsa_circRNA_102336), a plausible biomarker, promotes the tumorigenesis by sponging miR-515-5p in human bladder cancer. Biomed. Pharm. 2020, 126, 110059. [Google Scholar] [CrossRef]
- Chen, J.; Sun, Y.; Ou, Z.; Yeh, S.; Huang, C.P.; You, B.; Tsai, Y.C.; Sheu, T.J.; Zu, X.; Chang, C. Androgen receptor-regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020, 21, e48467. [Google Scholar] [CrossRef]
- Tian, J.; Fan, J.; Xu, J.; Ren, T.; Guo, H.; Zhou, L. circ-FNTA accelerates proliferation and invasion of bladder cancer. Oncol. Lett. 2020, 19, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiao, X.; Wei, W.; Huang, C.; Wang, M.; Wang, L.; He, Y.; Sun, J.; Jiang, Y.; Jiang, G.; et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73 axis in bladder cancer. Mol. Cancer 2021, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, B.; Chen, C.; Chen, J. Circular RNA circLIFR regulates the proliferation, migration, invasion and apoptosis of human vascular smooth muscle cells via the miR-1299/KDR axis. Metab. Brain Dis. 2021, 37, 253–263. [Google Scholar] [CrossRef]
- Jiang, C.; Zeng, X.; Shan, R.; Wen, W.; Li, J.; Tan, J.; Li, L.; Wan, R. The Emerging Picture of the Roles of CircRNA-CDR1as in Cancer. Front. Cell Dev. Biol. 2020, 8, 590478. [Google Scholar] [CrossRef]
- Yuan, W.; Zhou, R.; Wang, J.; Han, J.; Yang, X.; Yu, H.; Lu, H.; Zhang, X.; Li, P.; Tao, J.; et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol. Oncol. 2019, 13, 1559–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Yang, X.; Yuan, W.; Yang, C.; Zhang, X.; Han, J.; Wang, J.; Deng, X.; Yang, H.; Li, P.; et al. CircRNA-Cdr1as Exerts Anti-Oncogenic Functions in Bladder Cancer by Sponging MicroRNA-135a. Cell Physiol. Biochem. 2018, 46, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zheng, R.; Chen, J.; Ning, D. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 2020, 20, 289. [Google Scholar] [CrossRef]
- Zhao, Z.; Ji, M.; Wang, Q.; He, N.; Li, Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol. Ther. Nucleic Acids 2019, 18, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Xu, R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed. Pharm. 2020, 123, 109771. [Google Scholar] [CrossRef]
- Yang, W.; Gu, J.; Wang, X.; Wang, Y.; Feng, M.; Zhou, D.; Guo, J.; Zhou, M. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J. Cell Mol. Med. 2019, 23, 3166–3177. [Google Scholar] [CrossRef] [PubMed]
- Chi, B.J.; Zhao, D.M.; Liu, L.; Yin, X.Z.; Wang, F.F.; Bi, S.; Gui, S.L.; Zhou, S.B.; Qin, W.B.; Wu, D.M.; et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma 2019, 66, 197–202. [Google Scholar] [CrossRef]
- Roehl, K.A.; Han, M.; Ramos, C.G.; Antenor, J.A.; Catalona, W.J. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3,478 consecutive patients: Long-term results. J. Urol. 2004, 172, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freedland, S.J.; Humphreys, E.B.; Mangold, L.A.; Eisenberger, M.; Dorey, F.J.; Walsh, P.C.; Partin, A.W. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA 2005, 294, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Kupelian, P.A.; Mahadevan, A.; Reddy, C.A.; Reuther, A.M.; Klein, E.A. Use of different definitions of biochemical failure after external beam radiotherapy changes conclusions about relative treatment efficacy for localized prostate cancer. Urology 2006, 68, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Mosillo, C.; Iacovelli, R.; Ciccarese, C.; Fantinel, E.; Bimbatti, D.; Brunelli, M.; Bisogno, I.; Kinspergher, S.; Buttigliero, C.; Tucci, M.; et al. De novo metastatic castration sensitive prostate cancer: State of art and future perspectives. Cancer Treat. Rev. 2018, 70, 67–74. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, S.; Guo, L.R.; Zhao, X.J. Diagnostic strategies and the incidence of prostate cancer: Reasons for the low reported incidence of prostate cancer in China. Asian J. Androl. 2009, 11, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; van der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014, 65, 467–479. [Google Scholar] [CrossRef]
- Yap, T.A.; Smith, A.D.; Ferraldeschi, R.; Al-Lazikani, B.; Workman, P.; de Bono, J.S. Drug discovery in advanced prostate cancer: Translating biology into therapy. Nat. Rev. Drug Discov. 2016, 15, 699–718. [Google Scholar] [CrossRef]
- Litwin, M.S.; Tan, H.J. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017, 317, 2532–2542. [Google Scholar] [CrossRef]
- Nuhn, P.; De Bono, J.S.; Fizazi, K.; Freedland, S.J.; Grilli, M.; Kantoff, P.W.; Sonpavde, G.; Sternberg, C.N.; Yegnasubramanian, S.; Antonarakis, E.S. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-resistant Prostate Cancer in the Era of Precision Oncology. Eur. Urol. 2019, 75, 88–99. [Google Scholar] [CrossRef]
- Clarke, N.W.; Ali, A.; Ingleby, F.C.; Hoyle, A.; Amos, C.L.; Attard, G.; Brawley, C.D.; Calvert, J.; Chowdhury, S.; Cook, A.; et al. Addition of docetaxel to hormonal therapy in low—And high-burden metastatic hormone sensitive prostate cancer: Long-term survival results from the STAMPEDE trial. Ann. Oncol. 2019, 30, 1992–2003. [Google Scholar] [CrossRef] [Green Version]
- Fizazi, K.; Tran, N.; Fein, L.; Matsubara, N.; Rodriguez-Antolin, A.; Alekseev, B.Y.; Özgüroğlu, M.; Ye, D.; Feyerabend, S.; Protheroe, A.; et al. Abiraterone acetate plus prednisone in patients with newly diagnosed high-risk metastatic castration-sensitive prostate cancer (LATITUDE): Final overall survival analysis of a randomised, double-blind, phase 3 trial. Lancet Oncol. 2019, 20, 686–700. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Szmulewitz, R.Z.; Petrylak, D.P.; Holzbeierlein, J.; Villers, A.; Azad, A.; Alcaraz, A.; Alekseev, B.; Iguchi, T.; Shore, N.D.; et al. ARCHES: A Randomized, Phase III Study of Androgen Deprivation Therapy With Enzalutamide or Placebo in Men With Metastatic Hormone-Sensitive Prostate Cancer. J. Clin. Oncol. 2019, 37, 2974–2986. [Google Scholar] [CrossRef]
- Chi, K.N.; Agarwal, N.; Bjartell, A.; Chung, B.H.; Pereira de Santana Gomes, A.J.; Given, R.; Juárez Soto, Á.; Merseburger, A.S.; Özgüroğlu, M.; Uemura, H.; et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2019, 381, 13–24. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [Green Version]
- Greene, J.; Baird, A.M.; Lim, M.; Flynn, J.; McNevin, C.; Brady, L.; Sheils, O.; Gray, S.G.; McDermott, R.; Finn, S.P. Differential CircRNA Expression Signatures May Serve as Potential Novel Biomarkers in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 605686. [Google Scholar] [CrossRef]
- Luo, J.; Li, Y.; Zheng, W.; Xie, N.; Shi, Y.; Long, Z.; Xie, L.; Fazli, L.; Zhang, D.; Gleave, M.; et al. Characterization of a Prostate- and Prostate Cancer-Specific Circular RNA Encoded by the Androgen Receptor Gene. Mol. Ther. Nucleic Acids 2019, 18, 916–926. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Ma, T.; Ungerleider, N.; Roberts, C.; Kobelski, M.; Jin, L.; Concha, M.; Wang, X.; Baddoo, M.; Nguyen, H.M.; et al. Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene 2019, 38, 7060–7072. [Google Scholar] [CrossRef]
- Petrylak, D.P.; Macarthur, R.B.; O’Connor, J.; Shelton, G.; Judge, T.; Balog, J.; Pfaff, C.; Bagiella, E.; Heitjan, D.; Fine, R.; et al. Phase I trial of docetaxel with estramustine in androgen-independent prostate cancer. J. Clin. Oncol. 1999, 17, 958–967. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Huan, J.; Che, F. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020, 20, 334. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Tai, Y.C. Hsa_circ_0006404 and hsa_circ_0000735 Regulated Ovarian Cancer Response to Docetaxel Treatment via Regulating p-GP Expression. Biochem Genet. 2021, 60, 395–414. [Google Scholar] [CrossRef]
- Huang, W.; Xu, X.; Liu, M.; Cui, W.; Peng, G. Downregulation of Hsa_circ_0000735 Inhibits the Proliferation, Migration, Invasion, and Glycolysis in Non-small-cell Lung Cancer by Targeting miR-940/BMPER Axis. Oncol. Targets Ther. 2020, 13, 8427–8439. [Google Scholar] [CrossRef]
- Ding, T.; Zhu, Y.; Jin, H.; Zhang, P.; Guo, J.; Zheng, J. Circular RNA circ_0057558 Controls Prostate Cancer Cell Proliferation Through Regulating miR-206/USP33/c-Myc Axis. Front. Cell Dev. Biol. 2021, 9, 644397. [Google Scholar] [CrossRef]
- Chen, X.; Tan, Q.Q.; Tan, X.R.; Li, S.J.; Zhang, X.X. Circ_0057558 promotes nonalcoholic fatty liver disease by regulating ROCK1/AMPK signaling through targeting miR-206. Cell Death Dis. 2021, 12, 809. [Google Scholar] [CrossRef]
- Zhang, H.; Li, M.; Zhang, J.; Shen, Y.; Gui, Q. Exosomal Circ-XIAP Promotes Docetaxel Resistance in Prostate Cancer by Regulating miR-1182/TPD52 Axis. Drug Des. Devel Ther. 2021, 15, 1835–1849. [Google Scholar] [CrossRef]
- Feng, Y.; Yang, Y.; Zhao, X.; Fan, Y.; Zhou, L.; Rong, J.; Yu, Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019, 10, 792. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Zhou, L.; Zhang, C.; Xu, J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020, 468, 88–101. [Google Scholar] [CrossRef]
- Drula, R.; Pirlog, R.; Trif, M.; Slaby, O.; Braicu, C.; Berindan-Neagoe, I. circFOXO3: Going around the mechanistic networks in cancer by interfering with miRNAs regulatory networks. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166045. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, K.; Miao, Z.; Wang, Q.; Miao, Y.; Guo, Z.; Qiu, Y.; Chen, B.; Ren, L.; Wei, Z.; et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019, 21, 1284–1296. [Google Scholar] [CrossRef]
- Xiang, T.; Jiang, H.S.; Zhang, B.T.; Liu, G. CircFOXO3 functions as a molecular sponge for miR-143-3p to promote the progression of gastric carcinoma via upregulating USP44. Gene 2020, 753, 144798. [Google Scholar] [CrossRef]
- Ai, Y.; Wu, S.; Zou, C.; Wei, H. Circular RNA circFOXO3 regulates KDM2A by targeting miR-214 to promote tumor growth and metastasis in oral squamous cell carcinoma. J. Cell Mol. Med. 2021; Ahead of Print. [Google Scholar] [CrossRef]
- Xing, Y.; Zha, W.J.; Li, X.M.; Li, H.; Gao, F.; Ye, T.; Du, W.Q.; Liu, Y.C. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J. Cell Biochem. 2020, 121, 2595–2605. [Google Scholar] [CrossRef]
- Li, Y.; Qiao, L.; Zang, Y.; Ni, W.; Xu, Z. Circular RNA FOXO3 Suppresses Bladder Cancer Progression and Metastasis by Regulating MiR-9-5p/TGFBR2. Cancer Manag. Res. 2020, 12, 5049–5056. [Google Scholar] [CrossRef]
- Wang, C.; Tao, W.; Ni, S.; Chen, Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. 2019, 12, 8085–8094. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Huang, F.; Feng, C. CircFoxo3 Promotes Adriamycin Resistance through Regulation of miR-199a-5p/ATP Binding Cassette Subfamily C Member 1 Axis in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 5113–5122. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Zhu, C.; Wang, B.; Zheng, H.; McAlister, V.; Lacefield, J.C.; Quan, D.; Mele, T.; Greasley, A.; Liu, K.; et al. Circular RNA Foxo3 in cardiac ischemia-reperfusion injury in heart transplantation: A new regulator and target. Am. J. Transplant. 2021, 21, 2992–3004. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, C.; Wen, X.; Liu, W.; Huang, X.; Li, Y.; Zang, J.; Weng, Z.; Lu, D.; Tsang, C.K.; et al. Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol. Ther. 2021; Ahead of Print. [Google Scholar] [CrossRef]
- Lin, S.P.; Hu, J.; Wei, J.X.; Ye, S.; Bu, J.; Xu, W.; Chen, S.; Wu, Y.; Wu, G.; Zhu, L.; et al. Silencing of circFoxO3 Protects HT22 Cells from Glutamate-Induced Oxidative Injury via Regulating the Mitochondrial Apoptosis Pathway. Cell Mol. Neurobiol. 2020, 40, 1231–1242. [Google Scholar] [CrossRef]
- Feyerabend, S.; Saad, F.; Li, T.; Ito, T.; Diels, J.; Van Sanden, S.; De Porre, P.; Roiz, J.; Abogunrin, S.; Koufopoulou, M.; et al. Survival benefit, disease progression and quality-of-life outcomes of abiraterone acetate plus prednisone versus docetaxel in metastatic hormone-sensitive prostate cancer: A network meta-analysis. Eur. J. Cancer 2018, 103, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Buttigliero, C.; Tucci, M.; Bertaglia, V.; Vignani, F.; Bironzo, P.; Di Maio, M.; Scagliotti, G.V. Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat. Rev. 2015, 41, 884–892. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Li, D.; Ma, X.; Xu, K.; Ding, B.; Li, H.; Wang, Z.; Ouyang, W.; Long, G.; et al. Androgen Receptor Splice Variant 7 Predicts Shorter Response in Patients with Metastatic Hormone-sensitive Prostate Cancer Receiving Androgen Deprivation Therapy. Eur. Urol. 2021, 79, 879–886. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Xiao, W.; Yan, L.; Guan, W.; Hu, Z.; Wu, L.; Huang, Q.; Wang, J.; Xu, H.; et al. Androgen-receptor splice variant-7-positive prostate cancer: A novel molecular subtype with markedly worse androgen-deprivation therapy outcomes in newly diagnosed patients. Mod. Pathol. 2018, 31, 198–208. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Sun, Y.; Xiang, Z.; Wang, K.; Liu, B.; Xiao, G.; Niu, Y.; Wu, D.; Chang, C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019, 10, 37. [Google Scholar] [CrossRef]
- Greene, J.; Baird, A.M.; Casey, O.; Brady, L.; Blackshields, G.; Lim, M.; O’Brien, O.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci. Rep. 2019, 9, 10739. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Sun, S.; Mao, W.; Xu, B.; Chen, M. Identification of Enzalutamide Resistance-Related circRNA-miRNA-mRNA Regulatory Networks in Patients with Prostate Cancer. Onco Targets Ther. 2021, 14, 3833–3848. [Google Scholar] [CrossRef]
- Xiang, Z.; Xu, C.; Wu, G.; Liu, B.; Wu, D. CircRNA-UCK2 Increased TET1 Inhibits Proliferation and Invasion of Prostate Cancer Cells via Sponge MiRNA-767-5p. Open Med. 2019, 14, 833–842. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Feng, J.; Chen, L. Overexpression of circRNA circUCK2 Attenuates Cell Apoptosis in Cerebral Ischemia-Reperfusion Injury via miR-125b-5p/GDF11 Signaling. Mol. Ther. Nucleic Acids 2020, 22, 673–683. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, X.; Wu, D.; Zhang, X.; Shen, X.; Mi, K.; Qu, Z.; Jiang, Y.; Shang, D. Comprehensive characterization of a drug-resistance-related ceRNA network across 15 anti-cancer drug categories. Mol. Ther. Nucleic Acids 2021, 24, 11–24. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Qiu, F.; Yang, L.; Ling, X.; Yang, R.; Yang, X.; Zhang, L.; Fang, W.; Xie, C.; Huang, D.; Zhou, Y.; et al. Sequence Variation in Mature MicroRNA-499 Confers Unfavorable Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Clin. Cancer Res. 2015, 21, 1602–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef] [Green Version]
- Epis, M.R.; Barker, A.; Giles, K.M.; Beveridge, D.J.; Leedman, P.J. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J. Biol. Chem. 2011, 286, 41442–41454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Sun, Y.; Tao, W.; Fei, X.; Chang, C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017, 394, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Wang, W.T.; Zeng, Z.C.; Chen, T.Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.Y.; Zhou, Y.F.; et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dong, Y.; Zhao, L.; Su, L.; Luo, J. Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int. J. Oncol. 2018, 53, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, M.S. Autophagy—A key player in cellular and body metabolism. Nat. Rev. Endocrinol. 2014, 10, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.A.; Kwok, B.; Dragowska, W.H.; To, K.H.; Le, D.; Bally, M.B.; Gorski, S.M. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 2008, 112, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.G.; Yang, J.C.; Kung, H.J.; Shi, X.B.; Tilki, D.; Lara, P.N., Jr.; DeVere White, R.W.; Gao, A.C.; Evans, C.P. Targeting autophagy overcomes Enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene 2014, 33, 4521–4530. [Google Scholar] [CrossRef] [Green Version]
- Kong, R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol. Int. 2020, 44, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ai, G.; Zhou, J.; Mao, W.; Li, H.; Guo, J. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed. Pharm. 2019, 117, 109064. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, J.; Qin, L.; Yang, Z.; Xiong, J.; Zhang, Y.; Li, R.; Li, S.; Wang, H.; Yu, B.; et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging 2018, 10, 3806–3820. [Google Scholar] [CrossRef]
- Shang, J.; Chen, W.M.; Liu, S.; Wang, Z.H.; Wei, T.N.; Chen, Z.Z.; Wu, W.B. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk. Res. 2019, 85, 106198. [Google Scholar] [CrossRef]
- Cao, H.X.; Miao, C.F.; Sang, L.N.; Huang, Y.M.; Zhang, R.; Sun, L.; Jiang, Z.X. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020, 243, 117255. [Google Scholar] [CrossRef]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; He, H.; Zhu, J.; Zhang, Q.; Zheng, Z.; Liang, X.; Chen, L.; Yang, M.; Peng, K.; Zhang, Z.; et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer 2020, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, Q.; Cheng, X.; Wang, S.; Wu, R.; Leng, X.; Shao, L. miR-449c-5p availability is antagonized by circ-NOTCH1 for MYC-induced NOTCH1 upregulation as well as tumor metastasis and stemness in gastric cancer. J. Cell Biochem. 2020, 121, 4052–4063. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Stephens, M.A.; Pathak, H.; Rangarajan, A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011, 2, e179. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Song, X.; Li, Y.; Ma, T.; Su, P.; Guo, R.; Chen, B.; Zhang, H.; Sang, Y.; Liu, Y.; et al. Targeting the circBMPR2/miR-553/USP4 Axis as a Potent Therapeutic Approach for Breast Cancer. Mol. Ther. Nucleic Acids 2019, 17, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Song, X.; Li, Y.; Su, P.; Han, D.; Ma, T.; Guo, R.; Chen, B.; Zhao, W.; Sang, Y.; et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene 2019, 38, 6850–6866. [Google Scholar] [CrossRef]
- Huang, W.; Yang, Y.; Wu, J.; Niu, Y.; Yao, Y.; Zhang, J.; Huang, X.; Liang, S.; Chen, R.; Chen, S.; et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 2020, 27, 1709–1727. [Google Scholar] [CrossRef] [Green Version]
- Joseph, N.A.; Chiou, S.H.; Lung, Z.; Yang, C.L.; Lin, T.Y.; Chang, H.W.; Sun, H.S.; Gupta, S.K.; Yen, L.; Wang, S.D.; et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 2018, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Liu, N.; Liang, Y.; He, Q.; Yang, X.; Lei, Y.; Zhang, P.; Zhao, Y.; He, S.; Wang, Y.; et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. Cancer 2020, 19, 33. [Google Scholar] [CrossRef]
- Liu, J.; Xue, N.; Guo, Y.; Niu, K.; Gao, L.; Zhang, S.; Gu, H.; Wang, X.; Zhao, D.; Fan, R. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging 2019, 11, 12412–12427. [Google Scholar] [CrossRef]
- He, Y.; Mingyan, E.; Wang, C.; Liu, G.; Shi, M.; Liu, S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int. J. Biol. Macromol. 2019, 125, 116–123. [Google Scholar] [CrossRef]
- Xu, Y.; Kong, S.; Qin, S.; Shen, X.; Ju, S. Exosomal circRN.NAs: Sorting Mechanisms, Roles and Clinical Applications in Tumors. Front. Cell Dev. Biol. 2020, 8, 581558. [Google Scholar] [CrossRef]
- Li, L.; Li, W.; Chen, N.; Zhao, H.; Xu, G.; Zhao, Y.; Pan, X.; Zhang, X.; Zhou, L.; Yu, D.; et al. FLI1 Exonic Circular RNAs as a Novel Oncogenic Driver to Promote Tumor Metastasis in Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 1302–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hon, K.W.; Ab-Mutalib, N.S.; Abdullah, N.M.A.; Jamal, R.; Abu, N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci. Rep. 2019, 9, 16497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qiu, A.; Peng, F.; Tan, X.; Wang, J.; Gong, X. Exosomal transfer of circular RNA FBXW7 ameliorates the chemoresistance to oxaliplatin in colorectal cancer by sponging miR-18b-5p. Neoplasma 2021, 68, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Teng, F.; Chang, L.; Wang, J.; Liu, D.L.; Cui, Y.S.; Li, G.H. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging 2021, 13, 13264–13286. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Wu, X.; Bu, X.; Wang, D.; Wu, Z.; Zhang, G.; Gu, J.; Kang, D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020, 479, 1–12. [Google Scholar] [CrossRef]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal. Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef]
- Luo, Y.; Gui, R. Circulating Exosomal CircMYC Is Associated with Recurrence and Bortezomib Resistance in Patients with Multiple Myeloma. Turk. J. Haematol. 2020, 37, 248–262. [Google Scholar] [CrossRef]
- Luo, Y.; Gui, R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J. Gynecol. Oncol. 2020, 31, e75. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lin, Y.; Mi, C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol. Int. 2021, 45, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhao, Y.; Chen, Q.; Zhu, S.; Niu, Y.; Ye, Z.; Hu, P.; Chen, D.; Xu, P.; Chen, J.; et al. Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 2021, 40, 5505–5517. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.W.; Zhu, S.Q.; Pei, X.; Qiu, B.Q.; Xiong, D.; Long, X.; Lin, K.; Lu, F.; Xu, J.J.; Wu, Y.B. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol. Cancer 2021, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Arén Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Thana, M.; Wood, L. Immune Checkpoint Inhibitors in Genitourinary Malignancies. Curr. Oncol. 2020, 27, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Balar, A.V.; Castellano, D.; O’Donnell, P.H.; Grivas, P.; Vuky, J.; Powles, T.; Plimack, E.R.; Hahn, N.M.; de Wit, R.; Pang, L.; et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 1483–1492. [Google Scholar] [CrossRef]
- Balar, A.V.; Galsky, M.D.; Rosenberg, J.E.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman-Censits, J.; Perez-Gracia, J.L.; et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef]
- Ge, W.; Chi, H.; Tang, H.; Xu, J.; Wang, J.; Cai, W.; Ma, H. Circular RNA CELF1 drives immunosuppression and anti-PD1 therapy resistance in non-small cell lung cancer via the miR-491-5p/EGFR axis. Aging 2021, 13, 24560–24579. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Zhang, P.F.; Wei, C.Y.; Peng, R.; Lu, J.C.; Gao, C.; Cai, J.B.; Yang, X.; Fan, J.; Ke, A.W.; et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 2020, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Crocetto, F.; Buonerba, C.; Caputo, V.; Ferro, M.; Persico, F.; Trama, F.; Iliano, E.; Rapisarda, S.; Bada, M.; Facchini, G.; et al. Urologic malignancies: Advances in the analysis and interpretation of clinical findings. Future Sci. OA 2021, 7, FSO674. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Abraham, J.M.; Cheng, Y.; Wang, Z.; Wang, Z.; Zhang, G.; Ashktorab, H.; Smoot, D.T.; Cole, R.N.; Boronina, T.N.; et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol. Ther. Nucleic Acids 2018, 13, 312–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [Green Version]
- Han, K.; Wang, F.W.; Cao, C.H.; Ling, H.; Chen, J.W.; Chen, R.X.; Feng, Z.H.; Luo, J.; Jin, X.H.; Duan, J.L.; et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol. Cancer 2020, 19, 60. [Google Scholar] [CrossRef] [Green Version]
- Qiu, M.; Xia, W.; Chen, R.; Wang, S.; Xu, Y.; Ma, Z.; Xu, W.; Zhang, E.; Wang, J.; Fang, T.; et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res. 2018, 78, 2839–2851. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [Green Version]
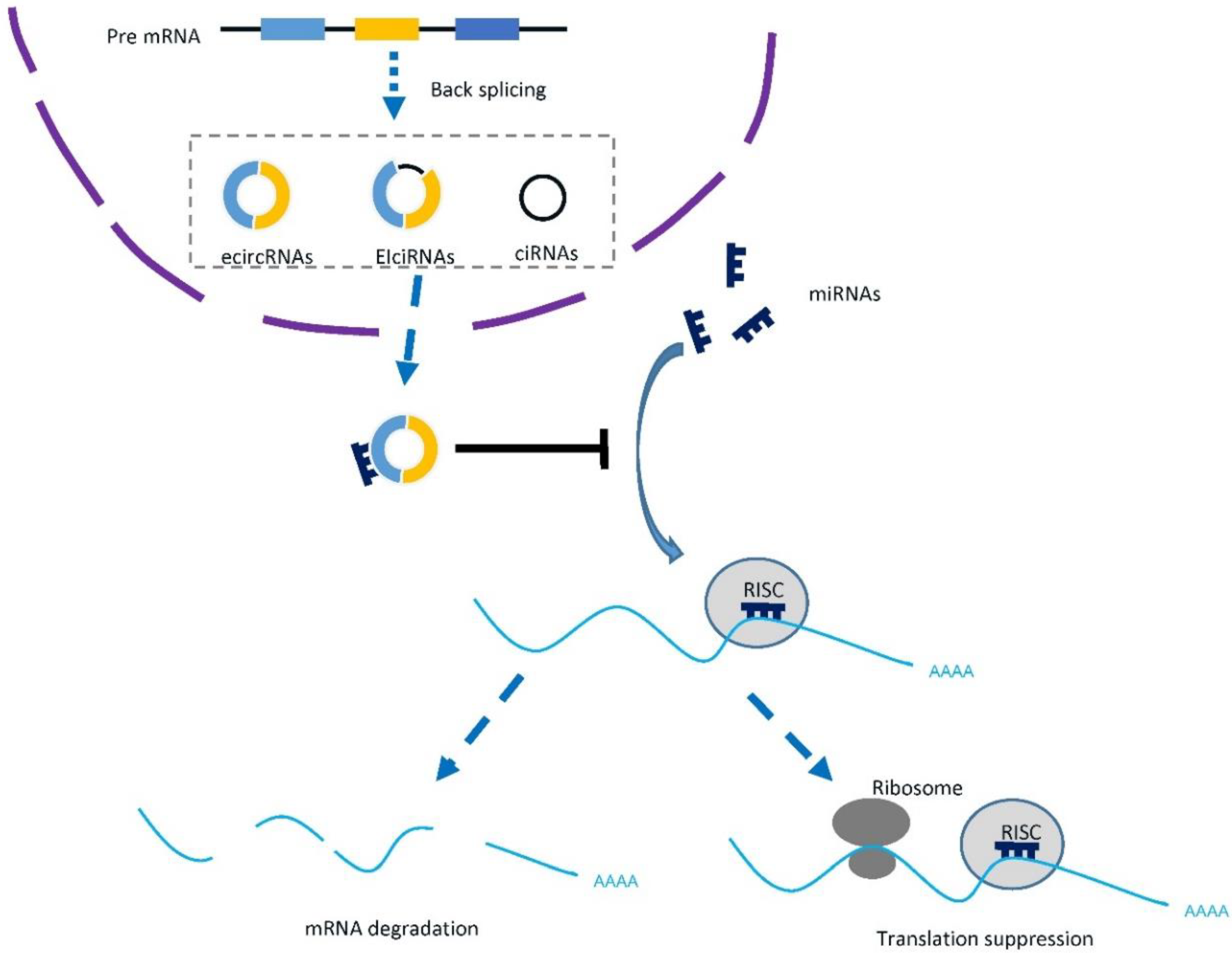

| Study | Cancer | Comparison | Differently Expressed circRNA | Expression Alteration | Further Investigation |
|---|---|---|---|---|---|
| [91] | Bladder cancer | T vs. N and cisplatin-resistant vs. naive | hsa_circ_0000285 | ↓ | Prognosis |
| [108] | Prostate cancer | CRPC vs. HSPC | circAR3 | ↓ | Invasion, proliferation, downstream pathways |
| [109] | Prostate cancer | CRPC vs. HSPC | 13 circARs | ↑ | Association with linear AR transcript |
| [137] | Prostate cancer | EnzR vs. naive | 111 circRNAs | NA | NA |
| [139] | Prostate cancer | EnzR vs. naive | circUCK2 | ↓ | Invasion, proliferation |
| [138] | Prostate cancer | EnzR vs. naive | 4 circRNAs | ↑ | |
| 9 circRNAs | ↓ | ||||
| hsa_circ_0047641 | ↑ | Invasion, proliferation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, G.; Ma, S.; Shi, R.; Sun, Y.; Hu, Z.; Chen, K. Circular RNAs and Drug Resistance in Genitourinary Cancers: A Literature Review. Cancers 2022, 14, 866. https://doi.org/10.3390/cancers14040866
Long G, Ma S, Shi R, Sun Y, Hu Z, Chen K. Circular RNAs and Drug Resistance in Genitourinary Cancers: A Literature Review. Cancers. 2022; 14(4):866. https://doi.org/10.3390/cancers14040866
Chicago/Turabian StyleLong, Gongwei, Siquan Ma, Runlin Shi, Yi Sun, Zhiquan Hu, and Ke Chen. 2022. "Circular RNAs and Drug Resistance in Genitourinary Cancers: A Literature Review" Cancers 14, no. 4: 866. https://doi.org/10.3390/cancers14040866
APA StyleLong, G., Ma, S., Shi, R., Sun, Y., Hu, Z., & Chen, K. (2022). Circular RNAs and Drug Resistance in Genitourinary Cancers: A Literature Review. Cancers, 14(4), 866. https://doi.org/10.3390/cancers14040866







