NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients/Subjects/Samples
2.2. NanoString Digital Spatial Profiling
2.3. NanoString MicroRNA Profiling
2.4. Immunohistochemistry
3. Results
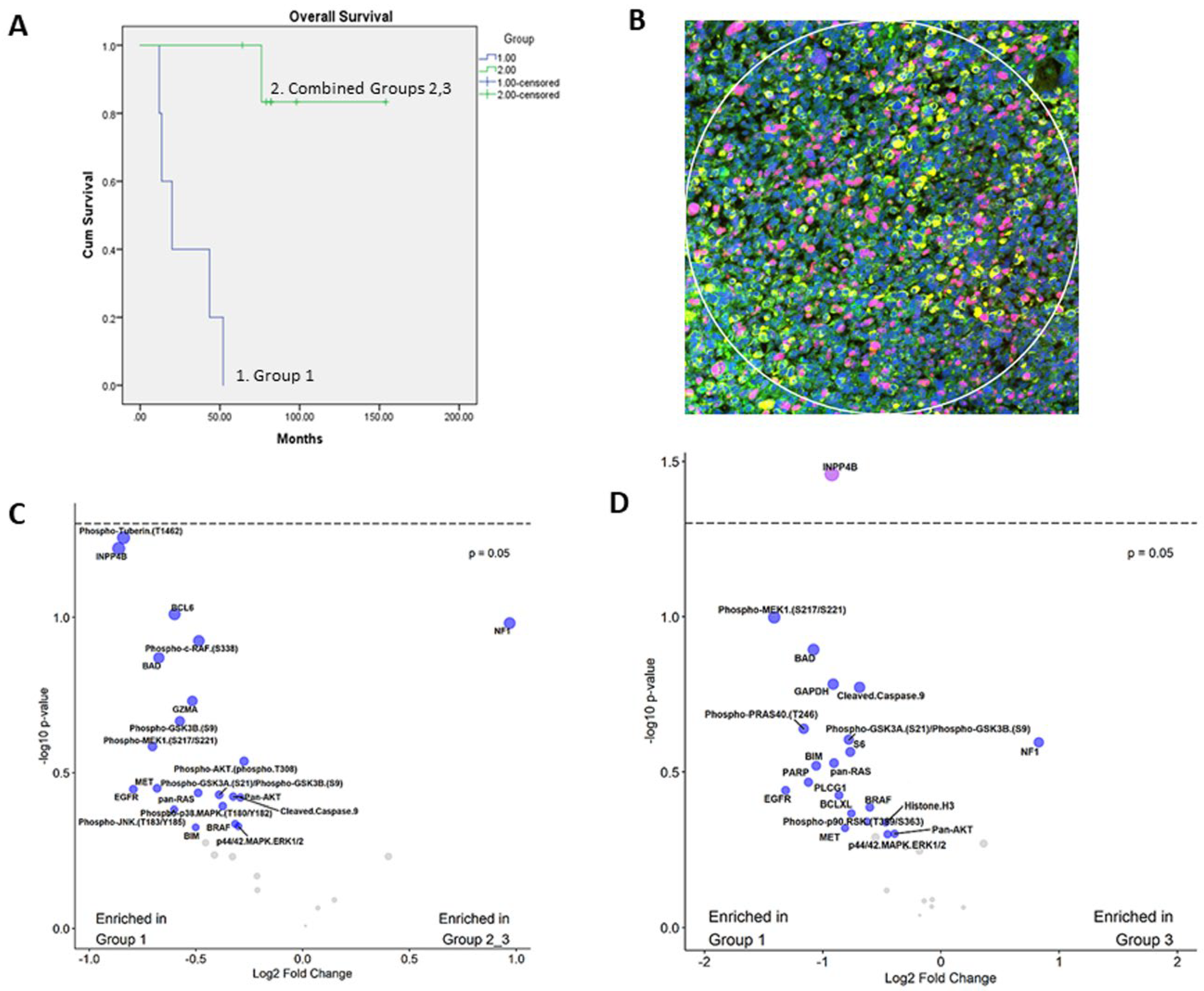
3.1. Prognostic Classification of Patients and Tumor Specimens
3.2. Expression of PI3K-AKT, MAPK and Apoptosis Pathways
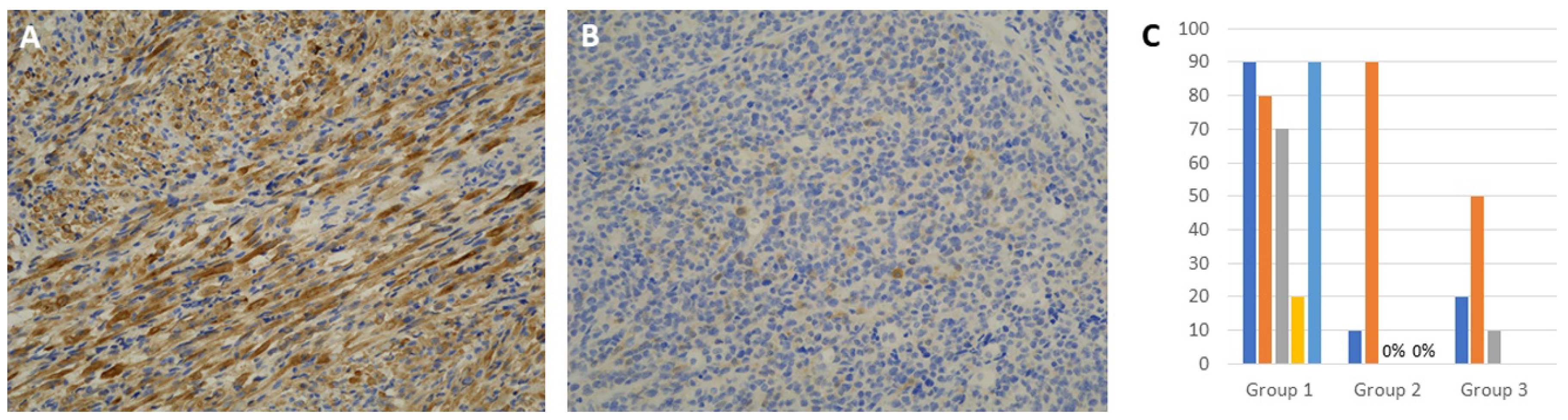
3.3. Immunohistochemical Expression of INPP4B
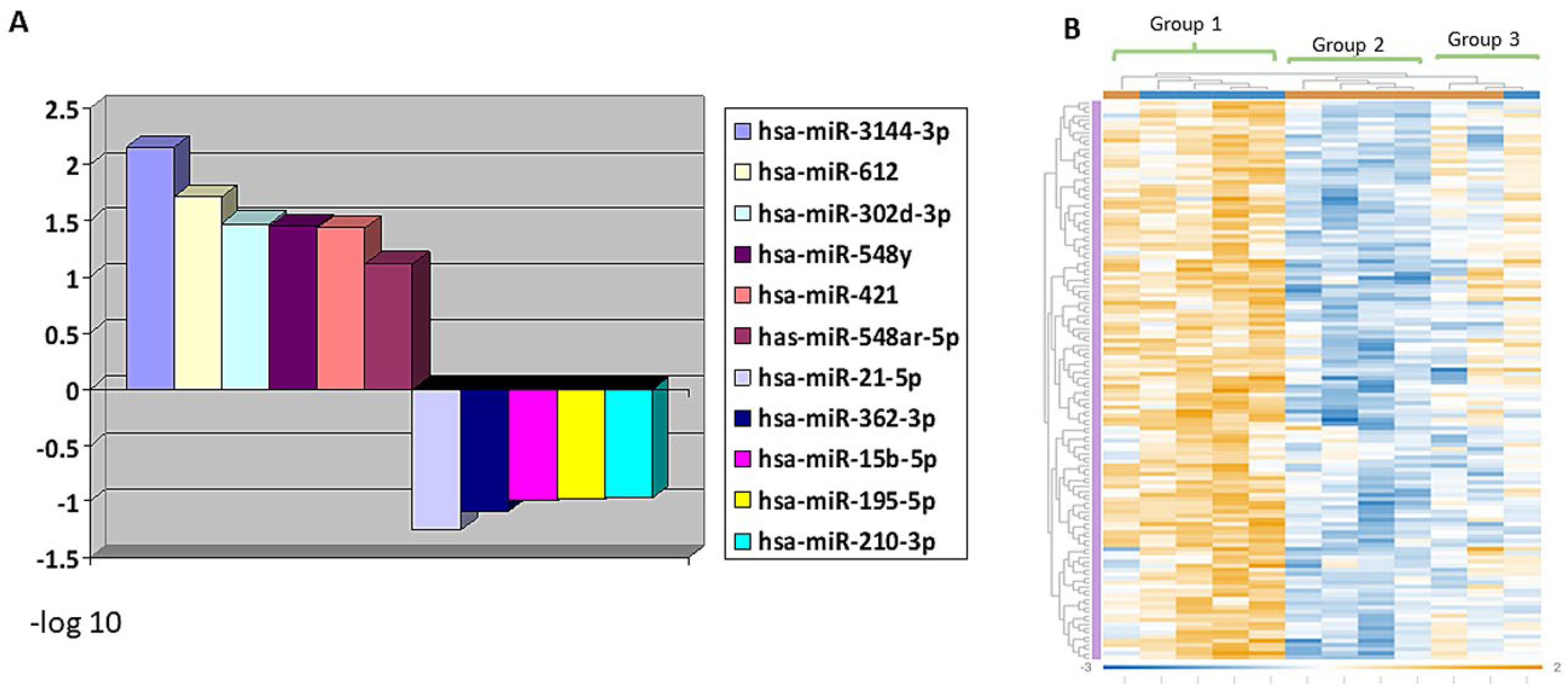
3.4. MiRNA Profiling Showing Differential Expression in RMS Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dasgupta, R.; Fuchs, J.; Rodeberg, D. Rhabdomyosarcoma. Semin. Pediatr. Surg. 2016, 25, 276–283. [Google Scholar] [CrossRef]
- Shern, J.F.; Yohe, M.E.; Khan, J. Pediatric rhabdomyosarcoma. Crit. Rev. Oncog. 2015, 20, 227–243. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Guo, W.; Shen, J.K.; Mankin, H.J.; Hornicek, F.J.; Duan, Z. Rhabdomyosarcoma: Advances in molecular and cellular biology. Sarcoma 2015, 2015, 232010. [Google Scholar] [CrossRef] [Green Version]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef] [Green Version]
- Ramadan, F.; Fahs, A.; Ghayad, S.E.; Saab, R. Signaling pathways in Rhabdomyosarcoma invasion and metastasis. Cancer Metastasis Rev. 2020, 39, 287–301. [Google Scholar] [CrossRef]
- Shukla, N.; Ameur, N.; Yilmaz, I.; Nafa, K.; Lau, C.Y.; Marchetti, A.; Borsu, L.; Barr, F.G.; Ladanyi, M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin. Cancer Res. 2012, 18, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, W.; Nie, Y.; Qaher, M.; Horton, H.E.; Yue, F.; Asakura, A.; Kuang, S. Loss of MyoD promotes fate transdifferentiation of myoblasts into brown adipocytes. EBioMedicine 2017, 16, 212–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasparini, P.; Ferrari, A.; Casanova, M.; Limido, F.; Massimino, M.; Sozzi, G.; Fortunato, O. MiRNAs as players in rhabdomyosarcoma development. Int. J. Mol. Sci. 2019, 20, 5818. [Google Scholar] [CrossRef] [Green Version]
- Tsang, H.F.; Xue, V.W.; Koh, S.P.; Chiu, Y.M.; Ng, L.P.; Wong, S.C. NanoString, a novel digital color-coded barcode technology: Current and future applications in molecular diagnostics. Expert Rev. Mol. Diagn. 2017, 17, 95–103. [Google Scholar] [CrossRef]
- Li, X.; Warren, S.; Pelekanou, V.; Wali, V.; Cesano, A.; Liu, M.; Danaher, P.; Elliott, N.; Nahleh, Z.A.; Hayes, D.F.; et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. J. Immunother. Cancer 2019, 7, 88. [Google Scholar] [CrossRef] [Green Version]
- Goytain, A.; Ng, T. NanoString nCounter technology: High-throughput RNA validation. Methods Mol. Biol. 2020, 2079, 125–139. [Google Scholar] [PubMed]
- Nerurkar, S.N.; Goh, D.; Cheung, C.C.L.; Nga, P.Q.Y.; Lim, J.C.T.; Yeong, J.P.S. Transcriptional spatial profiling of cancer tissues in the era of immunotherapy: The potential and promise. Cancers 2020, 12, 2572. [Google Scholar] [CrossRef]
- Valera, V.A.; Parra-Medina, R.; Walter, B.A.; Pinto, P.; Merino, M.J. microRNA expression profiling in young prostate cancer patients. J. Cancer 2020, 11, 4106–4114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Habeebu, S.; Farooqi, M.S.; Gamis, A.S.; Gonzalez, E.; Flatt, T.; Sherman, A.; Surrey, L.; Arnold, M.A.; Conces, M.; et al. MYOD1 as a prognostic indicator in rhabdomyosarcoma. Pediatr. Blood Cancer 2021, 68, e29085. [Google Scholar] [CrossRef]
- Petricoin, E.F., 3rd; Espina, V.; Araujo, R.P.; Midura, B.; Yeung, C.; Wan, X.; Eichler, G.S.; Johann, D.J., Jr.; Qualman, S.; Tsokos, M.; et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007, 67, 3431–3440. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.; McEvoy, J.; Wang, H.; Chen, X.; Honnell, V.; Ocarz, M.; Gordon, B.; Dapper, J.; Blankenship, K.; Yang, Y.; et al. St. Jude Children’s Research Hospital—Washington University Pediatric Cancer Genome Project. Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell 2018, 34, 411–426.e19. [Google Scholar] [CrossRef] [Green Version]
- Guenther, M.K.; Graab, U.; Fulda, S. Synthetic lethal interaction between PI3K/Akt/mTOR and RAS/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Lett. 2013, 337, 200–209. [Google Scholar] [CrossRef]
- Fulda, S. Targeting apoptosis resistance in rhabdomyosarcoma. Curr. Cancer Drug Targets 2008, 8, 536–544. [Google Scholar] [CrossRef]
- Thimmaiah, K.N.; Easton, J.B.; Houghton, P.J. Protection from rapamycin-induced apoptosis by insulin-like growth factor-I is partially dependent on protein kinase C signaling. Cancer Res. 2010, 70, 2000–2009. [Google Scholar] [CrossRef] [Green Version]
- Ruan, X.H.; Liu, X.M.; Yang, Z.X.; Zhang, S.P.; Li, Q.Z.; Lin, C.S. INPP4B promotes colorectal cancer cell proliferation by activating mTORC1 signaling and cap-dependent translation. Onco Targets Ther. 2019, 12, 3109–3117. [Google Scholar] [CrossRef] [Green Version]
- Mangialardi, E.M.; Chen, K.; Salmon, B.; Vacher, J.; Salmena, L. Investigating the duality of Inpp4b function in the cellular transformation of mouse fibroblasts. Oncotarget 2019, 10, 6378–6390. [Google Scholar] [CrossRef]
- Toki, M.I.; Merritt, C.R.; Wong, P.F.; Smithy, J.W.; Kluger, H.M.; Syrigos, K.N.; Ong, G.T.; Warren, S.E.; Beechem, J.M.; Rimm, D.L. High-Plex predictive marker discovery for melanoma immunotherapy-treated patients using digital spatial profiling. Clin. Cancer Res. 2019, 25, 5503–5512. [Google Scholar] [CrossRef]
- Dobashi, Y.; Suzuki, S.; Sato, E.; Hamada, Y.; Yanagawa, T.; Ooi, A. EGFR-dependent and independent activation of Akt/mTOR cascade in bone and soft tissue tumors. Mod. Pathol. 2009, 22, 1328–1340. [Google Scholar] [CrossRef]
- Yan, D.; Dong Xda, E.; Chen, X.; Wang, L.; Lu, C.; Wang, J.; Qu, J.; Tu, L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J. Biol. Chem. 2009, 284, 29596–29604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miekus, K.; Lukasiewicz, E.; Jarocha, D.; Sekula, M.; Drabik, G.; Majka, M. The decreased metastatic potential of rhabdomyosarcoma cells obtained through MET receptor downregulation and the induction of differentiation. Cell Death Dis. 2013, 4, e459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yohe, M.E.; Gryder, B.E.; Shern, J.F.; Song, Y.K.; Chou, H.C.; Sindiri, S.; Mendoza, A.; Patidar, R.; Zhang, X.; Guha, R.; et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci. Transl. Med. 2018, 10, eaan4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehr, S.; Haydn, T.; Bierbrauer, A.; Irmer, B.; Vogler, M.; Fulda, S. Targeting BCL-2 proteins in pediatric cancer: Dual inhibition of BCL-X(L) and MCL-1 leads to rapid induction of intrinsic apoptosis. Cancer Lett. 2020, 482, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hashimi, S.M.; Good, D.A.; Cao, S.; Duan, W.; Plummer, P.N.; Mellick, A.S.; Wei, M.Q. Apoptosis and microRNA aberrations in cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Kong, D.; He, M.; Gong, J.; Nie, Y.; Tai, S.; Teng, C.B. MiR-7 mediates mitochondrial impairment to trigger apoptosis and necroptosis in rhabdomyosarcoma. Biochim. Biophys. Acta Mol. Cell. Res. 2020, 1867, 118826. [Google Scholar] [CrossRef]
- Zhang, L.; Pang, Y.; Cui, X.; Jia, W.; Cui, W.; Liu, Y.; Liu, C.; Li, F. MicroRNA-410-3p upregulation suppresses proliferation, invasion and migration, and promotes apoptosis in rhabdomyosarcoma cells. Oncol. Lett. 2019, 18, 936–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rota, R.; Ciarapica, R.; Giordano, A.; Miele, L.; Locatelli, F. MicroRNAs in rhabdomyosarcoma: Pathogenetic implications and translational potentiality. Mol. Cancer 2011, 10, 120. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Cai, Q.; Zhang, X.; Zhang, H.; Zhong, Y.; Xu, C.; Li, Y. Two less common human microRNAs miR-875 and miR-3144 target a conserved site of E6 oncogene in most high-risk human papillomavirus subtypes. Protein Cell 2015, 6, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Ying, X.; Wang, J.; Piriyapongsa, J.; Jordan, I.K.; Sheng, J.; Yu, F.; Zhao, P.; Li, Y.; Wang, H.; et al. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. Cancer Res. 2014, 74, 2283–2294. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.S.; Lv, Z.W.; Yu, F.; Chang, Z.Y.; Cong, X.L.; Zhong, X.M.; Lu, G.X.; Zhu, J.; Fu, D. MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J. Exp. Clin. Cancer Res. 2018, 37, 252. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Tao, Z.H.; Wen, D.; Wan, J.L.; Liu, D.L.; Zhang, S.; Cui, J.F.; Sun, H.C.; Wang, L.; Zhou, J.; et al. MiR-612 suppresses the stemness of liver cancer via Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2014, 447, 210–215. [Google Scholar] [CrossRef]
- Lin, X.F.; Zhang, C.Q.; Dong, B.R. MiR-421 expression independently predicts unfavorable overall survival in patients with esophageal adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3790–3798. [Google Scholar] [PubMed]
- Morrow, M.; Donaldson, J. Nanostring’s nCounter—A true digital target profiling technology. PDA J. Pharm. Sci. Technol. 2011, 65, 692. [Google Scholar] [CrossRef] [PubMed]
| Group 1 (n = 5) | Group 2 (n = 4) | Group 3 (n = 3) | |
|---|---|---|---|
| Age (years) | 2–15 | 5–18 | 5–8 |
| Male: female ratio | 4:1 | 3:1 | 3:0 |
| Tumor histology | 3 SRMS, 2 ERMS | 4 SRMS | 3 ERMS |
| Overall survival (months) | Range: 5–52 Mean: 28.2 | Range: 64–82 Mean: 141 | Range: 82–154 Mean: 111 |
| Mortality rate | 5/5 | 1/7 | 0/3 |
| INPP4B, IHC positive rate | Range: 20–90% Mean: 70 | Range: 0–90% Mean: 29.15 | Range: 0–50 Mean: 33.09 |
| SNR Expression Levels | PI3K/AKT | MAPK | Apoptosis |
|---|---|---|---|
| SNR > 3 | INPP4B | Pan-Ras | BAD |
| Pan-AKT | EGFR | Cleaved caspase-9 | |
| MET | phospho-p90 RSK | BCLXL | |
| p44/42 MAPK ERK1/2 | NF1 | ||
| PARP | |||
| P53 | |||
| SNR > 1 | phospho-PRAS40 | BRAF | BIM |
| phospho-GSK3A | Phospho-MEK1 | ||
| Phospho-AKT1 | |||
| PLCG1 | |||
| SNR < 1 | Phospho-AKT | Phospho-cRAF | BCL-6 |
| Phospho-tuberin | Phospho-p44/42 MAPK ERK1/2 | CD95/Fas | |
| phospho-GSK3B | Phospho-JNK | ||
| Phospho-p38 MAPK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, A.A.; Farooqi, M.S.; Habeebu, S.S.; Gonzalez, E.; Flatt, T.G.; Wilson, A.L.; Barr, F.G. NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma. Cancers 2022, 14, 522. https://doi.org/10.3390/cancers14030522
Ahmed AA, Farooqi MS, Habeebu SS, Gonzalez E, Flatt TG, Wilson AL, Barr FG. NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma. Cancers. 2022; 14(3):522. https://doi.org/10.3390/cancers14030522
Chicago/Turabian StyleAhmed, Atif A., Midhat S. Farooqi, Sultan S. Habeebu, Elizabeth Gonzalez, Terrie G. Flatt, Ashley L. Wilson, and Frederic G. Barr. 2022. "NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma" Cancers 14, no. 3: 522. https://doi.org/10.3390/cancers14030522
APA StyleAhmed, A. A., Farooqi, M. S., Habeebu, S. S., Gonzalez, E., Flatt, T. G., Wilson, A. L., & Barr, F. G. (2022). NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma. Cancers, 14(3), 522. https://doi.org/10.3390/cancers14030522







