Small Gastric Stromal Tumors: An Underestimated Risk
Abstract
Simple Summary
Abstract
1. Introduction
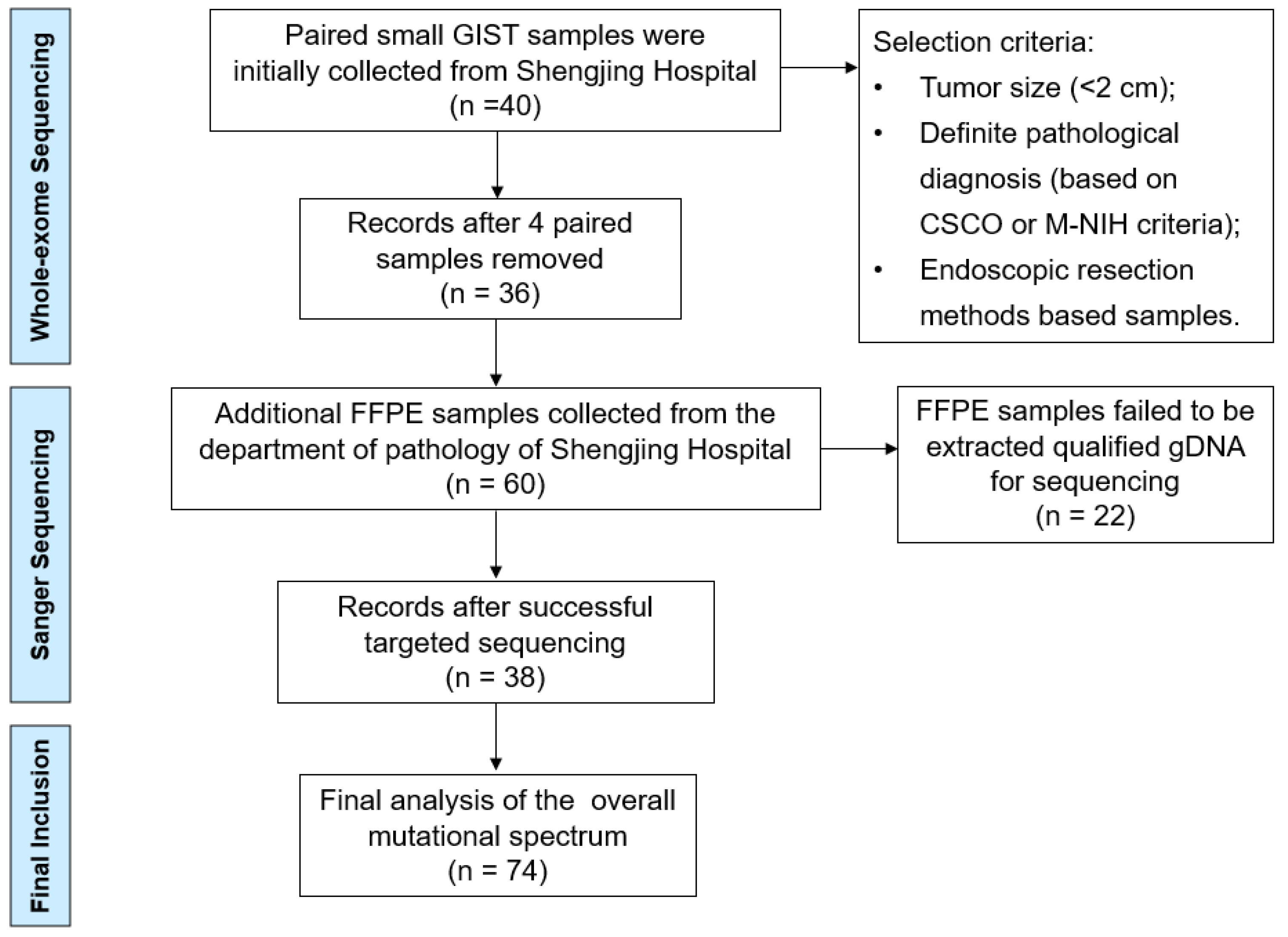
2. Patients and Methods
2.1. Clinical Samples
2.2. Ethics Statement
2.3. Whole-Exome Sequencing
2.4. Validation of Variants by Sanger Sequencing
2.5. In Silico Analysis
3. Results
3.1. Clinical Features
3.2. Molecular Analysis
Small GISTs with KIT/PDGFRA Mutations
3.3. KIT/PDGFRA Wild-Type (WT) Small GISTs
3.4. Suspicious Oncogenic Mutations in Small GISTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blay, J.-Y.; Kang, Y.-K.; Nishida, T.; von Mehren, M. Gastrointestinal Stromal Tumours. Nat. Rev. Dis. Prim. 2021, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, H.; Hohenberger, P.; Corless, C.L. Gastrointestinal Stromal Tumour. Lancet 2013, 382, 973–983. [Google Scholar] [CrossRef]
- Ma, G.L.; Murphy, J.D.; Martinez, M.E.; Sicklick, J.K. Epidemiology of Gastrointestinal Stromal Tumors in the Era of Histology Codes: Results of a Population-Based Study. Cancer Epidemiol. Biomark. Prev. 2015, 24, 298–302. [Google Scholar] [CrossRef]
- Nishida, T.; Goto, O.; Raut, C.P.; Yahagi, N. Diagnostic and Treatment Strategy for Small Gastrointestinal Stromal Tumors: Small Gastrointestinal Stromal Tumors. Cancer 2016, 122, 3110–3118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ye, Y.; Wang, J.; Zhang, B.; Qin, S.; Shi, Y.; He, Y.; Liang, X.; Liu, X.; Zhou, Y.; et al. Chinese Consensus Guidelines for Diagnosis and Management of Gastrointestinal Stromal Tumor. Chin. J. Cancer Res. 2017, 29, 281–293. [Google Scholar] [CrossRef] [PubMed]
- von Mehren, M.; Randall, R.L.; Benjamin, R.S.; Boles, S.; Bui, M.M.; Ganjoo, K.N.; George, S.; Gonzalez, R.J.; Heslin, M.J.; Kane, J.M.; et al. Soft Tissue Sarcoma, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 536–563. [Google Scholar] [CrossRef]
- Poveda, A.; García del Muro, X.; López-Guerrero, J.A.; Cubedo, R.; Martínez, V.; Romero, I.; Serrano, C.; Valverde, C.; Martín-Broto, J. GEIS Guidelines for Gastrointestinal Sarcomas (GIST). Cancer Treat. Rev. 2017, 55, 107–119. [Google Scholar] [CrossRef]
- Joensuu, H. Risk Stratification of Patients Diagnosed with Gastrointestinal Stromal Tumor. Hum. Pathol. 2008, 39, 1411–1419. [Google Scholar] [CrossRef]
- Miettinen, M.; Lasota, J. Gastrointestinal Stromal Tumors: Pathology and Prognosis at Different Sites. Semin. Diagn. Pathol. 2006, 23, 70–83. [Google Scholar] [CrossRef]
- Coe, T.M.; Fero, K.E.; Fanta, P.T.; Mallory, R.J.; Tang, C.-M.; Murphy, J.D.; Sicklick, J.K. Population-Based Epidemiology and Mortality of Small Malignant Gastrointestinal Stromal Tumors in the USA. J. Gastrointest. Surg. 2016, 20, 1132–1140. [Google Scholar] [CrossRef]
- Kawanowa, K.; Sakuma, Y.; Sakurai, S.; Hishima, T.; Iwasaki, Y.; Saito, K.; Hosoya, Y.; Nakajima, T.; Funata, N. High Incidence of Microscopic Gastrointestinal Stromal Tumors in the Stomach. Hum. Pathol. 2006, 37, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Khan, S.; Hui, Y.; Zhao, J.; Li, B.; Ma, S.; Guo, J.; Chen, X.; Wang, B. Treatment Recommendations for Small Gastric Gastrointestinal Stromal Tumors: Positive Endoscopic Resection. Scand. J. Gastroenterol. 2019, 54, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Gasparotto, D.; Toffolatti, L.; Pastrello, C.; Gallina, G.; Marzotto, A.; Sartor, C.; Barbareschi, M.; Cantaloni, C.; Messerini, L.; et al. Molecular and Clinicopathologic Characterization of Gastrointestinal Stromal Tumors (GISTs) of Small Size. Am. J. Surg. Pathol. 2010, 34, 1480–1491. [Google Scholar] [CrossRef]
- Feng, X.; Yang, Z.; Zhang, P.; Chen, T.; Qiu, H.; Zhou, Z.; Li, G.; Tao, K.; Wang, H.; Li, Y. Which Size Is the Best Cutoff for Primary Small Gastric Gastrointestinal Stromal Tumor? J. Gastrointest. Oncol. 2020, 11, 402–410. [Google Scholar] [CrossRef]
- Tran, T.; Davila, J.A.; El-Serag, H.B. The Epidemiology of Malignant Gastrointestinal Stromal Tumors: An Analysis of 1,458 Cases from 1992 to 2000. Am. J. Gastroenterol. 2005, 100, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Agaimy, A.; Blaszyk, H.; Dietmaier, W. Minute Gastric Sclerosing Stromal Tumors (GIST Tumorlets) Are Common in Adults and Frequently Show c-KIT Mutations. Am. J. Surg. Pathol. 2007, 31, 8. [Google Scholar] [CrossRef] [PubMed]
- Søreide, K. Cancer Biology of Small Gastrointestinal Stromal Tumors (<2 cm): What Is the Risk of Malignancy? Eur. J. Surg. Oncol. 2017, 43, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Basse, C.; Italiano, A.; Penel, N.; Mir, O.; Chemin, C.; Toulmonde, M.; Duffaud, F.; Le Cesne, A.; Chevreau, C.; Maynou, C.; et al. Sarcomas in Patients over 90: Natural History and Treatment-A Nationwide Study over 6 Years. Int. J. Cancer 2019, 145, 2135–2143. [Google Scholar] [CrossRef]
- Fang, Y.-J.; Cheng, T.-Y.; Sun, M.-S.; Yang, C.-S.; Chen, J.-H.; Liao, W.-C.; Wang, H.-P. Suggested Cutoff Tumor Size for Management of Small EUS-Suspected Gastric Gastrointestinal Stromal Tumors. J. Formos. Med. Assoc. 2012, 111, 88–93. [Google Scholar] [CrossRef]
- Ge, Q.-C.; Wu, Y.-F.; Liu, Z.-M.; Wang, Z.; Wang, S.; Liu, X.; Ge, N.; Guo, J.-T.; Sun, S.-Y. Efficacy of Endoscopic Ultrasound in the Evaluation of Small Gastrointestinal Stromal Tumors. World J. Gastroenterol. 2022, 28, 5457–5468. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, C.; Xue, Q.; Wang, J.; Shen, Z.; Jiang, K.; Shen, K.; Liang, B.; Yang, X.; Xie, Q.; et al. The Cut-off Value of Tumor Size and Appropriate Timing of Follow-up for Management of Minimal EUS-Suspected Gastric Gastrointestinal Stromal Tumors. BMC Gastroenterol. 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xue, A.; Yuan, W.; Gao, X.; Fu, M.; Fang, Y.; Wang, L.; Shu, P.; Li, H.; Hou, Y.; et al. Clinicopathological Features and Prognosis of Small Gastric Gastrointestinal Stromal Tumors (GISTs). J. Gastrointest. Surg. 2019, 23, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Kuwatani, M.; Sakamoto, N. Evolution and a Promising Role of EUS-FNA in Gene and Future Analyses. Endosc. Ultrasound 2020, 9, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Sofuni, A.; Tsuchiya, T.; Kono, S.; Ishii, K.; Tanaka, R.; Tonozuka, R.; Mukai, S.; Yamamoto, K.; Matsunami, Y.; et al. Efficacy of the Franseen Needle for Diagnosing Gastrointestinal Submucosal Lesions Including Small Tumors. Endosc. Ultrasound 2021, 10, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Boye, K.; Berner, J.-M.; Hompland, I.; Bruland, Ø.S.; Stoldt, S.; Sundby Hall, K.; Bjerkehagen, B.; Hølmebakk, T. Genotype and Risk of Tumour Rupture in Gastrointestinal Stromal Tumour. Br. J. Surg. 2018, 105, e169–e175. [Google Scholar] [CrossRef]
- Rizzo, A.; Pantaleo, M.A.; Astolfi, A.; Indio, V.; Nannini, M. The Identity of PDGFRA D842V-Mutant Gastrointestinal Stromal Tumors (GIST). Cancers 2021, 13, 705. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Jones, R.L.; von Mehren, M.; Schöffski, P.; Serrano, C.; Kang, Y.-K.; Cassier, P.A.; Mir, O.; Eskens, F.; Tap, W.D.; et al. Avapritinib in Advanced PDGFRA D842V-Mutant Gastrointestinal Stromal Tumour (NAVIGATOR): A Multicentre, Open-Label, Phase 1 Trial. Lancet Oncol. 2020, 21, 935–946. [Google Scholar] [CrossRef]
- Abbaspour Babaei, M.; Kamalidehghan, B.; Saleem, M.; Zaman Huri, H.; Ahmadipour, F. Receptor Tyrosine Kinase (c-Kit) In Drug Des. Dev. Ther. 2016, 10, 2443–2459. [Google Scholar] [CrossRef]
- Duan, Y.; Haybaeck, J.; Yang, Z. Therapeutic Potential of PI3K/AKT/MTOR Pathway in Gastrointestinal Stromal Tumors: Rationale and Progress. Cancers 2020, 12, 2972. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, J.; García-Valverde, A.; Serrano, C.; Flynn, D.L.; Smith, B.D. Ripretinib and MEK Inhibitors Synergize to Induce Apoptosis in Preclinical Models of GIST and Systemic Mastocytosis. Mol. Cancer Ther. 2021, 20, 1234–1245. [Google Scholar] [CrossRef]
- Haefliger, S.; Marston, K.; Juskevicius, D.; Meyer-Schaller, N.; Forster, A.; Nicolet, S.; Komminoth, P.; Stauffer, E.; Cathomas, G.; Hoeller, S.; et al. Molecular Profile of Gastrointestinal Stromal Tumors in Sixty-Eight Patients from a Single Swiss Institution. Pathobiology 2020, 87, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Huss, S.; Pasternack, H.; Ihle, M.A.; Merkelbach-Bruse, S.; Heitkötter, B.; Hartmann, W.; Trautmann, M.; Gevensleben, H.; Büttner, R.; Schildhaus, H.-U.; et al. Clinicopathological and Molecular Features of a Large Cohort of Gastrointestinal Stromal Tumors (GISTs) and Review of the Literature: BRAF Mutations in KIT/PDGFRA Wild-Type GISTs Are Rare Events. Hum. Pathol. 2017, 62, 206–214. [Google Scholar] [CrossRef]
- Alqathama, A. BRAF in Malignant Melanoma Progression and Metastasis: Potentials and Challenges. Am. J. Cancer Res. 2020, 10, 1103–1114. [Google Scholar] [PubMed]
- Ran, L.; Murphy, D.; Sher, J.; Cao, Z.; Wang, S.; Walczak, E.; Guan, Y.; Xie, Y.; Shukla, S.; Zhan, Y.; et al. ETV1-Positive Cells Give Rise to BRAF V600E -Mutant Gastrointestinal Stromal Tumors. Cancer Res. 2017, 77, 3758–3765. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Huh, W.J.; Franklin, J.L.; Heinrich, M.C.; Rubin, B.P.; Coffey, R.J. A Smooth Muscle-derived, BRAF -driven Mouse Model of Gastrointestinal Stromal Tumor (GIST): Evidence for an Alternative GIST Cell-of-origin. J. Pathol. 2020, 252, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Wang, X.; Xia, Q.; Rao, Q.; Shen, Q.; Ye, S.; Li, R.; Shi, Q.; Lu, Z.; Ma, H.; et al. P16 Overexpression in BRAF -Mutated Gastrointestinal Stromal Tumors. Expert Rev. Mol. Diagn. 2017, 17, 195–201. [Google Scholar] [CrossRef]
- Geng, A.; Tang, H.; Huang, J.; Qian, Z.; Qin, N.; Yao, Y.; Xu, Z.; Chen, H.; Lan, L.; Xie, H.; et al. The Deacetylase SIRT6 Promotes the Repair of UV-Induced DNA Damage by Targeting DDB2. Nucleic Acids Res. 2020, 48, 9181–9194. [Google Scholar] [CrossRef]
- Kiapour, A.M.; Cao, J.; Young, M.; Capellini, T.D. The Role of Gdf5 Regulatory Regions in Development of Hip Morphology. PLoS ONE 2018, 13, e0202785. [Google Scholar] [CrossRef]



| Primers | Sequence |
|---|---|
| KIT-Exon9-F | CCTTTAGATGCTCTGCTTC |
| KIT-Exon9-R | GGTAGACAGAGCCTAAACATC |
| KIT-Exon11-F | GTGCTCTAATGACTGAGACAAT |
| KIT-Exon11-R | AGGAAGCCACTGGAGTTC |
| KIT-Exon13-F | TGCATGCGCTTGACATCAGTTTG |
| KIT-Exon13-R | AGGCAGCTTGGACACGGCTT |
| KIT-Exon14-F | GTCTGATCCACTGAAGCTG |
| KIT-Exon14-R | ACCCCATGAACTGCCTGTC |
| KIT-Exon17-F | TGGTTTTCTTTTCTCCTCCAACC |
| KIT-Exon17-R | GCAGGACTGTCAAGCAGAG |
| PDGFRA-Exon12-F | TCCAGTCACTGTGCTGCTTC |
| PDGFRA-Exon 12-R | GCAAGGGAAAAGGGAGTCTT |
| PDGFRA-Exon14-F | GGTAGCTCAGCTGGACTGAT |
| PDGFRA-Exon14-R | GGATGGAGAGTGGAGGATTT |
| PDGFRA-Exon18-F | TCAGCTACAGATGGCTTGATC |
| PDGFRA-Exon18-R | TGAAGGAGGATGAGCCTGACC |
| BRAF Exon15-F | CTTCATAATGCTTGCTCTG |
| BRAF-Exon15-R | GTAACTCAGCAGCATCTCAG |
| Clinical Pathological Characteristics | Number (%) |
|---|---|
| Sex | |
| Male | 27 (36.5) |
| Female | 47 (63.5) |
| Age | |
| Median, years | 56 |
| Range, years | 30–75 |
| 30–50 years | 22 (29.7) |
| 51–60 years | 24 (32.4) |
| 61–75 years | 28 (37.9) |
| Primary site | |
| Fundus | 38 (51.3) |
| Junction of the fundus and body | 5 (6.8) |
| Body | 29 (39.2) |
| Antrum | 2 (2.7) |
| Tumor size | |
| <1 cm (micro-GIST) | 28 (37.8) |
| 1–2 cm (mini-GIST) | 46 (62.2) |
| Classification of risk | |
| Very low | 58 (78.4) |
| Low | 12 (16.2) |
| Intermediate | 3 (4.1) |
| High | 1 (1.3) |
| Gene | Size (cm) | Nucleotide Change (c.Notation) | Amino Acid Change (p.Notation) | SIFT | Polyphen2_HVAR | Malignancy Potential |
|---|---|---|---|---|---|---|
| SIRT6 | 2 ×1.5 | c.A956C | p.K319T | 0.007,D | 0.987,D | Low |
| GDF5 | 2 ×1.5 | c.A630T | p.Q210H | 0.248,T | 0.395,B | Low |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Ge, Q.; Yang, F.; Wang, S.; Ge, N.; Liu, X.; Shi, J.; Fusaroli, P.; Liu, Y.; Sun, S. Small Gastric Stromal Tumors: An Underestimated Risk. Cancers 2022, 14, 6008. https://doi.org/10.3390/cancers14236008
Guo J, Ge Q, Yang F, Wang S, Ge N, Liu X, Shi J, Fusaroli P, Liu Y, Sun S. Small Gastric Stromal Tumors: An Underestimated Risk. Cancers. 2022; 14(23):6008. https://doi.org/10.3390/cancers14236008
Chicago/Turabian StyleGuo, Jintao, Qichao Ge, Fan Yang, Sheng Wang, Nan Ge, Xiang Liu, Jing Shi, Pietro Fusaroli, Yang Liu, and Siyu Sun. 2022. "Small Gastric Stromal Tumors: An Underestimated Risk" Cancers 14, no. 23: 6008. https://doi.org/10.3390/cancers14236008
APA StyleGuo, J., Ge, Q., Yang, F., Wang, S., Ge, N., Liu, X., Shi, J., Fusaroli, P., Liu, Y., & Sun, S. (2022). Small Gastric Stromal Tumors: An Underestimated Risk. Cancers, 14(23), 6008. https://doi.org/10.3390/cancers14236008





