The Pseudokinase TRIB1 in Immune Cells and Associated Disorders
Abstract
:Simple Summary
Abstract
1. Introduction
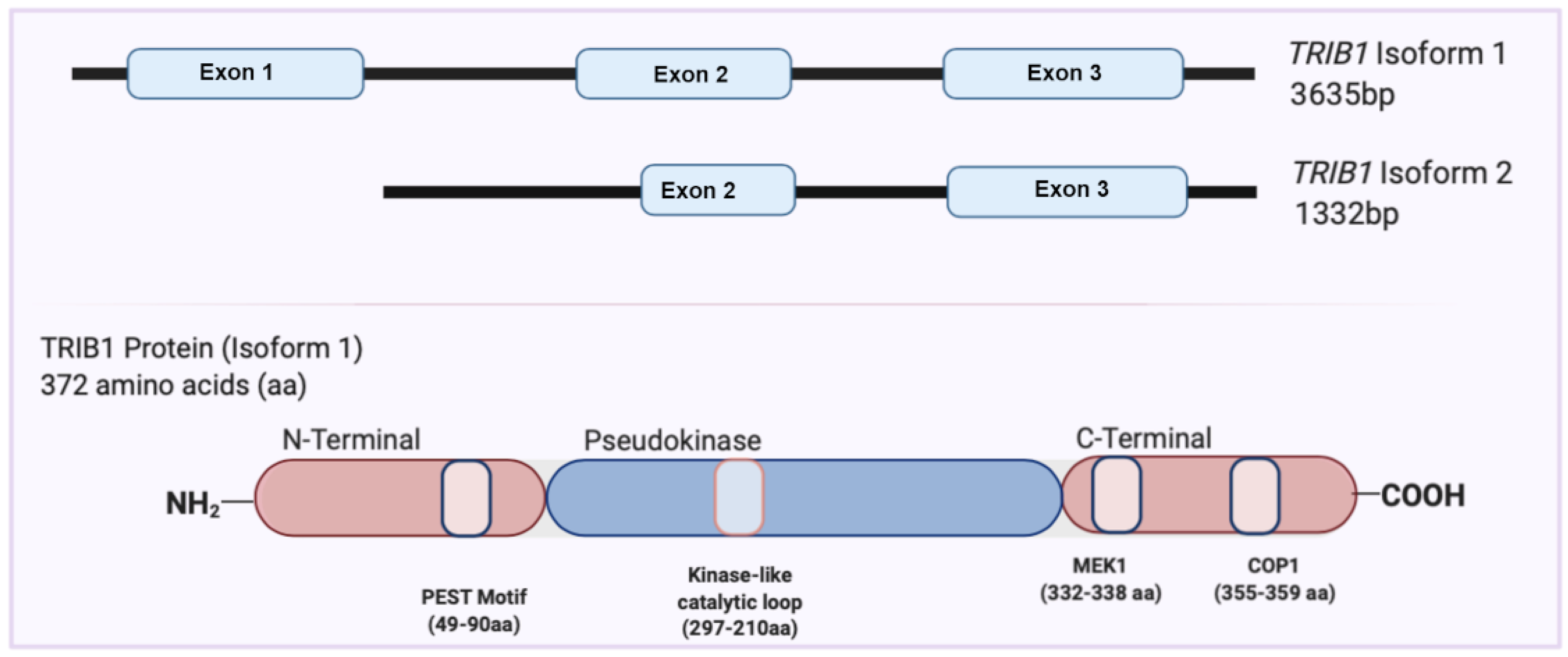
2. Overview of the TRIB1 Gene and Protein
3. A Role for TRIB1 in Physiopathology
| Disease | Description | Reference |
|---|---|---|
| Renal transplantation | Increased TRIB1 expression in PBMCs and biopsies from patients with CAMR | Ashton-Chess et al. 2008 [26] |
| Human Immunodeficiency Virus (HIV) | Overexpression of TRIB1 in PBMCs from HIV patients | Rome et al. 2020 [30] |
| Systemic Lupus Erythematosus (SLE) | Increased expression of TRIB1 in B cells from SLE patients | Garaud et al. 2011; Simoni et al. 2018 [29,43] |
| Inflammatory Bowel Disease (IBD) | TRIB1 SNP associated with IBD in patients | Liu et al. 2015; Jostins et al. 2012 [31,32] |
| Eczema | TRIB1 SNP associated with eczema | Grosche et al. 2021 [33] |
| Coronary Artery Disease | TRIB1 locus association with plasma triglycerides and coronary artery disease | Burkhardt et al. 2010; Douvris et al. 2014 [36,44] |
| Acute Myeloid Leukemia (AML) | TRIB1 induces inappropriate C/EBPα protein degradation. TRIB1 cooperates with BCL11A which represses several PU.1 target genes | Yokoyama et al. 2011; Yoshida et al. 2013; Yoshino et al. 2021; Sunami et al. 2002 [23,45,46,47] |
| Multiple Myeloma (MM) | Higher TRIB1 expression in bone marrow mononuclear cells from MM patients favoring M2 macrophage polarization | Chen et al. 2020 [48] |
| Hepatocellular Carcinoma (HCC) | TRIB1 promotes (HCC) tumorigenesis and invasiveness via the downregulation of p53 with the possible involvement of the β-catenin signaling pathway | Ye et al. 2017 [49] |
| Colorectal cancer | TRIB1 is overexpressed, similarly to MYC. TRIB1 promotes upregulation of MMP-2 through the activation of FAK/Src and ERK pathways | Y. Wang et al. 2017; Briffa et al. 2015; Camps et al. 2009 [25,50,51] |
| Prostate cancer | TRIB1 amplification and over-expression are associated with proliferation, cell survival, and metastasis. TRIB1 regulates the GRP78 endoplasmic reticulum chaperone protein, regulating Akt activation | Mashima et al. 2014; Shahrouzi et al. 2020; Moya et al. 2018 [24,52,53] |
| Non-small cell lung cancer (NSCLC) | Cisplatin treatment resulted in C/EBPβ-dependent increasing of TRIB1 which cooperates with HDAC1 to downregulate p53 activation. TRIB1 may be involved in chemotherapy resistance TRIB1 is modulated by the activation of the PI3K/AKT pathway, specifically by PIK3CA, in lung epithelial cells | L. Wang et al. 2017; De Marco et al. 2017 [54,55] |
| Glioma | TRIB1 binds to HDAC1 to inhibit p53 expression in glioma cells and participate in radioresistance | Tang et al. 2015 [56] |
| Several Amplicon 8q24 Associated Cancers | TRIB1 gene located at the same chromosomal locus 8q24.13 in close proximity to MYC; potentially co-amplified alongside | Röthlisberger et al. 2007 [21]. |
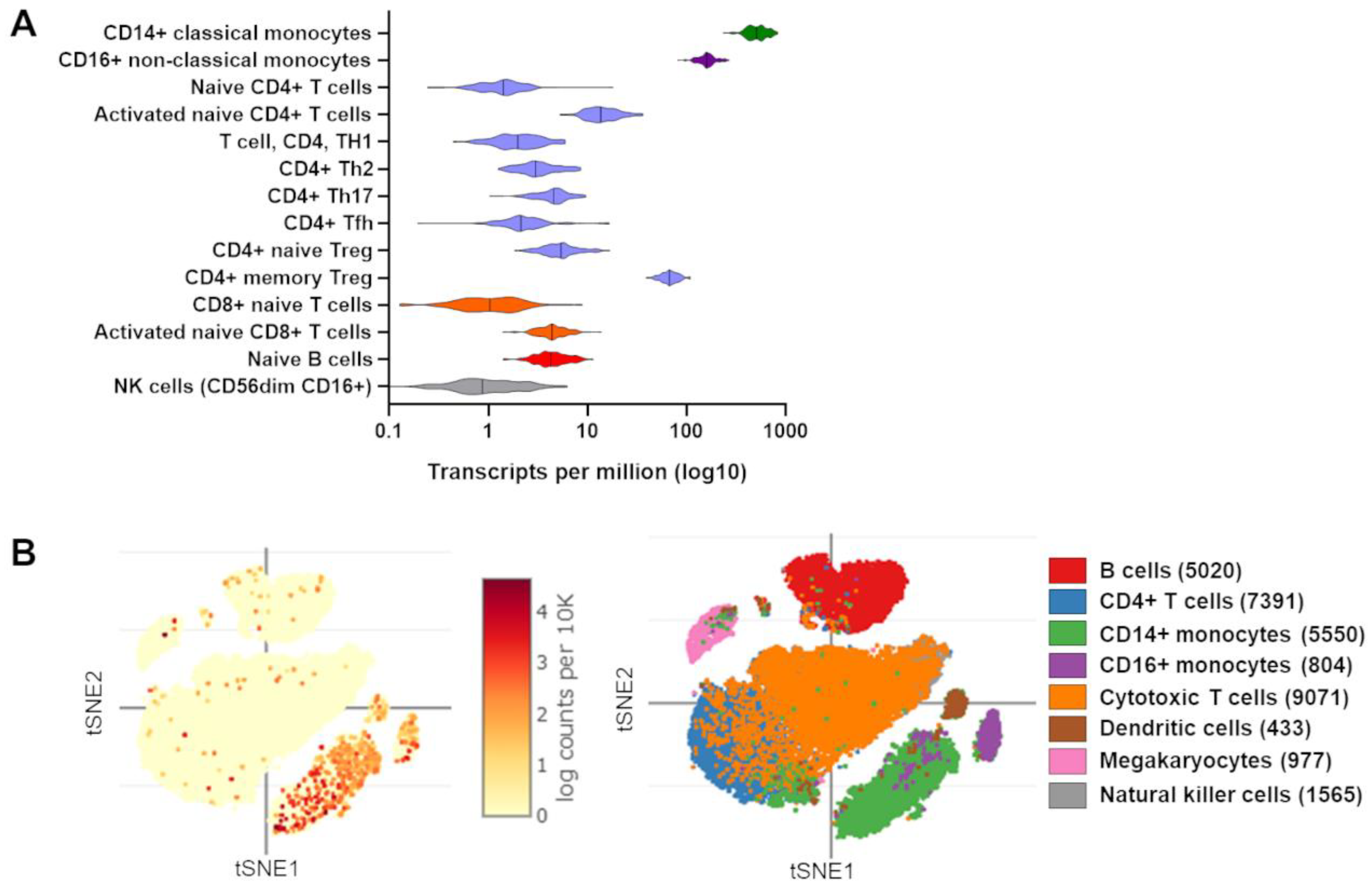
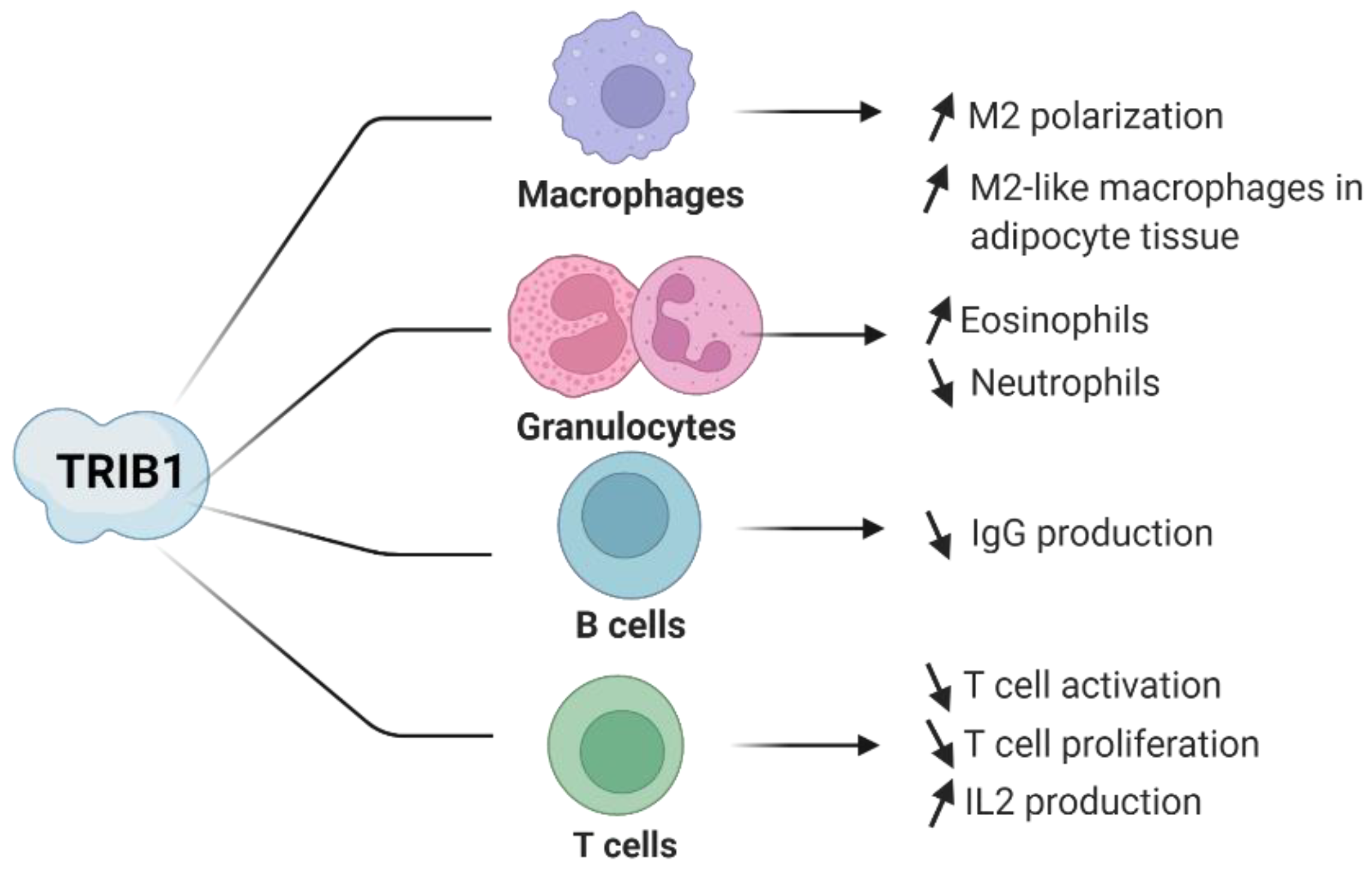
4. Evidence of TRIB1 Function in Immune Cell Subsets
4.1. TRIB1 in Macrophages
4.2. TRIB1 in CD4+ T Lymphocytes
4.3. TRIB1 in Regulatory T Cells
4.4. TRIB1 in B Lymphocytes
4.5. TRIB1 in Eosinophils and Neutrophils
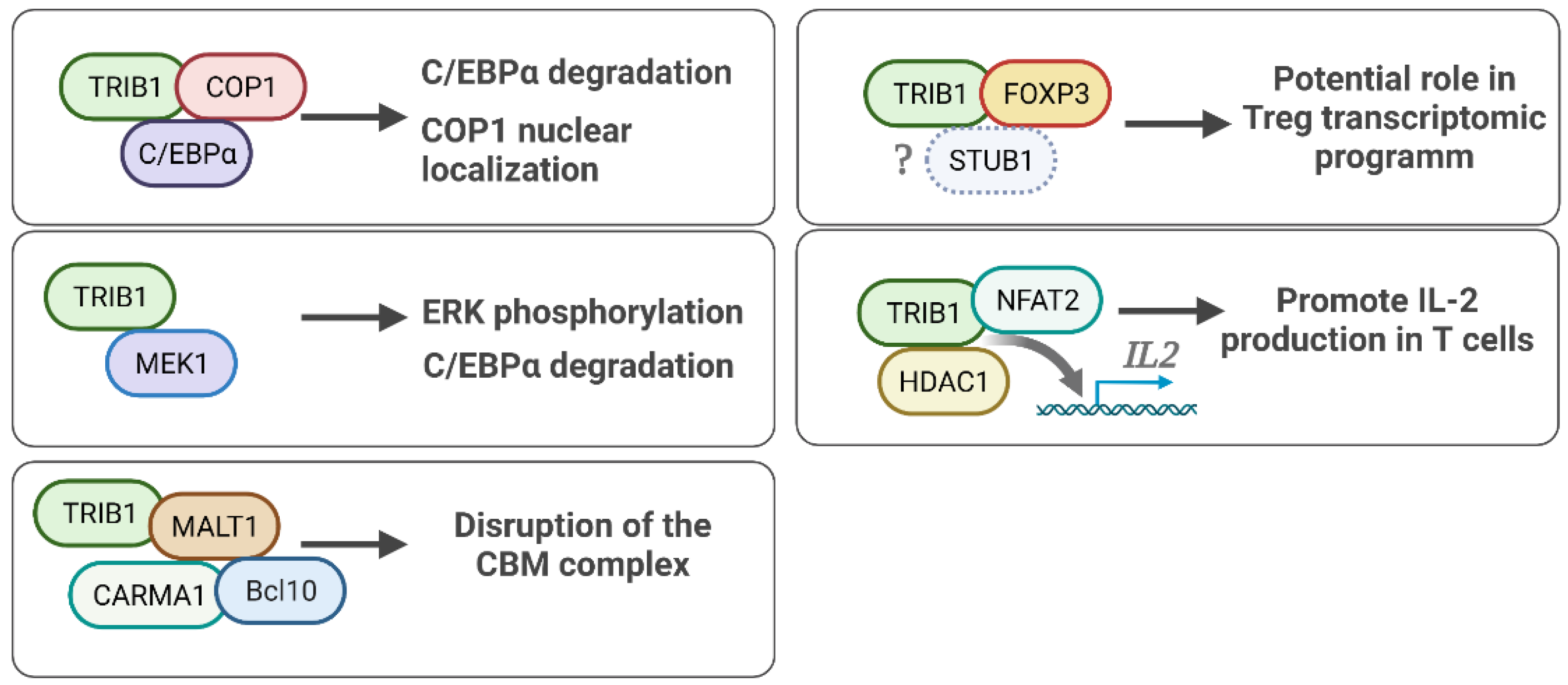
5. TRIB1 Cell Signaling and Protein Binding Partners
5.1. TRIB1-FOXP3 Protein Interaction
5.2. TRIB1-COP1 Regulation of the C/EBPα Protein
5.3. TRIB1-MEK1 in ERK/MAPK Signaling
5.4. TRIB1 Implication in NF/KB Pathway Activity
5.5. Other TRIB1 Protein Binding Partners
6. Regulation of TRIB1
6.1. Transcriptional Regulation of TRIB1
6.2. Posttranslational Modification of TRIB1
6.3. Cellular Colocalization of TRIB1
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilkin, F.; Savonet, V.; Radulescu, A.; Petermans, J.; Dumont, J.E.; Maenhaut, C. Identification and Characterization of Novel Genes Modulated in the Thyroid of Dogs Treated with Methimazole and Propylthiouracil. J. Biol. Chem. 1996, 271, 28451–28457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seher, T.C.; Leptin, M. Tribbles, a Cell-Cycle Brake That Coordinates Proliferation and Morphogenesis during Drosophila Gastrulation. Curr. Biol. 2000, 10, 623–629. [Google Scholar] [CrossRef] [Green Version]
- Mata, J.; Curado, S.; Ephrussi, A.; Rørth, P. Tribbles Coordinates Mitosis and Morphogenesis in Drosophila by Regulating String/CDC25 Proteolysis. Cell 2000, 101, 511–522. [Google Scholar] [CrossRef] [Green Version]
- Grosshans, J.; Wieschaus, E. A Genetic Link between Morphogenesis and Cell Division during Formation of the Ventral Furrow in Drosophila. Cell 2000, 101, 523–531. [Google Scholar] [CrossRef] [Green Version]
- Rørth, P.; Szabo, K.; Texido, G. The Level of C/EBP Protein Is Critical for Cell Migration during Drosophila Oogenesis and Is Tightly Controlled by Regulated Degradation. Mol. Cell 2000, 6, 23–30. [Google Scholar] [CrossRef]
- Mayumi-Matsuda, K.; Kojima, S.; Suzuki, H.; Sakata, T. Identification of a Novel Kinase-like Gene Induced during Neuronal Cell Death. Biochem. Biophys. Res. Commun. 1999, 258, 260–264. [Google Scholar] [CrossRef]
- Eyers, P.A.; Keeshan, K.; Kannan, N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2017, 27, 284–298. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Cantos, M.; Hutchison, C.E.; Shoulders, C.C. Musings from the Tribbles Research and Innovation Network. Cancers 2021, 13, 4517. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [Green Version]
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging Roles of Pseudokinases. Trends Cell Biol. 2006, 16, 443–452. [Google Scholar] [CrossRef]
- Hegedus, Z.; Czibula, A.; Kiss-Toth, E. Tribbles: A Family of Kinase-like Proteins with Potent Signalling Regulatory Function. Cell. Signal. 2007, 19, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Rogers, S.W. PEST Sequences and Regulation by Proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino Acid Sequences Common to Rapidly Degraded Proteins: The PEST Hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Wilkin, F.; Suarez-Huerta, N.; Robaye, B.; Peetermans, J.; Libert, F.; Dumont, J.E.; Maenhaut, C. Characterization of a Phosphoprotein Whose MRNA Is Regulated by the Mitogenic Pathways in Dog Thyroid Cells. Eur. J. Biochem. 1997, 248, 660–668. [Google Scholar] [CrossRef]
- Soubeyrand, S.; Martinuk, A.; Lau, P.; McPherson, R. TRIB1 Is Regulated Post-Transcriptionally by Proteasomal and Non-Proteasomal Pathways. PLoS ONE 2016, 11, e0152346. [Google Scholar] [CrossRef] [PubMed]
- Bowers, A.J.; Scully, S.; Boylan, J.F. SKIP3, a Novel Drosophila Tribbles Ortholog, Is Overexpressed in Human Tumors and Is Regulated by Hypoxia. Oncogene 2003, 22, 2823–2835. [Google Scholar] [CrossRef] [Green Version]
- Bailey, F.P.; Byrne, D.P.; Oruganty, K.; Eyers, C.E.; Novotny, C.J.; Shokat, K.M.; Kannan, N.; Eyers, P.A. The Tribbles 2 (TRB2) Pseudokinase Binds to ATP and Autophosphorylates in a Metal-Independent Manner. Biochem. J. 2015, 467, 47–62. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.M.; Nakatani, Y.; Jamieson, S.A.; Dai, W.; Lucet, I.S.; Mace, P.D. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure 2015, 23, 2111–2121. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, D.M.; Byrne, D.P.; Bailey, F.P.; Eyers, P.A. Tribbles Pseudokinases: Novel Targets for Chemical Biology and Drug Discovery? Biochem. Soc. Trans. 2015, 43, 1095–1103. [Google Scholar] [CrossRef]
- Jamieson, S.A.; Ruan, Z.; Burgess, A.E.; Curry, J.R.; McMillan, H.D.; Brewster, J.L.; Dunbier, A.K.; Axtman, A.D.; Kannan, N.; Mace, P.D. Substrate Binding Allosterically Relieves Autoinhibition of the Pseudokinase TRIB1. Sci. Signal. 2018, 11, eaau0597. [Google Scholar] [CrossRef] [Green Version]
- Röthlisberger, B.; Heizmann, M.; Bargetzi, M.J.; Huber, A.R. TRIB1 Overexpression in Acute Myeloid Leukemia. Cancer Genet. Cytogenet. 2007, 176, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Marsh, C.L.; Kurian, S.M.; Rice, J.C.; Whisenant, T.C.; David, J.; Rose, S.; Schieve, C.; Lee, D.; Case, J.; Barrick, B.; et al. Application of TruGraf v1: A Novel Molecular Biomarker for Managing Kidney Transplant Recipients With Stable Renal Function. Transplant. Proc. 2019, 51, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Kanno, Y.; Yamazaki, Y.; Takahara, T.; Miyata, S.; Dc, W.; Yokoyama, T.; Kanno, Y.; Yamazaki, Y.; Takahara, T.; et al. Trib1 Links the MEK1/ERK Pathway in Myeloid Leukemogenesis. Blood 2011, 116, 2768–2775. [Google Scholar] [CrossRef] [PubMed]
- Mashima, T.; Soma-Nagae, T.; Migita, T.; Kinoshita, R.; Iwamoto, A.; Yuasa, T.; Yonese, J.; Ishikawa, Y.; Seimiya, H. TRIB1 Supports Prostate Tumorigenesis and Tumor-Propagating Cell Survival by Regulation of Endoplasmic Reticulum Chaperone Expression. Cancer Res. 2014, 74, 4888–4897. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wu, N.; Pang, B.; Tong, D.; Sun, D.; Sun, H.; Zhang, C.; Sun, W.; Meng, X.; Bai, J.; et al. TRIB1 Promotes Colorectal Cancer Cell Migration and Invasion through Activation MMP-2 via FAK/Src and ERK Pathways. Oncotarget 2017, 8, 47931–47942. [Google Scholar] [CrossRef] [Green Version]
- Ashton-Chess, J.; Giral, M.; Mengel, M.; Renaudin, K.; Foucher, Y.; Gwinner, W.; Braud, C.; Dugast, E.; Quillard, T.; Thebault, P.; et al. Tribbles-1 as a Novel Biomarker of Chronic Antibody-Mediated Rejection. J. Am. Soc. Nephrol. JASN 2008, 19, 1116–1127. [Google Scholar] [CrossRef] [Green Version]
- Einecke, G.; Sis, B.; Reeve, J.; Mengel, M.; Campbell, P.M.; Hidalgo, L.G.; Kaplan, B.; Halloran, P.F. Antibody-Mediated Microcirculation Injury Is the Major Cause of Late Kidney Transplant Failure. Am. J. Transplant. 2009, 9, 2520–2531. [Google Scholar] [CrossRef]
- Alvarez, C.M.; Opelz, G.; Garcia, L.F.; Süsal, C. Expression of Regulatory T-Cell-Related Molecule Genes and Clinical Outcome in Kidney Transplant Recipients. Transplantation 2009, 87, 857–863. [Google Scholar] [CrossRef]
- Simoni, L.; Delgado, V.; Ruer-Laventie, J.; Bouis, D.; Soley, A.; Heyer, V.; Robert, I.; Gies, V.; Martin, T.; Korganow, A.S.; et al. Trib1 Is Overexpressed in Systemic Lupus Erythematosus, While It Regulates Immunoglobulin Production in Murine B Cells. Front. Immunol. 2018, 9, 373. [Google Scholar] [CrossRef]
- Rome, K.S.; Stein, S.J.; Kurachi, M.; Petrovic, J.; Schwartz, G.W.; Mack, E.A.; Uljon, S.; Wu, W.W.; Dehart, A.G.; Mcclory, S.E.; et al. Trib1 Regulates T Cell Differentiation during Chronic Infection by Restraining the Effector Program. J. Exp. Med. 2020, 217, e20190888. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Philip Schumm, L.; Sharma, Y.; Anderson, C.A.; et al. Host-Microbe Interactions Have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Z.; Van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association Analyses Identify 38 Susceptibility Loci for Inflammatory Bowel Disease and Highlight Shared Genetic Risk across Populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Grosche, S.; Marenholz, I.; Esparza-Gordillo, J.; Arnau-Soler, A.; Pairo-Castineira, E.; Rüschendorf, F.; Ahluwalia, T.S.; Almqvist, C.; Arnold, A.; Australian Asthma Genetics Consortium (AAGC); et al. Rare Variant Analysis in Eczema Identifies Exonic Variants in DUSP1, NOTCH4 and SLC9A4. Nat. Commun. 2021, 12, 6618. [Google Scholar] [CrossRef] [PubMed]
- Belarif, L.; Danger, R.; Kermarrec, L.; Nerrière-daguin, V.; Pengam, S.; Durand, T.; Mary, C.; Kerdreux, E.; Gauttier, V.; Kucik, A.; et al. IL-7 Receptor Influences Anti-TNF Responsiveness and T Cell Gut Homing in Inflammatory Bowel Disease. J. Clin. Investig. 2019, 129, 1910–1925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, J.M.; Angyal, A.; Bauer, R.C.; Hamby, S.; Suvarna, S.K.; Baidžajevas, K.; Hegedus, Z.; Dear, T.N.; Turner, M.; Wilson, H.L.; et al. Myeloid Tribbles 1 Induces Early Atherosclerosis via Enhanced Foam Cell Expansion. Sci. Adv. 2019, 5, eaax9183. [Google Scholar] [CrossRef] [Green Version]
- Burkhardt, R.; Toh, S.-A.; Lagor, W.R.; Birkeland, A.; Levin, M.; Li, X.; Robblee, M.; Fedorov, V.D.; Yamamoto, M.; Satoh, T.; et al. Trib1 Is a Lipid- and Myocardial Infarction—Associated Gene That Regulates Hepatic Lipogenesis and VLDL Production in Mice. J. Clin. Investig. 2010, 120, 4410–4414. [Google Scholar] [CrossRef]
- Iwamoto, S.; Boonvisut, S.; Makishima, S.; Ishizuka, Y.; Watanabe, K.; Nakayama, K. The Role of TRIB1 in Lipid Metabolism; from Genetics to Pathways. Biochem. Soc. Trans. 2015, 43, 1063–1068. [Google Scholar] [CrossRef]
- Tai, E.S.; Sim, X.L.; Ong, T.H.; Wong, T.Y.; Saw, S.M.; Aung, T.; Kathiresan, S.; Orho-Melander, M.; Ordovas, J.M.; Tan, J.T.; et al. Polymorphisms at Newly Identified Lipid-Associated Loci Are Associated with Blood Lipids and Cardiovascular Disease in an Asian Malay Population. J. Lipid Res. 2009, 50, 514–520. [Google Scholar] [CrossRef] [Green Version]
- Ishizuka, Y.; Nakayama, K.; Ogawa, A.; Makishima, S.; Boonvisut, S.; Hirao, A.; Iwasaki, Y.; Yada, T.; Yanagisawa, Y.; Miyashita, H.; et al. TRIB1 Downregulates Hepatic Lipogenesis and Glycogenesis via Multiple Molecular Interactions. J. Mol. Endocrinol. 2014, 52, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Bauer, R.C.; Sasaki, M.; Cohen, D.M.; Cui, J.; Smith, M.A.; Yenilmez, B.O.; Steger, D.J.; Rader, D.J. Tribbles-1 Regulates Hepatic Lipogenesis through Posttranscriptional Regulation of C/EBP α. J. Clin. Investig. 2015, 125, 3809–3818. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, B.I.; Santos, B.; Link, W.; De Sousa-Coelho, A.L. Tribbles Pseudokinases in Colorectal Cancer. Cancers 2021, 13, 2825. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.L.; O’Connor, C.; Veiga, J.P.; McCarthy, T.V.; Keeshan, K. TRIB2 Regulates Normal and Stress-Induced Thymocyte Proliferation. Cell Discov. 2016, 2, 15050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garaud, J.C.; Schickel, J.N.; Blaison, G.; Knapp, A.M.; Dembele, D.; Ruer-Laventie, J.; Korganow, A.S.; Martin, T.; Soulas-Sprauel, P.; Pasquali, J.L. B Cell Signature during Inactive Systemic Lupus Is Heterogeneous: Toward a Biological Dissection of Lupus. PLoS ONE 2011, 6, e23900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douvris, A.; Soubeyrand, S.; Naing, T.; Martinuk, A.; Nikpay, M.; Williams, A.; Buick, J.; Yauk, C.; McPherson, R. Functional Analysis of the TRIB1 Associated Locus Linked to Plasma Triglycerides and Coronary Artery Disease. J. Am. Heart Assoc. 2014, 3, e000884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, A.; Kato, J.-Y.; Nakamae, I.; Yoneda-Kato, N. COP1 Targets C/EBPα for Degradation and Induces Acute Myeloid Leukemia via Trib1. Blood 2013, 122, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, S.; Tanaka, M.; Sunami, Y.; Takahara, T.; Yamazaki, Y.; Homme, M.; Niibori-Nambu, A.; Osato, M.; Minami, T.; Ishihara, K.; et al. Trib1 Promotes the Development of Acute Myeloid Leukemia in a Ts1Cje Mouse Model of Down Syndrome. Leukemia 2021, 36, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Sunami, Y.; Yokoyama, T.; Yoshino, S.; Takahara, T.; Yamazaki, Y.; Harada, H.; Nakamura, T. BCL11A Promotes Myeloid Leukemogenesis by Repressing PU.1 Target Genes. Blood Adv. 2021. [Google Scholar] [CrossRef]
- Chen, H.; Li, M.; Sanchez, E.; Soof, C.M.; Bujarski, S.; Ng, N.; Cao, J.; Hekmati, T.; Zahab, B.; Nosrati, J.D.; et al. JAK1/2 Pathway Inhibition Suppresses M2 Polarization and Overcomes Resistance of Myeloma to Lenalidomide by Reducing TRIB1, MUC1, CD44, CXCL12, and CXCR4 Expression. Br. J. Haematol. 2020, 188, 283–294. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, G.; Wang, G.; Zhuang, J.; He, S.; Song, Y.; Ni, J.; Xia, W.; Wang, J. The Oncogenic Role of Tribbles 1 in Hepatocellular Carcinoma Is Mediated by a Feedback Loop Involving MicroRNA-23a and P53. Front. Physiol. 2017, 8, 789. [Google Scholar] [CrossRef] [Green Version]
- Briffa, R.; Um, I.; Faratian, D.; Zhou, Y.; Turnbull, A.K.; Langdon, S.P.; Harrison, D.J. Multi-Scale Genomic, Transcriptomic and Proteomic Analysis of Colorectal Cancer Cell Lines to Identify Novel Biomarkers. PLoS ONE 2015, 10, e0144708. [Google Scholar] [CrossRef] [Green Version]
- Camps, J.; Nguyen, Q.T.; Padilla-Nash, H.M.; Knutsen, T.; McNeil, N.E.; Wangsa, D.; Hummon, A.B.; Grade, M.; Ried, T.; Difilippantonio, M.J. Integrative Genomics Reveals Mechanisms of Copy Number Alterations Responsible for Transcriptional Deregulation in Colorectal Cancer. Genes Chromosomes Cancer 2009, 48, 1002–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahrouzi, P.; Astobiza, I.; Cortazar, A.R.; Torrano, V.; Macchia, A.; Flores, J.M.; Niespolo, C.; Mendizabal, I.; Caloto, R.; Ercilla, A.; et al. Genomic and Functional Regulation of TRIB1 Contributes to Prostate Cancer Pathogenesis. Cancers 2020, 12, 2593. [Google Scholar] [CrossRef] [PubMed]
- Moya, L.; Lai, J.; Hoffman, A.; Srinivasan, S.; Panchadsaram, J.; Chambers, S.; Clements, J.A.; Batra, J. Australian Prostate Cancer BioResource Association Analysis of a Microsatellite Repeat in the TRIB1 Gene with Prostate Cancer Risk, Aggressiveness and Survival. Front. Genet. 2018, 9, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Liu, X.; Ren, Y.; Zhang, J.; Chen, J.; Zhou, W.; Guo, W.; Wang, X.; Chen, H.; Li, M.; et al. Cisplatin-Enriching Cancer Stem Cells Confer Multidrug Resistance in Non-Small Cell Lung Cancer via Enhancing TRIB1/HDAC Activity. Cell Death Dis. 2017, 8, e2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marco, C.; Laudanna, C.; Rinaldo, N.; Oliveira, D.M.; Ravo, M.; Weisz, A.; Ceccarelli, M.; Caira, E.; Rizzuto, A.; Zoppoli, P.; et al. Specific Gene Expression Signatures Induced by the Multiple Oncogenic Alterations That Occur within the PTEN/PI3K/AKT Pathway in Lung Cancer. PLoS ONE 2017, 12, e0178865. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Wu, W.; Zhang, Q.; Sun, Y.; Cui, Y.; Wu, F.; Wei, X.; Qi, G.; Liang, X.; Tang, F.; et al. Inhibition of Tribbles Protein-1 Attenuates Radioresistance in Human Glioma Cells. Sci. Rep. 2015, 5, 15961. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Dugast, E.; Kiss-Toth, E.; Docherty, L.; Danger, R.; Chesneau, M.; Pichard, V.; Judor, J.P.; Pettré, S.; Conchon, S.; Soulillou, J.P.; et al. Identification of Tribbles-1 as a Novel Binding Partner of Foxp3 in Regulatory T Cells. J. Biol. Chem. 2013, 288, 10051–10060. [Google Scholar] [CrossRef] [Green Version]
- Satoh, T.; Kidoya, H.; Naito, H.; Yamamoto, M.; Takemura, N.; Nakagawa, K.; Yoshioka, Y.; Morii, E.; Takakura, N.; Takeuchi, O.; et al. Critical Role of Trib1 in Differentiation of Tissue-Resident M2-like Macrophages. Nature 2013, 495, 524–528. [Google Scholar] [CrossRef]
- Mack, E.A.; Stein, S.J.; Rome, K.S.; Xu, L.; Wertheim, G.B.; Melo, R.C.N.; Pear, W.S. Trib1 Regulates Eosinophil Lineage Commitment and Identity by Restraining the Neutrophil Program. Blood 2019, 133, 2413–2426. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.; Adiconis, X.; Simmons, S.K.; Kowalczyk, M.S.; Hession, C.C.; Marjanovic, N.D.; Hughes, T.K.; Wadsworth, M.H.; Burks, T.; Nguyen, L.T.; et al. Systematic Comparative Analysis of Single Cell RNA-Sequencing Methods. bioRxiv 2019, 632216. [Google Scholar] [CrossRef]
- Miyajima, C.; Itoh, Y.; Inoue, Y.; Hayashi, H. Positive Regulation of Interleukin-2 Expression by a Pseudokinase, Tribbles 1, in Activated T Cells. Biol. Pharm. Bull. 2015, 38, 1126–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndt, L.; Dokas, J.; Gericke, M.; Kutzner, C.E.; Müller, S.; Thiery, J.; Burkhardt, R. Tribbles Homolog 1 Deficiency Modulates Function and Polarization of Murine Bone Marrow-Derived Macrophages. J. Biol. Chem. 2018, 293, 11527–11536. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Uematsu, S.; Okamoto, T.; Matsuura, Y.; Sato, S.; Kumar, H.; Satoh, T.; Saitoh, T.; Takeda, K.; Ishii, K.J.; et al. Enhanced TLR-Mediated NF-IL6–Dependent Gene Expression by Trib1 Deficiency. J. Exp. Med. 2007, 204, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Uljon, S.; Xu, X.; Durzynska, I.; Stein, S.; Adelmant, G.; Marto, J.A.; Pear, W.S.; Blacklow, S.C. Structural Basis for Substrate Selectivity of the E3 Ligase COP1. Structure 2016, 24, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef]
- Ferraro, A.; Morena, A.; Alise, D.; Raj, T.; Asinovski, N.; Phillips, R.; Ergun, A.; D’Alise, A.M.; Raj, T.; Asinovski, N.; et al. Interindividual Variation in Human T Regulatory Cells. Proc. Natl. Acad. Sci. USA 2014, 111, 34–37. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Josefowicz, S.Z.; Kas, A.; Chu, T.-T.; Gavin, M.A.; Rudensky, A.Y. Genome-Wide Analysis of Foxp3 Target Genes in Developing and Mature Regulatory T Cells. Nature 2007, 445, 936–940. [Google Scholar] [CrossRef]
- Bhairavabhotla, R.; Kim, Y.C.; Glass, D.D.; Escobar, T.M.; Patel, M.C.; Zahr, R.; Nguyen, C.K.; Kilaru, G.K.; Muljo, S.A.; Shevach, E.M. Transcriptome Profiling of Human FoxP3+ Regulatory T Cells. Hum. Immunol. 2016, 77, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadlon, T.J.; Wilkinson, B.G.; Pederson, S.; Brown, C.Y.; Bresatz, S.; Gargett, T.; Melville, E.L.; Peng, K.; D’Andrea, R.J.; Glonek, G.G.; et al. Genome-Wide Identification of Human FOXP3 Target Genes in Natural Regulatory T Cells. J. Immunol. 2010, 185, 1071–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontenot, J.D.; Rasmussen, J.P.; Williams, L.M.; Dooley, J.L.; Farr, A.G.; Rudensky, A.Y. Regulatory T Cell Lineage Specification by the Forkhead Transcription Factor Foxp3. Immunity 2005, 22, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.; Gewies, A.; Pechloff, K.; Heuser, C.; Engleitner, T.; Gehring, T.; Hartjes, L.; Krebs, S.; Krappmann, D.; Kriegsmann, M.; et al. Bcl10-Controlled Malt1 Paracaspase Activity Is Key for the Immune Suppressive Function of Regulatory T Cells. Nat. Commun. 2019, 10, 2352. [Google Scholar] [CrossRef] [PubMed]
- Nakamae, I.; Kato, J.Y.; Yokoyama, T.; Ito, H.; Yoneda-Kato, N. Myeloid Leukemia Factor 1 Stabilizes Tumor Suppressor C/EBPα to Prevent Trib1-Driven Acute Myeloid Leukemia. Blood Adv. 2017, 1, 1682–1693. [Google Scholar] [CrossRef]
- Yokoyama, T.; Toki, T.; Aoki, Y.; Kanezaki, R.; Park, M.J.; Kanno, Y.; Takahara, T.; Yamazaki, Y.; Ito, E.; Hayashi, Y.; et al. Identification of TRIB1 R107L Gain-of-Function Mutation in Human Acute Megakaryocytic Leukemia. Blood 2012, 119, 2608–2611. [Google Scholar] [CrossRef]
- Hernández-Quiles, M.; Baak, R.; Borgman, A.; den Haan, S.; Sobrevals Alcaraz, P.; van Es, R.; Kiss-Toth, E.; Vos, H.; Kalkhoven, E. Comprehensive Profiling of Mammalian Tribbles Interactomes Implicates TRIB3 in Gene Repression. Cancers 2021, 13, 6318. [Google Scholar] [CrossRef]
- Kung, J.E.; Jura, N. The Pseudokinase TRIB 1 Toggles an Intramolecular Switch to Regulate COP 1 Nuclear Export. EMBO J. 2019, 38, e99708. [Google Scholar] [CrossRef]
- Chen, Z.; Barbi, J.; Bu, S.; Yang, H.-Y.; Li, Z.; Gao, Y.; Jinasena, D.; Fu, J.; Lin, F.; Chen, C.; et al. The Ubiquitin Ligase Stub1 Negatively Modulates Regulatory T Cell Suppressive Activity by Promoting Degradation of the Transcription Factor Foxp3. Immunity 2013, 39, 272–285. [Google Scholar] [CrossRef] [Green Version]
- Qi, M.; Elion, E.A. MAP Kinase Pathways. J. Cell Sci. 2005, 118, 3569–3572. [Google Scholar] [CrossRef] [Green Version]
- Raman, M.; Chen, W.; Cobb, M.H. Differential Regulation and Properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef] [Green Version]
- Eves, E.M.; Rosner, M.R.; Keshet, Y.; Seger, R. MAP Kinase Signaling of the Mitotic Spindle Checkpoint. Methods Mol. Biol. 2010, 661, 497–505. [Google Scholar] [PubMed]
- Kiss-Toth, E.; Bagstaff, S.M.; Sung, H.Y.; Jozsa, V.; Dempsey, C.; Caunt, J.C.; Oxley, K.M.; Wyllie, D.H.; Polgar, T.; Harte, M.; et al. Human Tribbles, a Protein Family Controlling Mitogen-Activated Protein Kinase Cascades. J. Biol. Chem. 2004, 279, 42703–42708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.Y.; Francis, S.E.; Crossman, D.C.; Kiss-Toth, E. Regulation of Expression and Signalling Modulator Function of Mammalian Tribbles Is Cell-Type Specific. Immunol. Lett. 2006, 104, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Ostertag, A.; Jones, A.; Rose, A.J.; Liebert, M.; Kleinsorg, S.; Reimann, A.; Vegiopoulos, A.; Diaz, M.B.; Strzoda, D.; Yamamoto, M.; et al. Control of Adipose Tissue Inflammation through TRB1. Diabetes 2010, 59, 1991–2000. [Google Scholar] [CrossRef] [Green Version]
- Gendelman, R.; Xing, H.; Mirzoeva, O.K.; Sarde, P.; Curtis, C.; Feiler, H.S.; McDonagh, P.; Gray, J.W.; Khalil, I.; Korn, W.M. Bayesian Network Inference Modeling Identifies TRIB1 as a Novel Regulator of Cell-Cycle Progression and Survival in Cancer Cells. Cancer Res. 2017, 77, 1575–1585. [Google Scholar] [CrossRef] [Green Version]
- Imajo, M.; Nishida, E. Human Tribbles Homolog 1 Functions as a Negative Regulator of Retinoic Acid Receptor. Genes Cells 2010, 15, 1089–1097. [Google Scholar] [CrossRef]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S.H. Database for MRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2009, 16, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.-Y.; Huang, Y.-Q.; Zhang, Y.-Q.; Han, Z.-D.; He, H.-C.; Ling, X.-H.; Fu, X.; Dai, Q.-S.; Cai, C.; Chen, J.-H.; et al. MicroRNA-224 Inhibits Progression of Human Prostate Cancer by Downregulating TRIB1. Int. J. Cancer 2014, 135, 541–550. [Google Scholar] [CrossRef]
- Bonzheim, I.; Irmler, M.; Klier-Richter, M.; Steinhilber, J.; Anastasov, N.; Schäfer, S.; Adam, P.; Beckers, J.; Raffeld, M.; Fend, F.; et al. Identification of C/EBPβ Target Genes in ALK+ Anaplastic Large Cell Lymphoma (ALCL) by Gene Expression Profiling and Chromatin Immunoprecipitation. PLoS ONE 2013, 8, e64544. [Google Scholar] [CrossRef]
- Orea-Soufi, A.; Castillo-Lluva, S.; Salvador-Tormo, N.; Martín-Cabrera, P.; Recuero, S.; Gabicagogeascoa, E.; Moreno-Valladares, M.; Mendiburu-Eliçabe, M.; Blanco-Gómez, A.; Ramos-Pittol, J.M.; et al. The Pseudokinase TRIB3 Negatively Regulates the HER2 Receptor Pathway and Is a Biomarker of Good Prognosis in Luminal Breast Cancer. Cancers 2021, 13, 5307. [Google Scholar] [CrossRef]
- Kiss-Toth, E.; Wyllie, D.H.; Holland, K.; Marsden, L.; Jozsa, V.; Oxley, K.M.; Polgar, T.; Qwarnstrom, E.E.; Dower, S.K. Functional Mapping and Identification of Novel Regulators for the Toll/Interleukin-1 Signalling Network by Transcription Expression Cloning. Cell. Signal. 2006, 18, 202–214. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danger, R.; Feseha, Y.; Brouard, S. The Pseudokinase TRIB1 in Immune Cells and Associated Disorders. Cancers 2022, 14, 1011. https://doi.org/10.3390/cancers14041011
Danger R, Feseha Y, Brouard S. The Pseudokinase TRIB1 in Immune Cells and Associated Disorders. Cancers. 2022; 14(4):1011. https://doi.org/10.3390/cancers14041011
Chicago/Turabian StyleDanger, Richard, Yodit Feseha, and Sophie Brouard. 2022. "The Pseudokinase TRIB1 in Immune Cells and Associated Disorders" Cancers 14, no. 4: 1011. https://doi.org/10.3390/cancers14041011
APA StyleDanger, R., Feseha, Y., & Brouard, S. (2022). The Pseudokinase TRIB1 in Immune Cells and Associated Disorders. Cancers, 14(4), 1011. https://doi.org/10.3390/cancers14041011







