Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
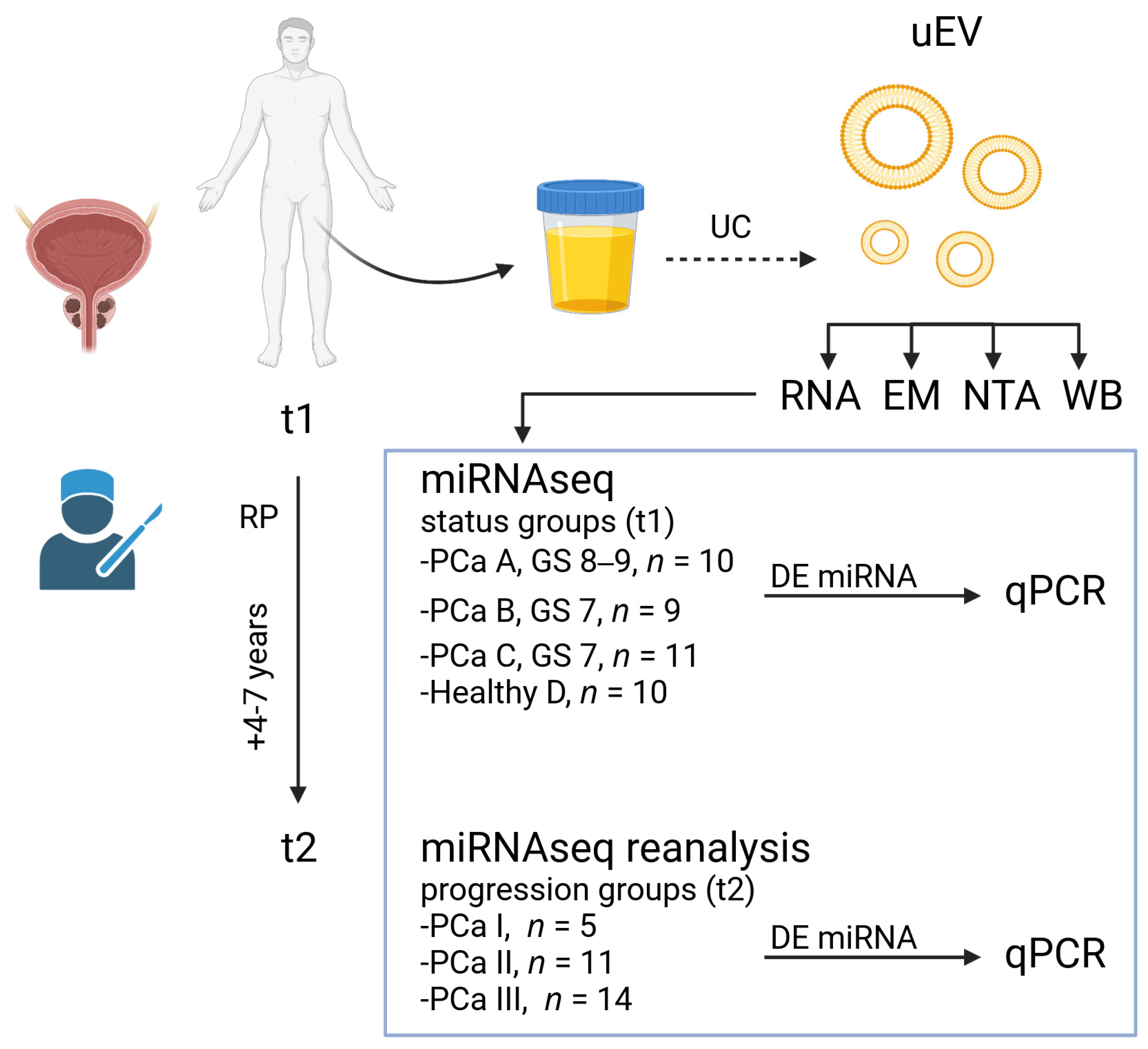
2.1. Study Participants and Groups
2.2. Urine and Plasma Sample Collections and EV Isolation
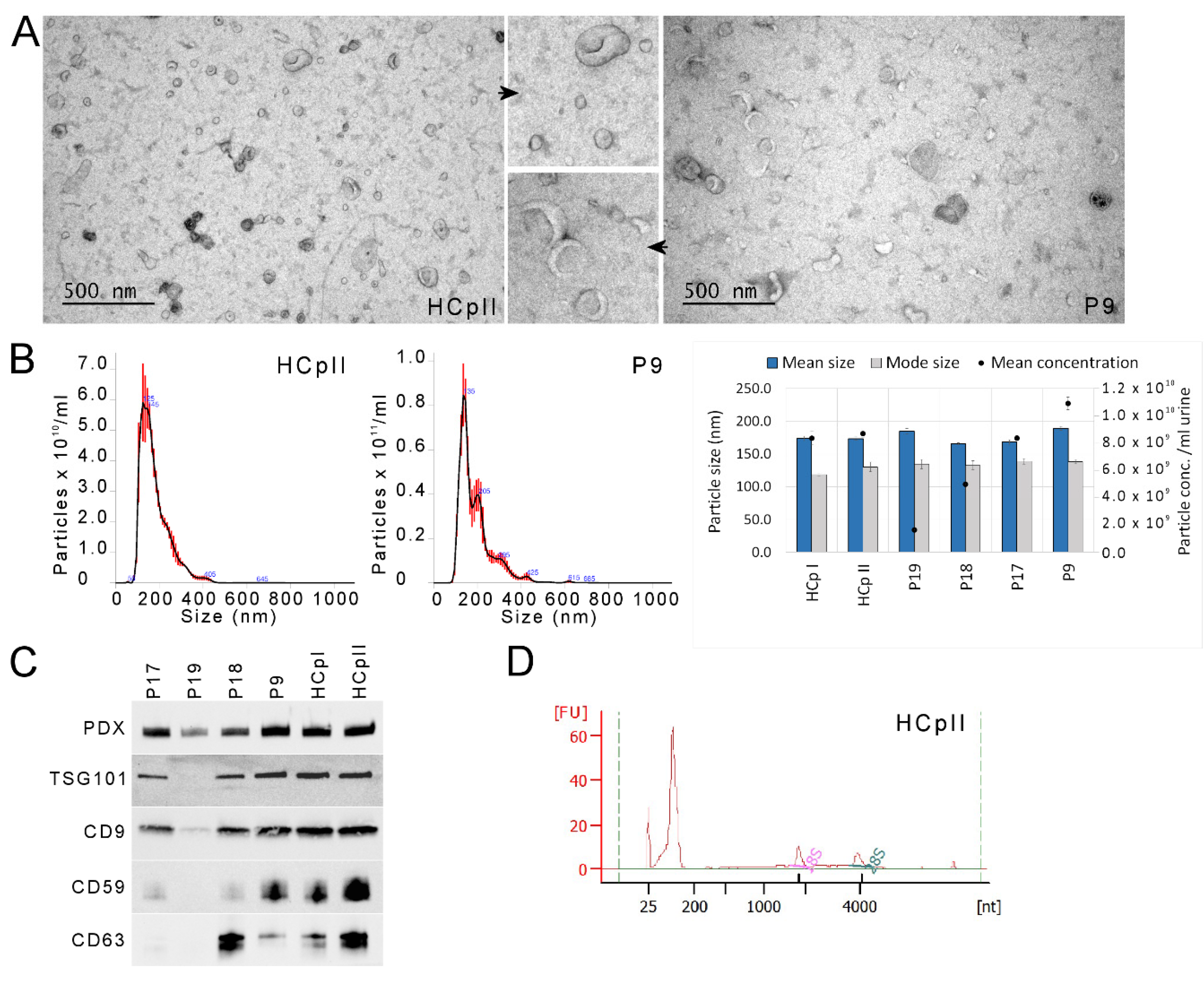
2.3. EV Characterization by EM, Western Blotting and NTA
2.4. RNA Extraction and miRNA Sequencing in the Main Study
2.5. Quantitative PCR and Analysis
2.6. RNA Extraction and miRNA and mRNA Sequencing for the Corelation Study of Three Patients
2.7. Statistical Testing and Venn Analysis
3. Results
3.1. Design of the Main Study
3.2. uEV and miRNA Sequencing Quality
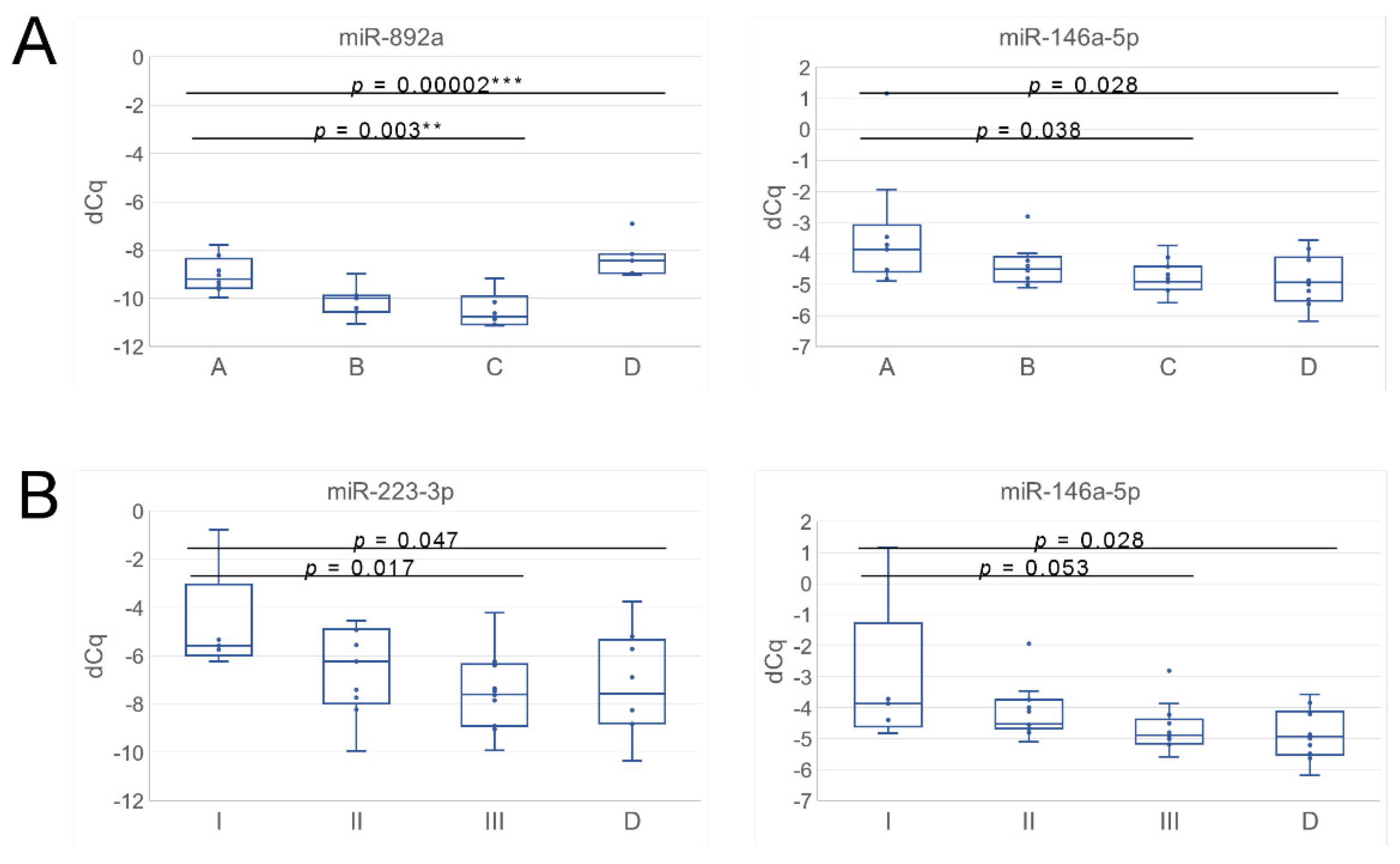
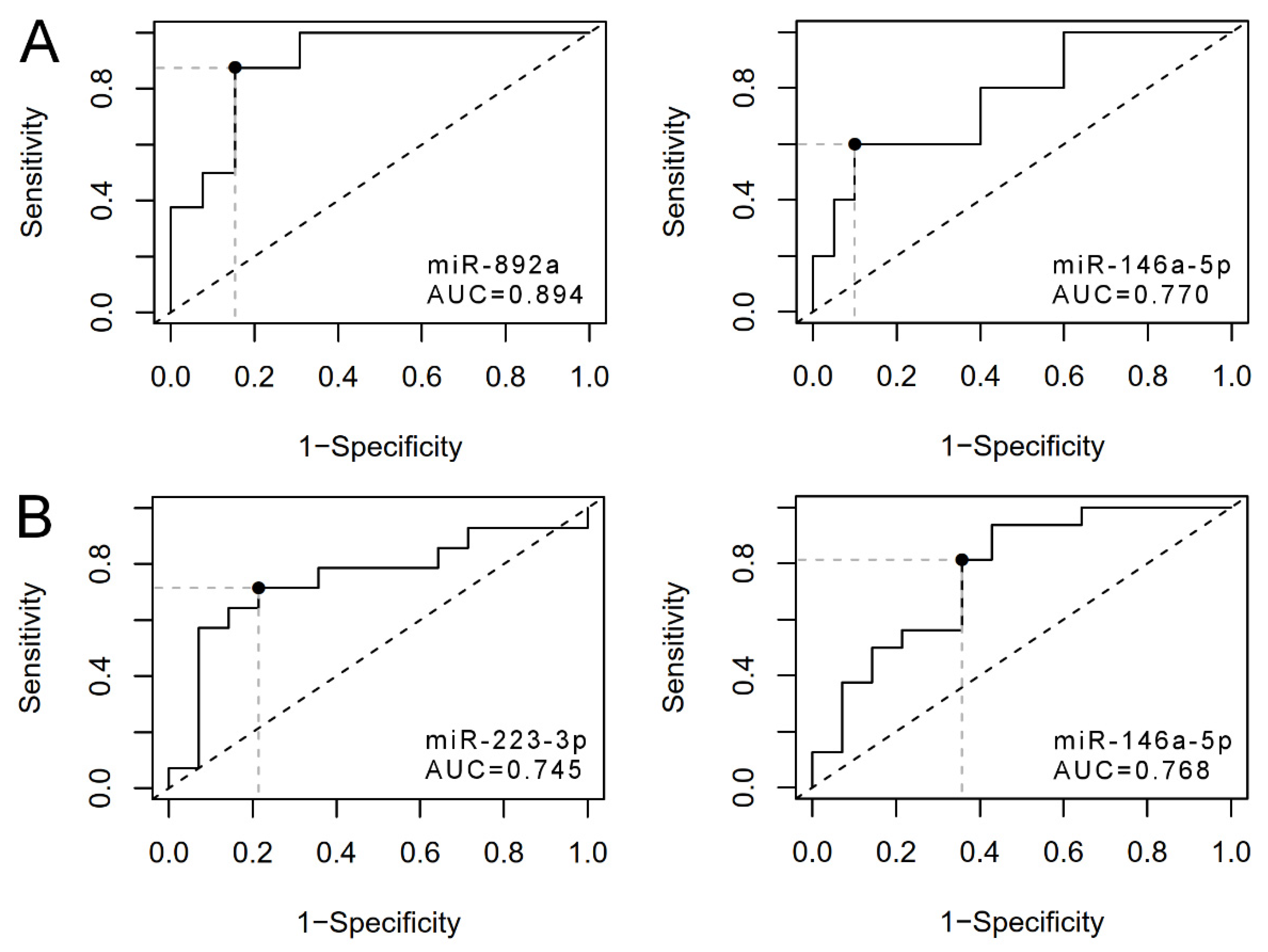
3.3. uEV from Prostate Cancer Patient Status Groups Differed in the Quantities of miRNAs Targeting Cancer and Progression-Linked Signaling, Resistance and Hormonal Pathways
3.4. Analysis of Prostate Cancer Progression Groups Uncovered Unique miRNA Signatures and Overlapping Cancer Progression-Linked Pathways
3.5. Correlation Study of miR-146a-5p, -892a and -223-3p Targets in Patient EV Reveals mRNAs of Interest for Detecting Prostate Cancer Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boerrigter, E.; Groen, L.N.; Van Erp, N.P.; Verhaegh, G.W.; Schalken, J.A. Clinical Utility of Emerging Biomarkers in Prostate Cancer Liquid Biopsies. Expert. Rev. Mol. Diagn. 2020, 20, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Auvinen, A.; Rannikko, A.; Taari, K.; Kujala, P.; Mirtti, T.; Kenttämies, A.; Rinta-Kiikka, I.; Lehtimäki, T.; Oksala, N.; Pettersson, K.; et al. A Randomized Trial of Early Detection of Clinically Significant Prostate Cancer (ProScreen): Study Design and Rationale. Eur. J. Epidemiol. 2017, 32, 521–527. [Google Scholar] [CrossRef]
- Nevo, A.; Navaratnam, A.; Andrews, P. Prostate cancer and the role of biomarkers. Abdom. Radiol. 2020, 45, 2120–2132. [Google Scholar] [CrossRef]
- Puhka, M.; Takatalo, M.; Nordberg, M.-E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T.; et al. Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics 2017, 7, 3824–3841. [Google Scholar] [CrossRef]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary Extracellular Vesicles: A Position Paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef]
- Duijvesz, D.; Luider, T.; Bangma, C.H.; Jenster, G. Exosomes as Biomarker Treasure Chests for Prostate Cancer. Eur. Urol. 2011, 59, 823–831. [Google Scholar] [CrossRef]
- Barreiro, K.; Dwivedi, O.P.; Leparc, G.; Rolser, M.; Delic, D.; Forsblom, C.; Groop, P.; Groop, L.; Huber, T.B.; Puhka, M.; et al. Comparison of Urinary Extracellular Vesicle Isolation Methods for Transcriptomic Biomarker Research in Diabetic Kidney Disease. J. Extracell. Vesicles 2020, 10, e12038. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, C.; Constâncio, V.; Henrique, R.; Carvalho, Â.; Jerónimo, C. Urinary Extracellular Vesicles as Potential Biomarkers for Urologic Cancers: An Overview of Current Methods and Advances. Cancers 2021, 13, 1529. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; de Menezes, R.; Misovic, B.; Wachalska, M.; Geldof, A.; Zini, N.; de Reijke, T.; Wurdinger, T.; Vis, A.; et al. Non-Invasive Prostate Cancer Detection by Measuring MiRNA Variants (IsomiRs) in Urine Extracellular Vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Royo, F.; Diwan, I.; Tackett, M.R.; Zuñiga, P.; Sanchez-Mosquera, P.; Loizaga-Iriarte, A.; Ugalde-Olano, A.; Lacasa, I.; Perez, A.; Unda, M.; et al. Comparative MiRNA Analysis of Urine Extracellular Vesicles Isolated through Five Different Methods. Cancers 2016, 8, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Linē, A.; Llorente, A. Identification of Non-Invasive MiRNAs Biomarkers for Prostate Cancer by Deep Sequencing Analysis of Urinary Exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y.; et al. Unveiling Massive Numbers of Cancer-Related Urinary-MicroRNA Candidates via Nanowires. Sci. Adv. 2017, 3, e1701133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-Free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.Y.; Shin, H.; Moon, H.W.; Park, Y.H.; Park, J.; Lee, J.Y. Urinary Exosomal MicroRNA Profiling in Intermediate-Risk Prostate Cancer. Sci. Rep. 2021, 11, 7355. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Glavich, G.J.; Pahuski, M.; Short, A.; Semmes, O.J.; Yang, L.; Galkin, V.; Drake, R.; Esquela-Kerscher, A. Characterization and Evidence of the MiR-888 Cluster as a Novel Cancer Network in Prostate. Mol. Cancer Res. 2018, 16, 669–681. [Google Scholar] [CrossRef] [Green Version]
- Ku, A.; Ravi, N.; Yang, M.; Evander, M.; Laurell, T.; Lilja, H.; Ceder, Y. A Urinary Extracellular Vesicle MicroRNA Biomarker Discovery Pipeline; from Automated Extracellular Vesicle Enrichment by Acoustic Trapping to MicroRNA Sequencing. PLoS ONE 2019, 14, e0217507. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, L.-X.; Diao, Y.-J.; Wang, J.; Ye, Y.; Hao, X.-K. Identification of Urinary Exosomal MiRNAs for the Non-Invasive Diagnosis of Prostate Cancer. Cancer Manag. Res. 2021, 13, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Kusuma, G.D.; Crucitta, S.; Lim, H.K.; Cooper, C.; Riches, J.E.; Azad, A.; Ochiya, T.; Boyle, G.M.; Southey, M.C.; et al. Androgens Alter the Heterogeneity of Small Extracellular Vesicles and the Small RNA Cargo in Prostate Cancer. J. Extracell. Vesicles 2021, 10, e12136. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer. 2021. [Google Scholar] [CrossRef]
- Barreiro, K.; Dwivedi, O.P.; Valkonen, S.; Groop, P.; Tuomi, T.; Holthofer, H.; Rannikko, A.; Yliperttula, M.; Siljander, P.; Laitinen, S.; et al. Urinary Extracellular Vesicles: Assessment of Pre-analytical Variables and Development of a Quality Control with Focus on Transcriptomic Biomarker Research. J. Extracell. Vesicles 2021, 10, e12158. [Google Scholar] [CrossRef]
- Hildebrandt, A.; Kirchner, B.; Nolte-’t Hoen, E.N.M.; Pfaffl, M.W. MiREV: An Online Database and Tool to Uncover Potential Reference RNAs and Biomarkers in Small-RNA Sequencing Data Sets from Extracellular Vesicles Enriched Samples. J. Mol. Biol. 2021, 433, 167070. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karttunen, J.; Stewart, S.E.; Kalmar, L.; Grant, A.J.; Karet Frankl, F.E.; Williams, T.L. Size-Exclusion Chromatography Separation Reveals That Vesicular and Non-Vesicular Small RNA Profiles Differ in Cell Free Urine. Int. J. Mol. Sci. 2021, 22, 4881. [Google Scholar] [CrossRef] [PubMed]
- Mussack, V.; Wittmann, G.; Pfaffl, M.W. Comparing Small Urinary Extracellular Vesicle Purification Methods with a View to RNA Sequencing-Enabling Robust and Non-Invasive Biomarker Research. Biomol. Detect. Quantif. 2019, 17, 100089. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, A.; Yliperttula, M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Vlaeminck-Guillem, V. Extracellular Vesicles in Prostate Cancer Carcinogenesis, Diagnosis, and Management. Front. Oncol. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [Green Version]
- Logozzi, M.; Angelini, D.F.; Iessi, E.; Mizzoni, D.; Di Raimo, R.; Federici, C.; Lugini, L.; Borsellino, G.; Gentilucci, A.; Pierella, F.; et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett. 2017, 403, 318–329. [Google Scholar] [CrossRef]
- Salciccia, S.; Capriotti, A.L.; Laganà, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367. [Google Scholar] [CrossRef]
- Puhka, M.; Nordberg, M.E.; Valkonen, S.; Rannikko, A.; Kallioniemi, O.; Siljander, P.; Af Hällström, T.M. KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. Eur. J. Pharm. Sci. 2017, 98, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of RNA-Seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE. 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Lu, M.; Shi, B.; Wang, J.; Cao, Q.; Cui, Q. TAM: A Method for Enrichment and Depletion Analysis of a MicroRNA Category in a List of MicroRNAs. BMC Bioinform. 2010, 11, 419. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA Set Analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N.; et al. MicroRNA Profiling in Prostate Cancer--the Diagnostic Potential of Urinary MiR-205 and MiR-214. PLoS ONE 2013, 8, e76994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, M.K.; Andreassen, R.; Haugen, T.B.; Furu, K. Identification of Endogenous Controls for Use in MiRNA Quantification in Human Cancer Cell Lines. Cancer Genom. Proteom. 2016, 13, 63–68. [Google Scholar]
- Gracia, T.; Wang, X.; Su, Y.; Norgett, E.E.; Williams, T.L.; Moreno, P.; Micklem, G.; Frankl, F.E.K. Urinary Exosomes Contain MicroRNAs Capable of Paracrine Modulation of Tubular Transporters in Kidney. Sci. Rep. 2017, 7, 40601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A Novel and Universal Method for MicroRNA RT-QPCR Data Normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [Green Version]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. easyROC: An Interactive Web-tool for ROC Curve Analysis Using R Language Environment. R J. 2016, 8, 213–230. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual comparisons by ranking methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell. Vesicles 2019, 9, 1697028. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Bujanda, Z.A.; Obradovic, A.; Nirschl, T.R.; Crowley, L.; Macedo, R.; Papachristodoulou, A.; O’Donnell, T.; Laserson, U.; Zarif, J.C.; Reshef, R.; et al. TGM4: An immunogenic prostate-restricted antigen. J. Immunother. Cancer 2021, 9, e001649. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Park, Y.H.; Jung, S.-H.; Jang, S.-H.; Kim, M.Y.; Lee, J.Y.; Chung, Y.-J. Urinary Exosome MicroRNA Signatures as a Noninvasive Prognostic Biomarker for Prostate Cancer. NPJ Genom. Med. 2021, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.A.; Kim, H.; Albeltagy, R.S.; El-Habit, O.H.; Abdel-Mageed, A.B. MicroRNAs in Prostate Cancer: From Function to Biomarker Discovery. Exp. Biol. Med. 2018, 243, 817–825. [Google Scholar] [CrossRef]
- Lin, S.-R.; Mokgautsi, N.; Liu, Y.-N. Ras and Wnt Interaction Contribute in Prostate Cancer Bone Metastasis. Molecules 2020, 25, 2380. [Google Scholar] [CrossRef]
- Davidsson, S.; Ohlson, A.-L.; Andersson, S.-O.; Fall, K.; Meisner, A.; Fiorentino, M.; Andrén, O.; Rider, J.R. CD4 Helper T Cells, CD8 Cytotoxic T Cells, and FOXP3(+) Regulatory T Cells with Respect to Lethal Prostate Cancer. Mod. Pathol. 2013, 26, 448–455. [Google Scholar] [CrossRef]
- Thompson, V.C.; Morris, T.G.W.; Cochrane, D.R.; Cavanagh, J.; Wafa, L.A.; Hamilton, T.; Wang, S.; Fazli, L.; Gleave, M.E.; Nelson, C.C. Relaxin Becomes Upregulated during Prostate Cancer Progression to Androgen Independence and Is Negatively Regulated by Androgens. Prostate 2006, 66, 1698–1709. [Google Scholar] [CrossRef]
- Wan, Y.; Zeng, Z.-C.; Xi, M.; Wan, S.; Hua, W.; Liu, Y.-L.; Zhou, Y.-L.; Luo, H.-W.; Jiang, F.-N.; Zhong, W. Dysregulated MicroRNA-224/Apelin Axis Associated with Aggressive Progression and Poor Prognosis in Patients with Prostate Cancer. Hum. Pathol. 2015, 46, 295–303. [Google Scholar] [CrossRef]
- Bonkhoff, H. Estrogen Receptor Signaling in Prostate Cancer: Implications for Carcinogenesis and Tumor Progression. Prostate 2018, 78, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Montano, X.; Djamgoz, M.B.A. Epidermal Growth Factor, Neurotrophins and the Metastatic Cascade in Prostate Cancer. FEBS Lett. 2004, 571, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imam, J.S.; Plyler, J.R.; Bansal, H.; Prajapati, S.; Bansal, S.; Rebeles, J.; Chen, H.-I.H.; Chang, Y.-F.; Panneerdoss, S.; Zoghi, B.; et al. Genomic Loss of Tumor Suppressor MiRNA-204 Promotes Cancer Cell Migration and Invasion by Activating AKT/MTOR/Rac1 Signaling and Actin Reorganization. PLoS ONE 2012, 7, e52397. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Yu, Y.; Song, Y.; Li, X.; Lan, D.; Zhang, P.; Xiao, Y.; Xing, Y. Activation of BDNF/TrkB Pathway Promotes Prostate Cancer Progression via Induction of Epithelial-Mesenchymal Transition and Anoikis Resistance. FASEB J. 2020, 34, 9087–9101. [Google Scholar] [CrossRef]
- Esper, R.M.; Loeb, J.A. Neurotrophins Induce Neuregulin Release through Protein Kinase Cdelta Activation. J. Biol. Chem. 2009, 284, 26251–26260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Karthaus, W.R.; Lee, Y.S.; Gao, V.R.; Wu, C.; Russo, J.W.; Liu, M.; Mota, J.M.; Abida, W.; Linton, E.; et al. Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer. Cancer Cell 2020, 38, 279–296.e9. [Google Scholar] [CrossRef] [PubMed]
- Mujoo, K.; Choi, B.-K.; Huang, Z.; Zhang, N.; An, Z. Regulation of ERBB3/HER3 Signaling in Cancer. Oncotarget 2014, 5, 10222–10236. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Yang, J.; Yi, L.; Wang, Y.; Dong, Z.; Liu, Z.; Ouyang, S.; Wu, H.; Zhong, Z.; Yin, Z.; et al. MiR-223-3p Targeting SEPT6 Promotes the Biological Behavior of Prostate Cancer. Sci. Rep. 2014, 4, 7546. [Google Scholar] [CrossRef]
- Barceló, M.; Castells, M.; Bassas, L.; Vigués, F.; Larriba, S. Semen MiRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci. Rep. 2019, 9, 13772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercadal, M.; Herrero, C.; López-Rodrigo, O.; Castells, M.; de la Fuente, A.; Vigués, F.; Bassas, L.; Larriba, S. Impact of Extracellular Vesicle Isolation Methods on Downstream Mirna Analysis in Semen: A Comparative Study. Int. J. Mol. Sci. 2020, 21, 5949. [Google Scholar] [CrossRef]
- Baffoe-Bonnie, A.B.; Smith, J.R.; Stephan, D.A.; Schleutker, J.; Carpten, J.D.; Kainu, T.; Gillanders, E.M.; Matikainen, M.; Teslovich, T.M.; Tammela, T.; et al. A Major Locus for Hereditary Prostate Cancer in Finland: Localization by Linkage Disequilibrium of a Haplotype in the HPCX Region. Hum. Genet. 2005, 117, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.; Schindler, M.; Isotalo, J.; Ikonen, T.; Vihinen, M.; Oja, H.; Tammela, T.L.J.; Wahlfors, T.; Schleutker, J. NMD and MicroRNA Expression Profiling of the HPCX1 Locus Reveal MAGEC1 as a Candidate Prostate Cancer Predisposition Gene. BMC Cancer 2011, 11, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacona, J.R.; Lutz, C.S. MiR-146a-5p: Expression, Regulation, and Functions in Cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, X.; Liu, X.; Wang, Y.; Huang, J.; Jiang, B.; Chen, Q.; Yu, J. MiR-146a Functions as a Tumor Suppressor in Prostate Cancer by Targeting Rac1. Prostate 2014, 74, 1613–1621. [Google Scholar] [CrossRef]
- Östling, P.; Leivonen, S.-K.; Aakula, A.; Kohonen, P.; Mäkelä, R.; Hagman, Z.; Edsjö, A.; Kangaspeska, S.; Edgren, H.; Nicorici, D.; et al. Systematic Analysis of MicroRNAs Targeting the Androgen Receptor in Prostate Cancer Cells. Cancer Res. 2011, 71, 1956–1967. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Wang, N.; Wang, X.; Tong, N.; Shao, N.; Tao, J.; Li, P.; Niu, X.; Feng, N.; Zhang, L.; et al. MiR-146a Suppresses Tumor Growth and Progression by Targeting EGFR Pathway and in a p-ERK-Dependent Manner in Castration-Resistant Prostate Cancer: Role of MiR-146a in CRPC. Prostate 2012, 72, 1171–1178. [Google Scholar] [CrossRef]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Beckler, M.D.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-dependent sorting of miRNA to exosomes. Elife 2015, 4, e07197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Liang, H.X.; Wu, S.H.; Jiang, H.Q.; Wang, Q.; Yu, Z.J. Overexpressed Tumor Suppressor Exosomal miR-15a-5p in Cancer Cells Inhibits PD1 Expression in CD8+T Cells and Suppresses the Hepatocellular Carcinoma Progression. Front. Oncol. 2021, 11, 622263. [Google Scholar] [CrossRef]
- Dong, L.; Huang, C.Y.; Johnson, E.J.; Yang, L.; Zieren, R.C.; Horie, K.; Kim, C.J.; Warren, S.; Amend, S.R.; Xue, W.; et al. High-Throughput Simultaneous mRNA Profiling Using nCounter Technology Demonstrates That Extracellular Vesicles Contain Different mRNA Transcripts Than Their Parental Prostate Cancer Cells. Anal. Chem. 2021, 93, 3717–3725. [Google Scholar] [CrossRef]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef]
- Tóth, E.Á.; Turiák, L.; Visnovitz, T.; Cserép, C.; Mázló, A.; Sódar, B.W.; Försönits, A.I.; Petővári, G.; Sebestyén, A.; Komlósi, Z.; et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J Extracell. Vesicles 2021, 10, e12140. [Google Scholar] [CrossRef]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Graves-Deal, R.; Trinh, V.Q.; Ramirez, M.A.; Sohn, Y.; Neininger, A.C.; Taneja, N.; McKinley, E.T.; et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021, 9, 1–15. [Google Scholar] [CrossRef]
- Narain, N.R.; Diers, A.R.; Lee, A.; Lao, S.; Chan, J.Y.; Schofield, S.; Andreazi, J.; Ouro-Djobo, R.; Jimenez, J.J.; Friss, T.; et al. Identification of Filamin-A and -B as potential biomarkers for prostate cancer. Future Sci. OA 2016, 3, FSO161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Wang, J.; Dong, C.; Wei, W.; Li, J.; Li, X. Membranous type matrix metalloproteinase 16 induces human prostate cancer metastasis. Oncol. Lett. 2017, 14, 3096–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, T.; Oue, N.; Sakamoto, N.; Anami, K.; Oo, H.Z.; Sentani, K.; Ohara, S.; Teishima, J.; Matsubara, A.; Yasui, W. Identification of transmembrane protein in prostate cancer by the Escherichia coli ampicillin secretion trap: Expression of CDON is involved in tumor cell growth and invasion. Pathobiology. 2011, 78, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Polavaram, N.S.; Dutta, S.; Islam, R.; Bag, A.K.; Roy, S.; Poitz, D.; Karnes, J.; Hofbauer, L.C.; Kohli, M.; Costello, B.A.; et al. Tumor- and osteoclast-derived NRP2 in prostate cancer bone metastases. Bone Res. 2021, 14, 9–24. [Google Scholar] [CrossRef]
- Zhang, N.; Li, Z.; Bai, F.; Zhang, S. PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis. 2019, 10, 734. [Google Scholar] [CrossRef]
- Samaržija, I. Site-Specific and Common Prostate Cancer Metastasis Genes as Suggested by Meta-Analysis of Gene Expression Data. Life 2021, 11, 636. [Google Scholar] [CrossRef]
- Sung, S.Y.; Wu, I.H.; Chuang, P.H.; Petros, J.A.; Wu, H.C.; Zeng, H.J.; Huang, W.C.; Chung, L.W.; Hsieh, C.L. Targeting L1 cell adhesion molecule expression using liposome-encapsulated siRNA suppresses prostate cancer bone metastasis and growth. Oncotarget 2014, 5, 9911–9929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, N.; Wu, Y.P.; Yin, H.B.; Chen, S.H.; Li, X.D.; Xue, X.Y.; Gou, X. SHCBP1 promotes tumor cell proliferation, migration, and invasion, and is associated with poor prostate cancer prognosis. J. Cancer Res. Clin. Oncol. 2020, 146, 1953–1969. [Google Scholar] [CrossRef]
- Nickerson, M.L.; Im, K.M.; Misner, K.J.; Tan, W.; Lou, H.; Gold, B.; Wells, D.W.; Bravo, H.C.; Fredrikson, K.M.; Harkins, T.T.; et al. Somatic alterations contributing to metastasis of a castration-resistant prostate cancer. Hum. Mutat. 2013, 34, 1231–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Hu, Q.; Liu, X.; Ji, Y.; Chao, H.P.; Liu, Y.; Tracz, A.; Kirk, J.; Buonamici, S.; Zhu, P.; et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2020, 11, 2089. [Google Scholar] [CrossRef]
- Verma, S.; Shankar, E.; Chan, E.R.; Gupta, S. Metabolic Reprogramming and Predominance of Solute Carrier Genes during Acquired Enzalutamide Resistance in Prostate Cancer. Cells 2020, 9, 2535. [Google Scholar] [CrossRef]
- Singh, A.P.; Bafna, S.; Chaudhary, K.; Venkatraman, G.; Smith, L.; Eudy, J.D.; Johansson, S.L.; Lin, M.F.; Batra, S.K. Genome-wide expression profiling reveals transcriptomic variation and perturbed gene networks in androgen-dependent and androgen-independent prostate cancer cells. Cancer Lett. 2008, 259, 28–38. [Google Scholar] [CrossRef] [Green Version]
- da Costa, F.H.B.; Lewis, M.S.; Truong, A.; Carson, D.D.; Farach-Carson, M.C. SULF1 suppresses Wnt3A-driven growth of bone metastatic prostate cancer in perlecan-modified 3D cancer-stroma-macrophage triculture models. PLoS ONE 2020, 15, e0230354. [Google Scholar] [CrossRef]
- Liao, D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol. Cancer Res. 2009, 7, 1893–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, M.; Su, Y.; Kang, W.; Gao, R.; Li, X.; Zhanget, G. Aberrant expression and functions of protocadherins in human malignant tumors. Tumor Biol. 2016, 37, 12969–12981. [Google Scholar] [CrossRef] [PubMed]
- Nyquist, M.D.; Prasad, B.; Mostaghel, E.A. Harnessing Solute Carrier Transporters for Precision Oncology. Molecules 2017, 22, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Status Groups | A | B | C | D | ANOVA p-Value (A–C) | Progression Groups | I | II | III | ANOVA p-Value (I–III) |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of subjects | 10 | 9 | 11 | 10 | Number of subjects | 5 | 11 | 14 | ||
| Previous classification in status group (number of subjects) | ||||||||||
| A | 4 | 4 | 2 | |||||||
| B | 1 | 4 | 4 | |||||||
| C | 0 | 3 | 8 | |||||||
| Age (years) | 0.209 | Age (years) | 0.777 | |||||||
| Mean | 69 | 64 | 63 | <45 | Mean | 67 | 65 | 64 | ||
| Range | 54–74 | 51–75 | 51–73 | Range | 62–74 | 51–75 | 54–73 | |||
| Gleason score (number of subjects) | 2.1 × 1016 | Gleason score (number of subjects) | 0.121 | |||||||
| 7 | 0 | 9 | 11 | 7 | 1 | 7 | 12 | |||
| 3 + 4 | 0 | 3 | 4 | 3 + 4 | 0 | 5 | 2 | |||
| 4 + 3 | 0 | 6 | 7 | 4 + 3 | 1 | 2 | 10 | |||
| 8 | 2 | 0 | 0 | 8 | 2 | 0 | 0 | |||
| 4 + 4 | 2 | 0 | 0 | 4 + 4 | 2 | 0 | 0 | |||
| 9 | 8 | 0 | 0 | 9 | 2 | 4 | 2 | |||
| 4 + 5 | 8 | 0 | 0 | 4 + 5 | 2 | 4 | 2 | |||
| 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | |||
| Stage (number of subjects) | 0.005 | Stage (number of subjects) | 0.026 | |||||||
| T2 | 3 | 3 | 10 | T2 | 1 | 4 | 11 | |||
| T3 | 7 | 6 | 1 | T3 | 4 | 7 | 3 | |||
| Pathological features-prostatectomy tissues (number of subjects) | Pathological features-prostatectomy tissues (number of subjects) | |||||||||
| Positive surgical margin | 3 | 7 | 0 | Positive surgical margin | 2 | 5 | 3 | |||
| Growth through capsule | 6 (9) | 6 | 1 | Growth through capsule | 3 (4) | 7 | 3 | |||
| Invasion to seminal vesicles | 3 (9) | 1 (8) | 0 | Invasion to seminal vesicles | 2 (3) | 1 | 1 | |||
| Lymph node positivity | 1 (8) | 2 (7) | 0 (4) | Lymph node positivity | 2 (4) | 0 (6) | 1 (9) | |||
| PSA (post-RP) (number of subjects or concentration) | 0.260 | PSA (post-RP) (number of subjects or concentration) | 0.017 | |||||||
| <0.05 (ng/mL) | 5 | 5 | 8 | <0.05 (ng/mL) | 0 | 5 | 13 | |||
| ≥0.05, <0.2, (ng/mL) | 2 | 2 | 3 | ≥0.05, <0.2, (ng/mL) | 1 | 5 | 1 | |||
| ≥0.2, (ng/mL) | 3 | 2 | 0 | ≥0.2, (ng/mL) | 4 | 1 | 0 | |||
| Range for ≥0.05 (ng/mL) | 0.12–8.42 | 0.05–0.53 | 0.06–0.08 | Range for ≥0.05 (ng/mL) | 0.14–8.42 | 0.05–0.53 | 0.08 | |||
| Events and treatments during follow-up (number of subjects) | ||||||||||
| Death due to Pca | 2 | 0 | 0 | |||||||
| Metastasis (M1) | 3 | 0 | 0 | |||||||
| Hormonal treatment | 5 | 0 | 0 | |||||||
| Secondary treatment | 5 | 10 | 0 (10) | |||||||
| Biochemical recurrence | 5 | 6 | 0 | |||||||
| MiRNA Name | ||||
|---|---|---|---|---|
| Comparison | miR-146a-5p | miR-892a | miR-223-3p | FC or p-Value |
| A vs. BC | 2.3 | 2.4 | FC | |
| A vs. B | 2.1 | FC | ||
| A vs. C | 2.6 | 2.8 | FC | |
| B vs. C | FC | |||
| AB vs. C | 1.9 | 2.0 | FC | |
| ANOVA A–C | 0.038 | 0.003 | p-value | |
| ANOVA A–D | 0.028 | 0.00002 | p-value | |
| I vs. II | FC | |||
| I vs. III | 3.0 | 6.9 | FC | |
| II vs. III | 1.5, p = 0.051 | FC | ||
| I vs. II + III | 5.4 | FC | ||
| I + II vs. III | 1.8 | 3.0 | FC | |
| ANOVA I–III | 0.053 | 0.017 | p-value | |
| ANOVA I–III and D | 0.028 | 0.007 | 0.047 | p-value |
| Comparison | Category | Term | Count | Fold | p-Value | Bonferroni | FDR | DE miRNAs |
|---|---|---|---|---|---|---|---|---|
| I vs. II | Disease | Carcinoma, Prostate | 6 | 2.2 | NaN | 0.0 × 10⁰ | 0.0 × 10⁰ | 382, 323a, 139, 22, 187, 485 |
| Function | Angiogenesis | 5 | 7.2 | 4.4 × 10−4 | 1.4 × 10−1 | 8.7 × 10−2 | 891a, 382, 22, 184, 363 | |
| I vs. III | Disease | Carcinoma, Prostate | 33 | 3.6 | NaN | 0.0 × 10⁰ | 0.0 × 10⁰ | 30d, 194-2, 1307, 135a-2, 138-1, 375, 497, 30c-1, 30a, 222, 378a, 196a-2, 1297, 195, 27b, 221, 4516, 503, 194-1, 135a-1, 138-2, 187, 1299, let-7c, 148a, 149, let-7e, 34c, 30c-2, 21, 141, 223, 29a |
| Function | Epithelial-to-Mesenchymal Transition | 19 | 6.5 | 7.9 × 10−¹² | 8.4 × 10−⁹ | 9.4 × 10−¹⁰ | 486-2, 221, let-7c, 1246, 30d, 194-2, let-7e, 486-1, 194-1, 30c-1, 30a, 30c-2, 542, 21, 141, 223, 29a, 375, 192 | |
| Function | Hematopoiesis | 15 | 7.4 | 2.2 × 10−¹⁰ | 2.4 × 10−⁷ | 2.0 × 10−⁸ | 486-2, 221, let-7c, let-7e, 486-1, 30c-1, 30c-2, 222, 378a, 196a-2, 363, 223, 196a-1, 29a, 142 | |
| Function | Angiogenesis | 15 | 6.5 | 1.7 × 10−⁹ | 1.8 × 10−⁶ | 9.8 × 10−⁸ | 486-2, 891a, 221, 1246, 149, 486-1, 30a, 1275, 222, 378a, 10b, 363, 21, 27b, 497 | |
| Function | Aging | 14 | 6.3 | 1.1 × 10−⁸ | 1.2 × 10−⁵ | 5.5 × 10−⁷ | 221, let-7c, 148a, 30d, 194-2, let-7e, 194-1, 30a, 222, 10a, 21, 141, 223, 195 | |
| Function | Inflammation | 18 | 4.5 | 1.6 × 10−⁸ | 1.7 × 10−⁵ | 7.3 × 10−⁷ | 194-2, 99b, 135a-2, 138-1, 222, 27b, 192, 221, 194-1, 135a-1, 138-2, 148a, 34c, 21, 141, 223, 29a, 142 | |
| Function | Osteogenesis | 13 | 6.2 | 4.5 × 10−⁸ | 4.8 × 10−⁵ | 1.8 × 10−⁶ | 221, 106a, 194-2, 194-1, 34c, 30a, 222, 378a, 138-2, 21, 1297, 195, 138-1 | |
| Function | Apoptosis | 17 | 4.5 | 4.5 × 10−⁸ | 4.7 × 10−⁵ | 1.8 × 10−⁶ | 135a-2, 138-1, 497, 1246, 30a, 222, 10a, 195, 221, 4516, 135a-1, 138-2, let-7c, 148a, 34c, 21, 29a | |
| Function | Cell Cycle | 14 | 4.8 | 4.6 × 10−⁷ | 4.9 × 10−⁴ | 1.3 × 10−⁵ | 221, 503, 34c, 222, 138-2, 141, 196a-2, 21, 223, 196a-1, 195, 138-1, 27b, 497 | |
| Function | Cell Proliferation | 13 | 4.6 | 2.0 × 10−⁶ | 2.1 × 10−³ | 4.3 × 10−⁵ | 221, let-7c, 503, let-7e, 509-1, 34c, 222, 378a, 21, 509-2, 509-3, 29a, 27b | |
| Function | Immune Response | 13 | 4.0 | 1.0 × 10−⁵ | 1.1 × 10−² | 1.7 × 10−⁴ | 486-1, 30a, 196a-2, 27b, 192, 532, 196a-1, 486-2, 148a, 34c, 21, 223, 29a | |
| Function | Brain Development | 8 | 6.3 | 2.2 × 10−⁵ | 2.3 × 10−² | 3.3 × 10−⁴ | 221, 106a, 135a-1, 222, 10a, 10b, 135a-2, 192 | |
| Function | T-helper 17 Cell Differentiation | 6 | 8.9 | 3.0 × 10−⁵ | 3.1 × 10−² | 4.1 × 10−⁴ | 30c-2, 141, 106a, 27b, 21, 30c-1 | |
| Function | Pancreas Development | 3 | 28.2 | 4.3 × 10−⁵ | 4.5 × 10−² | 5.6 × 10−⁴ | let-7e, 375, 30d | |
| Function | Cell Death | 11 | 4.0 | 5.6 × 10−⁵ | 5.9 × 10−² | 6.9 × 10−⁴ | 221, let-7c, 30d, let-7e, 30c-1, 30c-2, 222, 10b, 21, 29a, 497 | |
| Function | Regulation of Stem Cell | 11 | 3.9 | 6.3 × 10−⁵ | 6.6 × 10−² | 7.8 × 10−⁴ | 221, 106a, 148a, 222, 10a, 21, 141, 223, 195, 142, 192 | |
| Function | Myogensis | 4 | 14.1 | 9.0 × 10−⁵ | 9.6 × 10−² | 1.1 × 10−³ | 135a-1, 222, 135a-2, 221 | |
| Function | Lipid Metabolism | 8 | 5.0 | 1.2 × 10−⁴ | 1.3 × 10−¹ | 1.4 × 10−³ | 378a, 10b, 196a-2, 196a-1, 29a, 375, 27b, 192 | |
| Function | Cleavage Stage Development | 3 | 21.2 | 1.7 × 10−⁴ | 1.8 × 10−¹ | 1.8 × 10−³ | 375, 21, 34c | |
| Function | Nephrotoxicity | 5 | 8.3 | 2.1 × 10−⁴ | 2.3 × 10−¹ | 2.2 × 10−³ | 30a, 30d, 29a, 192, 21 | |
| Function | Onco-MiRNAs | 7 | 5.3 | 2.2 × 10−⁴ | 2.4 × 10−¹ | 2.3 × 10−³ | 221, 106a, 194-2, 194-1, 222, 196a-2, 196a-1 | |
| Function | Oxidative Stress | 4 | 11.3 | 2.6 × 10−⁴ | 2.7 × 10−¹ | 2.6 × 10−³ | 503, 222, 21, 141 | |
| Function | Smooth Muscle Cell Proliferation | 5 | 7.8 | 2.9 × 10−⁴ | 3.0 × 10−¹ | 2.8 × 10−³ | 222, 138-1, 10a, 138-2, 21 | |
| Function | Tumour Suppressor MiRNAs | 9 | 3.9 | 3.3 × 10−⁴ | 3.5 × 10−¹ | 3.1 × 10−³ | let-7c, let-7e, 34c, 138-2, 141, 195, 29a, 138-1, 27b | |
| Function | Cell Migration | 4 | 10.3 | 3.9 × 10−⁴ | 4.2 × 10−¹ | 3.7 × 10−³ | 142, 509-3, 509-1, 509-2 | |
| Function | Adipocyte Differentiation | 7 | 4.8 | 4.4 × 10−⁴ | 4.6 × 10−¹ | 3.8 × 10−³ | 221, let-7e, 222, 378a, 375, 27b, 192 | |
| Function | Adipogenesis | 5 | 7.1 | 4.9 × 10−⁴ | 5.2 × 10−¹ | 4.2 × 10−³ | 148a, 194-2, 29a, 194-1, 363 | |
| Function | Innate Immunity | 7 | 4.7 | 5.1 × 10−⁴ | 5.4 × 10−¹ | 4.3 × 10−³ | let-7c, 149, let-7e, 30a, 21, 223, 142 | |
| Function | Skeletal Muscle Cell Differentiation | 5 | 6.4 | 7.9 × 10−⁴ | 8.4 × 10−¹ | 6.1 × 10−³ | 30d, 30a, 542, 138-2, 138-1 | |
| Function | Cholesterol Efflux | 4 | 7.5 | 1.5 × 10−³ | 1.0 × 10⁰ | 9.3 × 10−³ | 486-2, 486-1, 27b, 378a | |
| Function | Regulation of Akt Pathway | 5 | 5.4 | 1.8 × 10−³ | 1.0 × 10⁰ | 1.1 × 10−² | 221, 222, 196a-2, 141, 196a-1 | |
| Function | T-Cell Differentiation | 4 | 7.1 | 1.9 × 10−³ | 1.0 × 10⁰ | 1.1 × 10−² | let-7e, let-7c, 10a, 21 | |
| Function | Cardiotoxicity | 4 | 6.6 | 2.4 × 10−³ | 1.0 × 10⁰ | 1.4 × 10−² | 34c, 486-2, 486-1, 187 | |
| Function | Glucose Metabolism | 5 | 5.0 | 2.5 × 10−³ | 1.0 × 10⁰ | 1.4 × 10−² | let-7c, let-7e, 223, 195, 375 | |
| Function | Cell Differentiation | 7 | 3.5 | 3.0 × 10−³ | 1.0 × 10⁰ | 1.7 × 10−² | let-7c, 194-2, 503, let-7e, 194-1, 34c, 222 | |
| Function | Cholesterol Homeostasis | 3 | 9.4 | 3.1 × 10−³ | 1.0 × 10⁰ | 1.7 × 10−² | 223, 30c-2, 30c-1 | |
| Function | Bone Regeneration | 5 | 4.7 | 3.4 × 10−³ | 1.0 × 10⁰ | 1.9 × 10−² | 221, 34c, 222, 196a-2, 196a-1 | |
| Function | Tumour Cell Radiation Sensitivity | 2 | 18.8 | 3.6 × 10−³ | 1.0 × 10⁰ | 1.9 × 10−² | 223, 21 | |
| Function | Hormone-mediated Signalling Pathway | 7 | 3.4 | 3.6 × 10−³ | 1.0 × 10⁰ | 1.9 × 10−² | 221, 30d, 363, 21, 223, 29a, 375 | |
| Function | Circadian Rhythm | 4 | 5.1 | 6.5 × 10−³ | 1.0 × 10⁰ | 3.1 × 10−² | 194-2, 194-1, 29a, 192 | |
| Function | Cardiomyocyte Proliferation | 2 | 14.1 | 7.1 × 10−³ | 1.0 × 10⁰ | 3.3 × 10−² | 222, 10a | |
| Function | Peritoneal Cavity Homeostasis | 4 | 4.9 | 7.7 × 10−³ | 1.0 × 10⁰ | 3.4 × 10−² | 148a, 30a, 497, 192 | |
| II vs. III | Disease | Carcinoma, Prostate | 34 | 3.6 | 0.0 × 10⁰ | 0.0 × 10⁰ | 0.0 × 10⁰ | 96, 200c, 574, let-7d, 409, 449a, 135a-2, 375, 497, 155, 182, 195, 204, 424, 4516, 503, 218-2, 135a-1, 146a, 187, 381, 455, 483, let-7c, 148a, 149, 130b, 487b, 191, 21, 141, 218-1, 92b, 29a |
| Function | Apoptosis | 20 | 5.1 | 2.4 × 10−¹⁰ | 2.5 × 10−⁷ | 3.2 × 10−⁸ | 96, 449a, 135a-2, 497, 155, 10a, 182, 195, 204, 424, 4516, 218-2, 135a-1, 146a, let-7c, 148a, 216a, 21, 218-1, 29a | |
| Function | Inflammation | 18 | 4.3 | 3.5 × 10−⁸ | 3.8 × 10−⁵ | 2.2 × 10−⁶ | 584, 20b, let-7d, 135a-2, 155, 182, 424, 218-2, 135a-1, 146a, 455, 148a, 130b, 21, 141, 218-1, 29a, 328 | |
| Function | Epithelial-to-Mesenchymal Transition | 14 | 4.5 | 8.5 × 10−⁷ | 9.1 × 10−⁴ | 3.4 × 10−⁵ | let-7c, 200c, 450a-2, let-7d, 191, 211, 542, 450a-1, 21, 141, 424, 29a, 375, 155 | |
| Function | Aging | 12 | 5.1 | 1.5 × 10−⁶ | 1.6 × 10−³ | 5.6 × 10−⁵ | 96, let-7c, 200c, 148a, let-7d, 146a, 10a, 21, 141, 195, 204, 155 | |
| Function | Cell Cycle | 13 | 4.2 | 5.4 × 10−⁶ | 5.7 × 10−³ | 1.6 × 10−⁴ | 96, 200c, 503, 191, 182, 141, 21, 449a, 195, 424, 92b, 155, 497 | |
| Function | Cell Differentiation | 10 | 4.8 | 2.3 × 10−⁵ | 2.5 × 10−² | 5.5 × 10−⁴ | 96, let-7c, 200c, 503, 218-2, let-7d, 182, 218-1, 424, 155 | |
| Function | Brain Development | 8 | 6.0 | 3.1 × 10−⁵ | 3.3 × 10−² | 7.0 × 10−⁴ | 218-2, 191, 135a-1, 10a, 10b, 218-1, 135a-2, 155 | |
| Function | Myofibroblast Differentiation | 3 | 26.9 | 4.9 × 10−⁵ | 5.2 × 10−² | 9.8 × 10−⁴ | 218-1, 218-2, 424 | |
| Function | Hematopoiesis | 9 | 4.3 | 1.7 × 10−⁴ | 1.8 × 10−¹ | 2.5 × 10−³ | let-7c, 20b, 218-2, let-7d, 146a, 363, 218-1, 29a, 155 | |
| Function | Cardiomyocyte Proliferation | 3 | 20.2 | 1.9 × 10−⁴ | 2.0 × 10−¹ | 2.8 × 10−³ | 204, 424, 10a | |
| Function | T-Cell Differentiation | 5 | 8.4 | 1.9 × 10−⁴ | 2.1 × 10−¹ | 2.7 × 10−³ | let-7c, 10a, 155, let-7d, 21 | |
| Function | Cardiotoxicity | 5 | 7.9 | 2.7 × 10−⁴ | 2.8 × 10−¹ | 3.5 × 10−³ | 424, 146a, 1303, 182, 187 | |
| Function | Nephrotoxicity | 5 | 7.9 | 2.7 × 10−⁴ | 2.8 × 10−¹ | 3.5 × 10−³ | let-7d, 29a, 130b, 200c, 21 | |
| Function | O×idative Stress | 4 | 10.8 | 3.1 × 10−⁴ | 3.3 × 10−¹ | 3.9 × 10−³ | 503, 146a, 21, 141 | |
| Function | Cell Death | 10 | 3.5 | 4.2 × 10−⁴ | 4.5 × 10−¹ | 5.0 × 10−³ | let-7c, 130b, let-7d, 146a, 10b, 182, 21, 29a, 497, 155 | |
| Function | Adipogenesis | 5 | 6.7 | 6.1 × 10−⁴ | 6.5 × 10−¹ | 6.8 × 10−³ | 148a, 204, 455, 29a, 363 | |
| Function | Toll-Like Receptor Signalling Pathway | 3 | 13.5 | 9.0 × 10−⁴ | 9.7 × 10−¹ | 9.8 × 10−³ | 149, 146a, 381 | |
| Function | Osteogenesis | 8 | 3.7 | 1.2 × 10−³ | 1.0 × 10⁰ | 1.2 × 10−² | 96, 200c, 218-2, 211, 21, 218-1, 195, 424 | |
| Function | Neuron Differentiation | 4 | 7.7 | 1.3 × 10−³ | 1.0 × 10⁰ | 1.3 × 10−² | 218-1, 218-2, 96, 182 | |
| Function | Regulation of Nf-Κb Pathway | 3 | 11.5 | 1.5 × 10−³ | 1.0 × 10⁰ | 1.3 × 10−² | 146a, 497, 21 | |
| Function | Regulation of Stem Cell | 9 | 3.1 | 2.0 × 10−³ | 1.0 × 10⁰ | 1.6 × 10−² | 200c, 148a, 146a, 10a, 21, 182, 141, 195, 155 | |
| Function | Cell Proliferation | 9 | 3.0 | 2.2 × 10−³ | 1.0 × 10⁰ | 1.7 × 10−² | let-7c, 200c, 503, let-7d, 146a, 21, 449a, 29a, 92b | |
| Function | T-Cell Activation | 3 | 10.1 | 2.4 × 10−³ | 1.0 × 10⁰ | 1.9 × 10−² | 146a, 155, 21 | |
| Function | Response to Estrogen | 3 | 10.1 | 2.4 × 10−³ | 1.0 × 10⁰ | 1.9 × 10−² | 146a, 21, 182 | |
| Function | Embryonic Development | 4 | 6.3 | 2.9 × 10−³ | 1.0 × 10⁰ | 2.2 × 10−² | 20b, 130b, 10a, 21 | |
| Function | Glucose Metabolism | 5 | 4.8 | 3.1 × 10−³ | 1.0 × 10⁰ | 2.3 × 10−² | let-7c, let-7d, 625, 195, 375 | |
| Function | Innate Immunity | 6 | 3.8 | 3.8 × 10−³ | 1.0 × 10⁰ | 2.7 × 10−² | let-7c, 149, let-7d, 146a, 21, 155 | |
| Function | Bone Regeneration | 5 | 4.5 | 4.2 × 10−³ | 1.0 × 10⁰ | 2.7 × 10−² | 20b, 130b, let-7d, 424, 155 | |
| Function | T-helper 17 Cell Differentiation | 4 | 5.7 | 4.4 × 10−³ | 1.0 × 10⁰ | 2.8 × 10−² | 141, 20b, 155, 21 | |
| Function | Granulopoiesis | 3 | 8.1 | 4.9 × 10−³ | 1.0 × 10⁰ | 3.1 × 10−² | let-7d, 155, 21 | |
| Function | Neurotoxicity | 4 | 5.4 | 5.4 × 10−³ | 1.0 × 10⁰ | 3.3 × 10−² | 92b, 96, 10a, 10b | |
| Function | Immune System(Xiao’s Cell 2010) | 4 | 5.1 | 6.5 × 10−³ | 1.0 × 10⁰ | 3.9 × 10−² | 20b, 146a, 363, 155 | |
| Function | Cell Motility | 4 | 5.1 | 6.5 × 10−³ | 1.0 × 10⁰ | 3.9 × 10−² | 584, 130b, 10b, 21 | |
| Function | Circadian Rhythm | 4 | 4.9 | 7.7 × 10−³ | 1.0 × 10⁰ | 4.4 × 10−² | 96, 191, 182, 29a | |
| Function | Cleavage Stage Development | 2 | 13.5 | 7.8 × 10−³ | 1.0 × 10⁰ | 4.4 × 10−² | 375, 21 | |
| Function | Type II Pneumocyte Differentiation | 2 | 13.5 | 7.8 × 10−³ | 1.0 × 10⁰ | 4.4 × 10−² | 200c, 29a | |
| Function | Adiponectin Signalling | 2 | 13.5 | 7.8 × 10−³ | 1.0 × 10⁰ | 4.4 × 10−² | 218-1, 218-2 | |
| I vs. II + III | Disease | Carcinoma, Prostate | 17 | 3.6 | NaN | 0.0 × 10⁰ | 0.0 × 10⁰ | 194-2, 500b, 376c, 132, 134, 378a, 139, 708, 29c, 503, 194-1, 187, 143, 1299, 483, 223, 29a |
| Function | Inflammation | 10 | 4.8 | 1.8 × 10−⁵ | 1.2 × 10−² | 2.3 × 10−³ | 194-2, 144, 132, 134, 708, 194-1, 143, 140, 223, 29a | |
| Function | Adipogenesis | 5 | 13.5 | 2.1 × 10−⁵ | 1.4 × 10−² | 2.4 × 10−³ | 194-2, 140, 29a, 194-1, 29c | |
| Function | Circadian Rhythm | 5 | 12.2 | 3.4 × 10−⁵ | 2.3 × 10−² | 3.6 × 10−³ | 29c, 194-2, 194-1, 29a, 132 | |
| Function | Epithelial-to-Mesenchymal Transition | 8 | 5.2 | 8.6 × 10−⁵ | 5.9 × 10−² | 5.3 × 10−³ | 29c, 194-2, 194-1, 542, 223, 29a, 144, 143 | |
| Function | Hematopoiesis | 6 | 5.7 | 4.7 × 10−⁴ | 3.2 × 10−¹ | 1.7 × 10−² | 29c, 378a, 223, 29a, 144, 143 | |
| Function | Cell Growth | 2 | 35.9 | 9.9 × 10−⁴ | 6.7 × 10−¹ | 3.0 × 10−² | 132, 143 | |
| Function | Stress Response | 2 | 26.9 | 2.0 × 10−³ | 1.0 × 10⁰ | 4.5 × 10−² | 29c, 143 | |
| I + II vs. III | Disease | Carcinoma, Prostate | 6 | 2.4 | NaN | 0.0 × 10⁰ | 0.0 × 10⁰ | 888, 323a, 134, 146a, 1299, 223 |
| Function | Regulation of Stem Cell | 5 | 6.6 | 6.2 × 10−⁴ | 3.0 × 10−¹ | 1.1 × 10−¹ | 134, 146a, 223, 323a, 142 |
| Individual | P33 | P34 | P35 | HC11 |
|---|---|---|---|---|
| Status group | B | E | E | D |
| Age (years) | ||||
| Primary sample | 57 | 67 | 85 | <45 |
| Post-RP | 58 | |||
| Stage | ||||
| T3 | T3-4 | T3-4 | ||
| N0M0 | NXM1 | NXM1 | ||
| Gleason score | ||||
| 7 (4+3) | 7 (4+3) | 8 (4+4) | ||
| PSA (ng/mL) | ||||
| Primary sample | 17 | 125 | 2.6 | |
| Post-RP | 0 | |||
| mRNA Targets (miR-146a-5p, -223-3p or -892a) | uEV P33 | uEV P34 | uEV P35 | pEV P33 | pEV P34 | pEV P35 |
|---|---|---|---|---|---|---|
| MAP2 | X | X | X | X | X | |
| SLC9A7 | X | X | X | X | X | |
| TLR2 | X | X | X | X | X | |
| LGSN | X | X | X | X | X | |
| VWC2 | X | X | X | X | X | |
| STARD4 | X | X | X | X | X | |
| VCAN | X | X | X | X | X | |
| FMNL3, FLNA | X | X | X | X | ||
| ALG9, GDPD1 | X | X | X | X | ||
| CFTR, PKD2L2, POFUT2, ST8SIA1, SYNPO2, ZNF714 | X | X | X | X | ||
| CADM2, MORC1, RGS5, SLCO3A1 | X | X | X | X | ||
| HAL | X | X | X | X | ||
| GABRB2 | X | X | X | X | ||
| SULT1B1 | X | X | X | X | ||
| INHBB | X | X | X | X | ||
| VNN1 | X | X | X | X | ||
| ATG9A, RBL1, SPATA13, TSHZ3, XPR1 | X | X | X | X | ||
| DSCC1 | X | X | X | |||
| NLRP3 | X | X | X | |||
| SLC35F1, VWA2 | X | X | X | |||
| CTNNA2 | X | X | X | |||
| SLC6A15 | X | X | X | |||
| STXBP5L | X | X | X | |||
| KCND3 | X | X | X | |||
| FZD1, TRDMT1, ZNF367 | X | X | X | |||
| SHOX2 | X | X | X | |||
| MDN1 | X | X | X | |||
| IL1RL2, GJC1 | X | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puhka, M.; Thierens, L.; Nicorici, D.; Forsman, T.; Mirtti, T.; af Hällström, T.; Serkkola, E.; Rannikko, A. Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression. Cancers 2022, 14, 532. https://doi.org/10.3390/cancers14030532
Puhka M, Thierens L, Nicorici D, Forsman T, Mirtti T, af Hällström T, Serkkola E, Rannikko A. Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression. Cancers. 2022; 14(3):532. https://doi.org/10.3390/cancers14030532
Chicago/Turabian StylePuhka, Maija, Lisse Thierens, Daniel Nicorici, Tarja Forsman, Tuomas Mirtti, Taija af Hällström, Elina Serkkola, and Antti Rannikko. 2022. "Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression" Cancers 14, no. 3: 532. https://doi.org/10.3390/cancers14030532
APA StylePuhka, M., Thierens, L., Nicorici, D., Forsman, T., Mirtti, T., af Hällström, T., Serkkola, E., & Rannikko, A. (2022). Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression. Cancers, 14(3), 532. https://doi.org/10.3390/cancers14030532






