GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Isolation, Library Construction, and High-throughput Sequencing
2.3. Screening of Promising Drug Targets
2.4. RNA Extraction and cDNA Synthesis
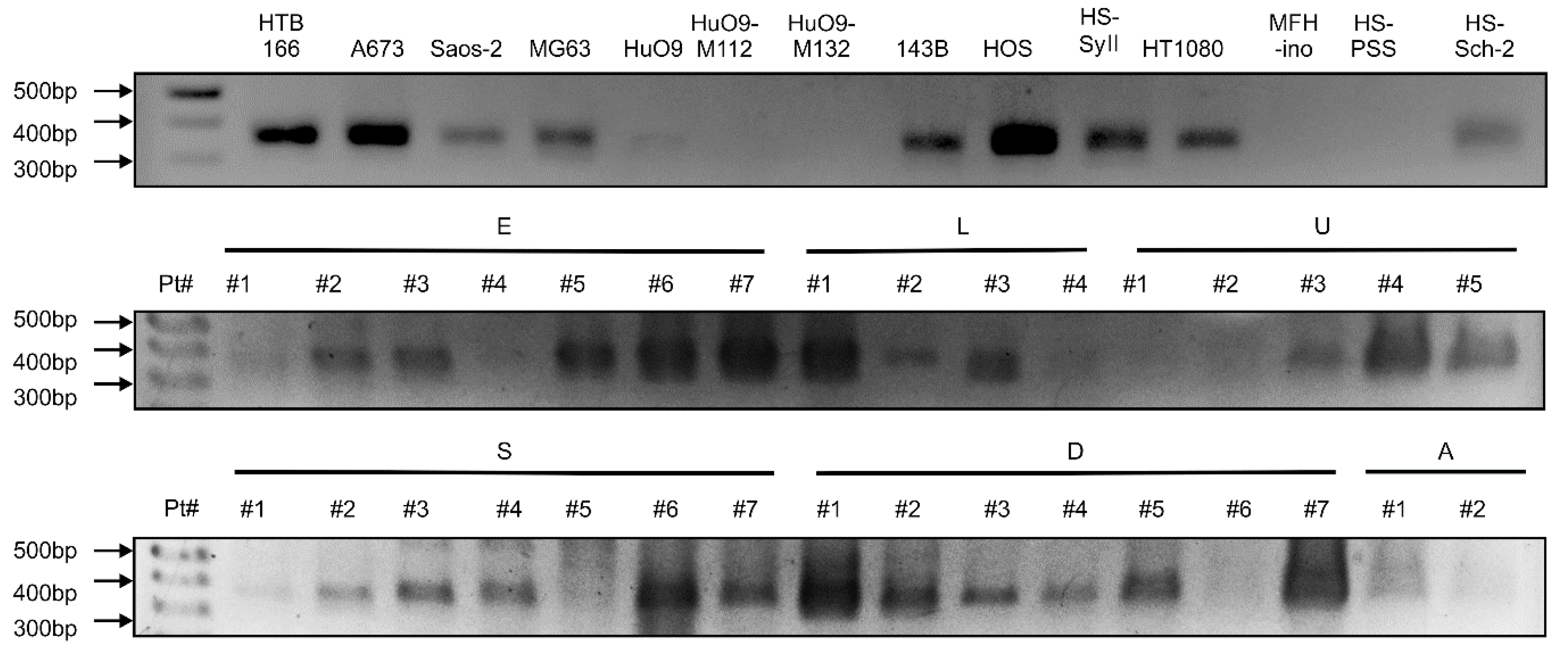
2.5. PCR Analysis
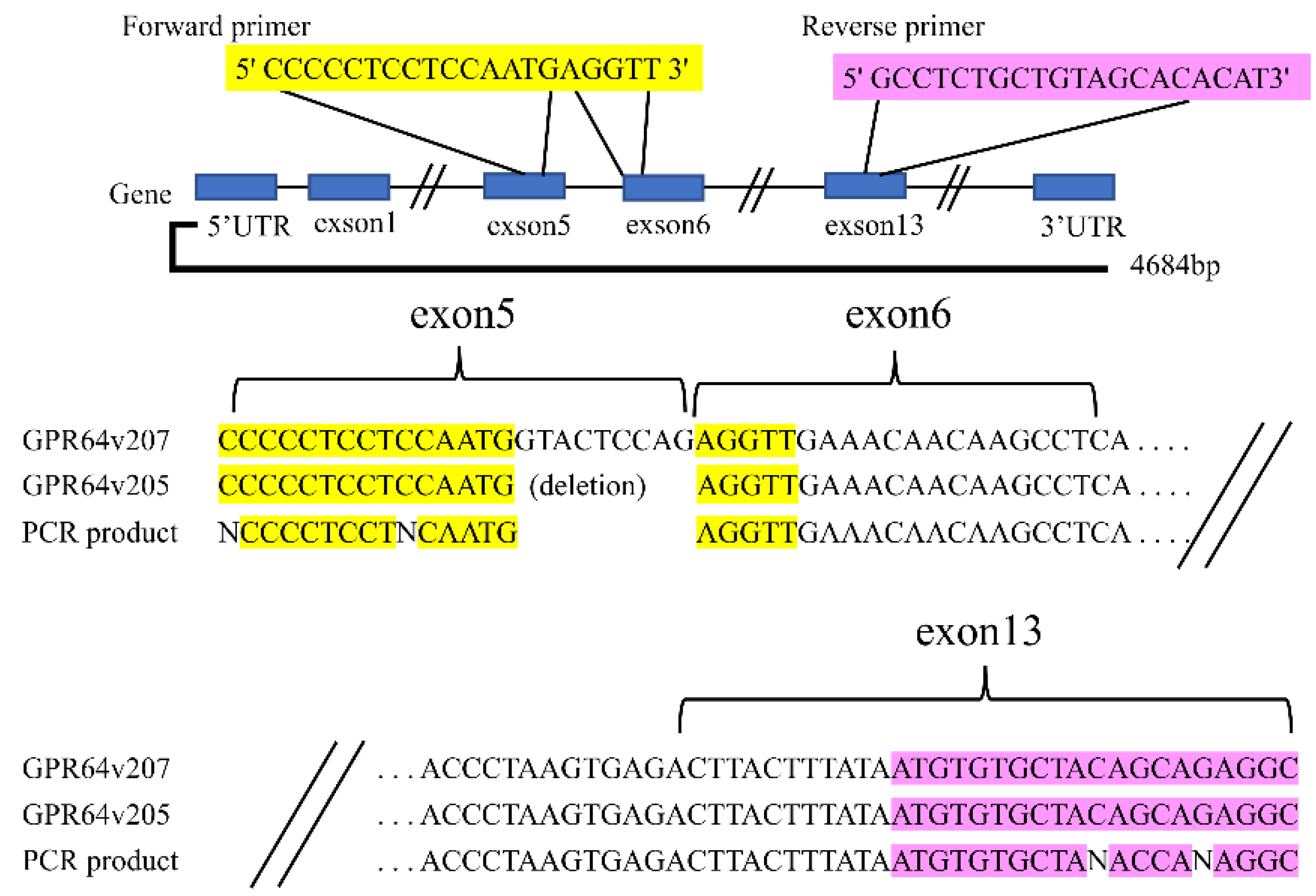
2.6. Direct Sequencing of the GPR64 Splicing Variant Cassette (Exons 5 to 13)
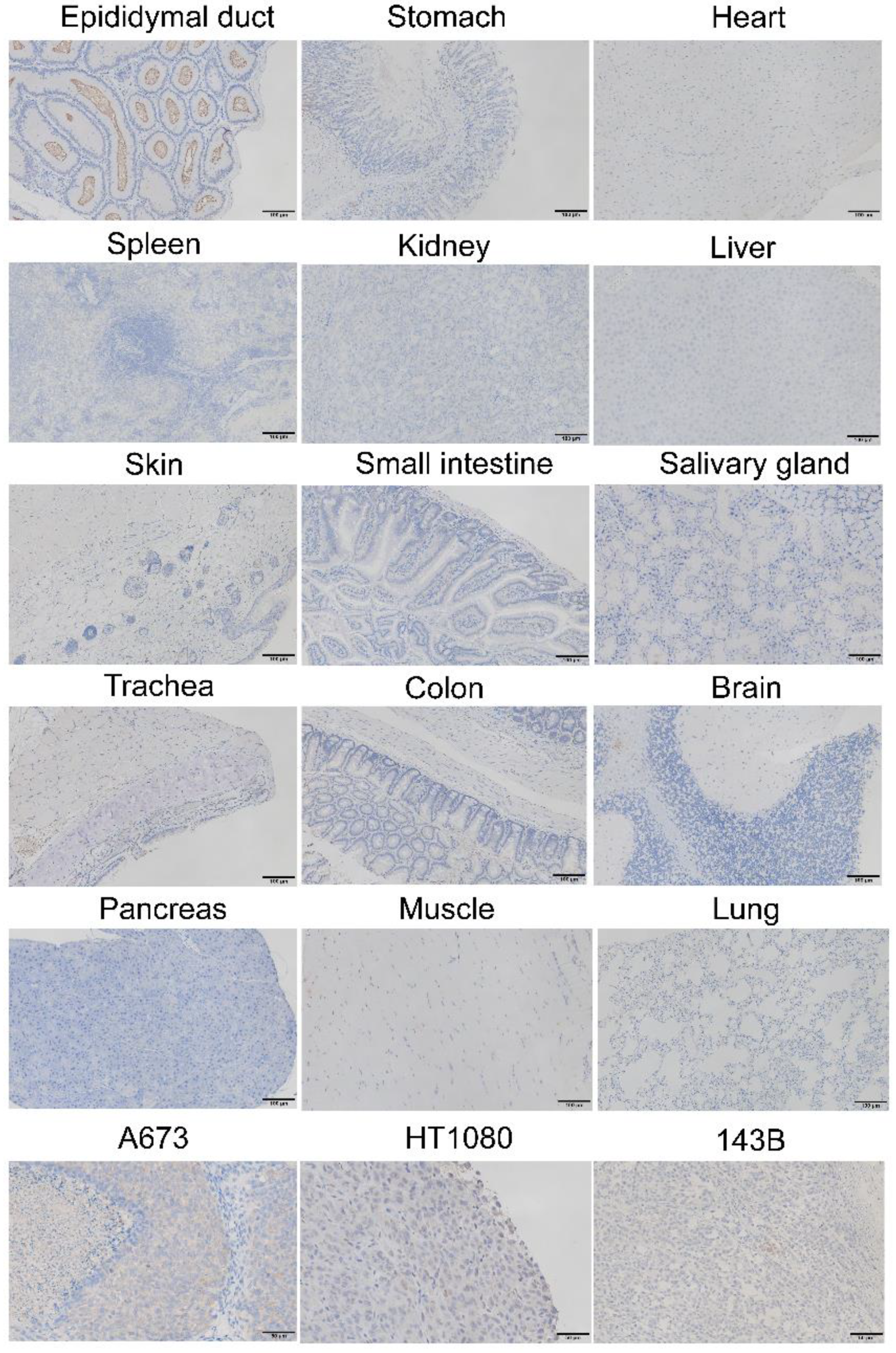
2.7. Immunohistochemistry of Surgical Specimens
2.8. Quantification of Immunohistochemical Staining for Surgical Specimens
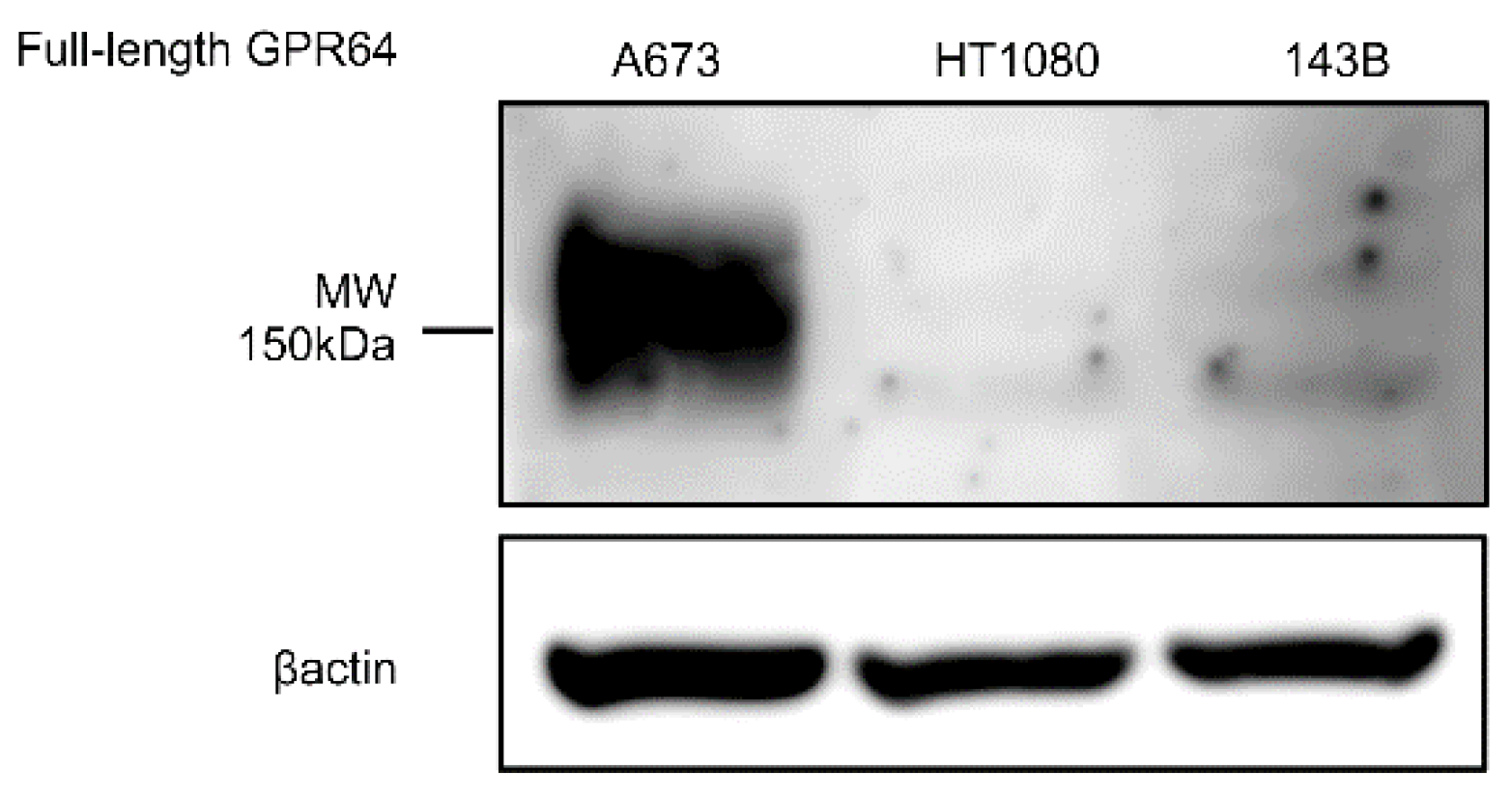
2.9. Western Blot Analysis
2.10. Immunocytochemistry
2.11. Xenograft Model
2.12. In Vivo Near-infrared (NIR) Optical Imaging
2.13. Ex Vivo NIR Optical Imaging and Immunohistochemistry
2.14. Fluorescence Image Analysis
2.15. Statistical Analysis
3. Results
3.1. Cell Surface Protein Data Reveal Therapeutic Targets for Ewing Sarcoma
3.2. Primer Design of GPR64v205 and Direct Sequencing of the PCR Product
3.3. GPR64v205 mRNA Is Expressed in Sarcomas
3.4. GPR64 Protein Is Expressed in Sarcoma Tissues
3.5. Sarcoma Cell Lines for In Vivo Study Express GPR64 Protein
3.6. Binding Capacity of the Anti-GPR64 Antibody under Non-Denaturing Conditions
3.7. In Vivo NIR Optical Imaging
3.8. Ex Vivo NIR Optical Imaging
3.9. Immunohistochemical Staining of the Resected Tissues
4. Discussion
4.1. G Protein-Coupled Receptors, GPR64, and Ewing Sarcoma
4.2. Alternative Splicing
4.3. RNA Expression Rate
4.4. Protein Expression Rate
4.5. Immunocytochemistry
4.6. NIR Imaging
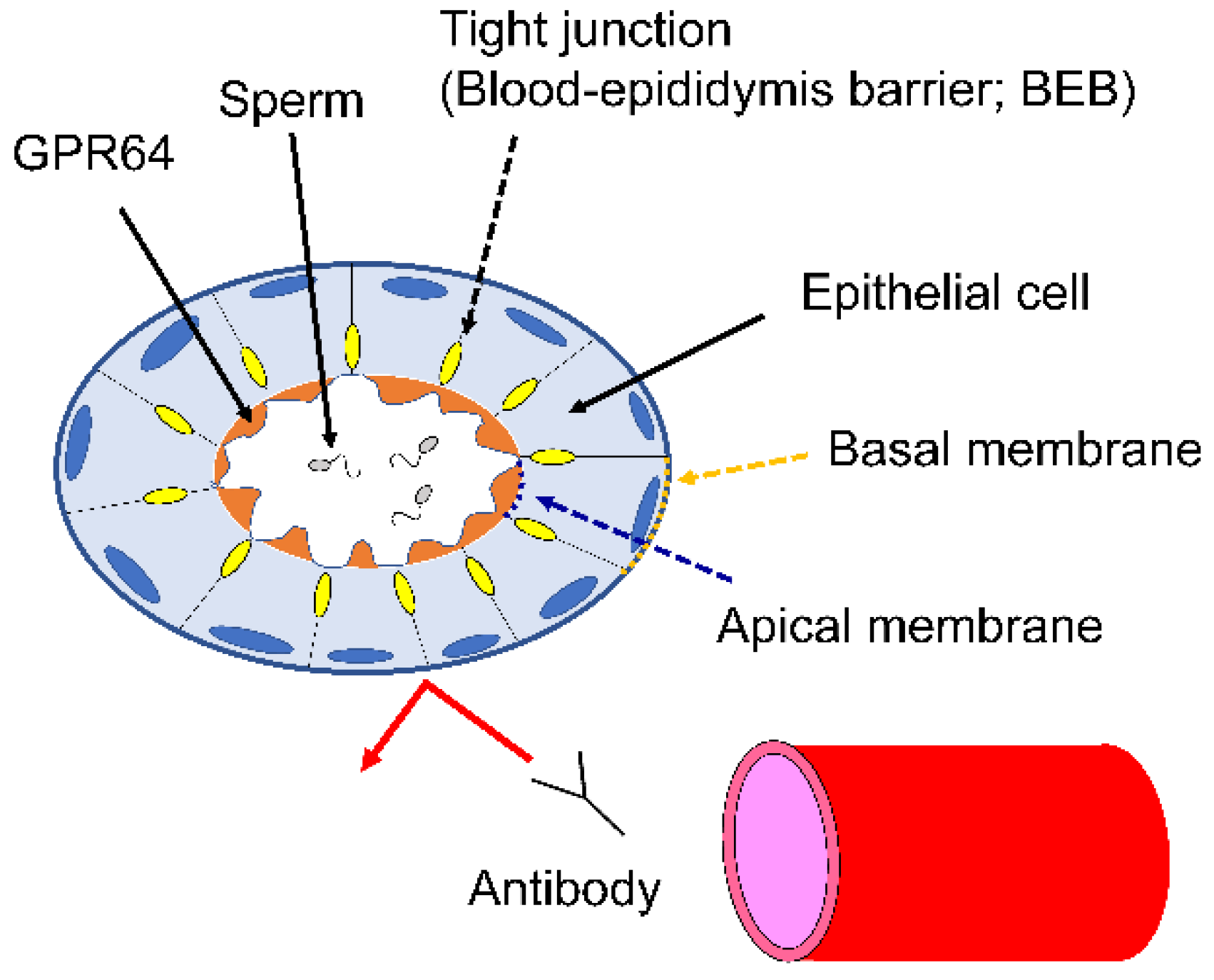
4.7. BEB
4.8. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sankar, S.; Lessnick, S.L. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011, 204, 351–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorlick, R.; Janeway, K.; Lessnick, S.; Randall, R.L.; Marina, N.; COG Bone Tumor Committee. Children’s Oncology Group’s 2013 blueprint for research: Bone tumors. Pediatr. Blood Cancer 2013, 60, 1009–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbiah, V.; Anderson, P.; Lazar, A.J.; Burdett, E.; Raymond, K.; Ludwig, J.A. Ewing’s sarcoma: Standard and experimental treatment options. Curr. Treat. Options Oncol. 2009, 10, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Town, J.; Pais, H.; Harrison, S.; Stead, L.F.; Bataille, C.; Bunjobpol, W.; Zhang, J.; Rabbitts, T.H. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc. Natl Acad. Sci. USA 2016, 113, 3603–3608. [Google Scholar] [CrossRef] [Green Version]
- Weiner, L.M.; Surana, R.; Wang, S. Monoclonal antibodies: Versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010, 10, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Filley, A.C.; Henriquez, M.; Dey, M. CART immunotherapy: Development, success, and translation to malignant gliomas and other solid tumors. Front. Oncol. 2018, 8, 453. [Google Scholar] [CrossRef] [Green Version]
- Mitsunaga, M.; Ogawa, M.; Kosaka, N.; Rosenblum, L.T.; Choyke, P.L.; Kobayashi, H. Cancer cell-selective in vivo near infrared photoimmunotherapy targeting specific membrane molecules. Nat. Med. 2011, 17, 1685–1691. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Wakiyama, H.; Furusawa, A.; Choyke, P.L.; Kobayashi, H. Near infrared photoimmunotherapy; a review of targets for cancer therapy. Cancers 2021, 13, 2535. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Zeng, J.; Yang, X.; Cheng, L.; Liu, R.; Lei, Y.; Dong, D.; Li, F.; Lau, Q.C.; Deng, L.; Nice, E.C.; et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J. Transl. Med. 2013, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Tang, Z.H.; Liu, S.; Guo, S.S. Clinicopathological significance of overexpression of interleukin-6 in colorectal cancer. World J. Gastroenterol. 2017, 23, 1780–1786. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osterhoff, C.; Ivell, R.; Kirchhoff, C. Cloning of a human epididymis-specific mRNA, HE6, encoding a novel member of the seven transmembrane-domain receptor superfamily. DNA Cell Biol. 1997, 16, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Hamann, J.; Aust, G.; Araç, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef]
- Richter, G.H.; Fasan, A.; Hauer, K.; Grunewald, T.G.; Berns, C.; Rössler, S.; Naumann, I.; Staege, M.S.; Fulda, S.; Esposito, I.; et al. G-protein coupled receptor 64 promotes invasiveness and metastasis in Ewing sarcomas through PGF and MMP1. J. Pathol. 2013, 230, 70–81. [Google Scholar] [CrossRef]
- Chen, J.; Weiss, W.A. Alternative splicing in cancer: Implications for biology and therapy. Oncogene 2015, 34, 1–14. [Google Scholar] [CrossRef]
- Cheng, C.; Yaffe, M.B.; Sharp, P.A. A Positive feedback loop couples ras activation and CD44 alternative splicing. Genes Dev. 2006, 20, 1715–1720. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Harada, M.; Shimozato, O.; Souda, H.; Takiguchi, N.; Nabeya, Y.; Kamijo, T.; Akita, H.; Anzai, N.; Chiba, K.; et al. Cancer-type OATP1B3 mRNA has the potential to become a detection and prognostic biomarker for human colorectal cancer. Biomark. Med. 2017, 11, 629–639. [Google Scholar] [CrossRef]
- Obermann, H.; Samalecos, A.; Osterhoff, C.; Schröder, B.; Heller, R.; Kirchhoff, C. HE6, a two-subunit heptahelical receptor associated with apical membranes of efferent and epididymal duct epithelia. Mol. Reprod. Dev. 2003, 64, 13–26. [Google Scholar] [CrossRef]
- Baird, K.; Davis, S.; Antonescu, C.R.; Harper, U.L.; Walker, R.L.; Chen, Y.; Glatfelter, A.A.; Duray, P.H.; Meltzer, P.S. Gene expression profiling of human sarcomas: Insights into sarcoma biology. Cancer Res. 2005, 65, 9226–9235. [Google Scholar] [CrossRef] [Green Version]
- Monoclonal Antibodies. Livertoxicol: Clinical and Research Information on Drug-Induced Liver Injury; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012. [Google Scholar]
- Davies, B.; Baumann, C.; Kirchhoff, C.; Ivell, R.; Nubbemeyer, R.; Habenicht, U.F.; Theuring, F.; Gottwald, U. Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol. Cell. Biol. 2004, 24, 8642–8648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, M.; Cyr, D.G. The blood-epididymis barrier and inflammation. Spermatogenesis 2014, 4, e979619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchhoff, C.; Osterhoff, C.; Samalecos, A. HE6/GPR64 Adhesion receptor co-localizes with apical and subapical f-actin scaffold in male excurrent duct epithelia. Reproduction 2008, 136, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchhoff, C.; Obermann, H.; Behnen, M.; Davies, B. Role of Epididymal receptor he6 in the regulation of sperm microenvironment. Mol. Cell. Endocrinol. 2006, 250, 43–48. [Google Scholar] [CrossRef] [PubMed]




| Splice Variant | HTB166 (FPKM) | WBC (FPKM) | Protein Isoform | Protein Length (aa) |
|---|---|---|---|---|
| GPR64v201 | 1.15869 | 0 | 10 | 898 |
| GPR64v202 | 1.69 × 10−8 | 1.58 × 10−5 | 10 | 898 |
| GPR64v203 | 3.4776 | 0 | 4 | 1003 |
| GPR64v204 | 2.26204 | 9.20 × 10−82 | 7 | 987 |
| GPR64v205 | 5.29482 | 0 | 2 | 1014 |
| GPR64v206 | 0.0028735 | 1.32 × 10−82 | 6 | 995 |
| GPR64v207 | 3.90 × 10−8 | 6.73 × 10−85 | 1 | 1017 |
| GPR64v208 | 3.50 × 10−6 | 0.0684216 | 9 | 966 |
| GPR64v209 | 4.29718 | 0.000105219 | 5 | 993 |
| GPR64v210 | 1.49 × 10−6 | 1.58 × 10−5 | 3 | 1001 |
| GPR64v211 | 0.419505 | 0 | No protein |
| Diagnosis | n | Positive Rate (%) | High Expression Rate (%) |
|---|---|---|---|
| Ewing sarcoma | 11 | 100 | 90.9 |
| Osteosarcoma | 13 | 76.9 | 30.7 |
| Chondrosarcoma | 11 | 90.9 | 36.3 |
| PLPS | 1 | 100 | 100 |
| Myxofibrosarcoma | 12 | 50 | 16.6 |
| Chordoma | 15 | 93.3 | 80.0 |
| UPS | 13 | 46.1 | 0 |
| Synovial sarcoma | 4 | 0 | 0 |
| DLPS | 13 | 53.8 | 15.4 |
| Leiomyosarcoma | 9 | 33.3 | 11.1 |
| Fibrosarcoma | 4 | 50 | 50 |
| MPNST | 3 | 0 | 0 |
| MLPS | 3 | 0 | 0 |
| Total | 112 | 62.5 | 33.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, K.; Asanuma, K.; Okamoto, T.; Yoshida, K.; Matsuyama, Y.; Kita, K.; Hagi, T.; Nakamura, T.; Sudo, A. GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas. Cancers 2022, 14, 814. https://doi.org/10.3390/cancers14030814
Nakamura K, Asanuma K, Okamoto T, Yoshida K, Matsuyama Y, Kita K, Hagi T, Nakamura T, Sudo A. GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas. Cancers. 2022; 14(3):814. https://doi.org/10.3390/cancers14030814
Chicago/Turabian StyleNakamura, Koichi, Kunihiro Asanuma, Takayuki Okamoto, Keisuke Yoshida, Yumi Matsuyama, Kouji Kita, Tomohito Hagi, Tomoki Nakamura, and Akihiro Sudo. 2022. "GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas" Cancers 14, no. 3: 814. https://doi.org/10.3390/cancers14030814
APA StyleNakamura, K., Asanuma, K., Okamoto, T., Yoshida, K., Matsuyama, Y., Kita, K., Hagi, T., Nakamura, T., & Sudo, A. (2022). GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas. Cancers, 14(3), 814. https://doi.org/10.3390/cancers14030814






