Systematic Investigation of the Diagnostic and Prognostic Impact of LINC01087 in Human Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomics Data Analysis
2.2. Survival Curve Analysis
2.3. ROC Curve Analysis
2.4. Network and Pathway Enrichment Analyses of LINC01087
3. Results
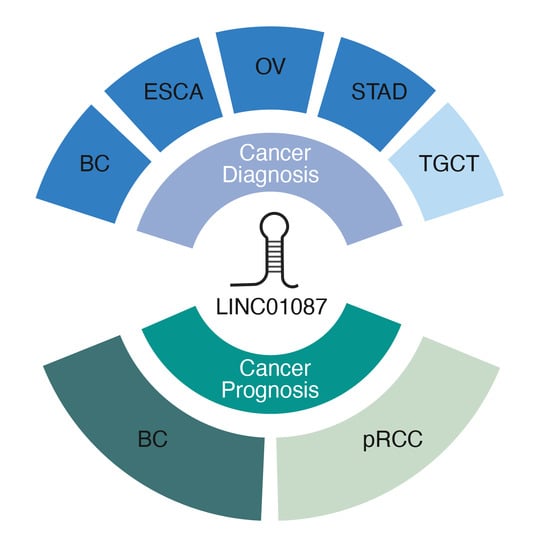
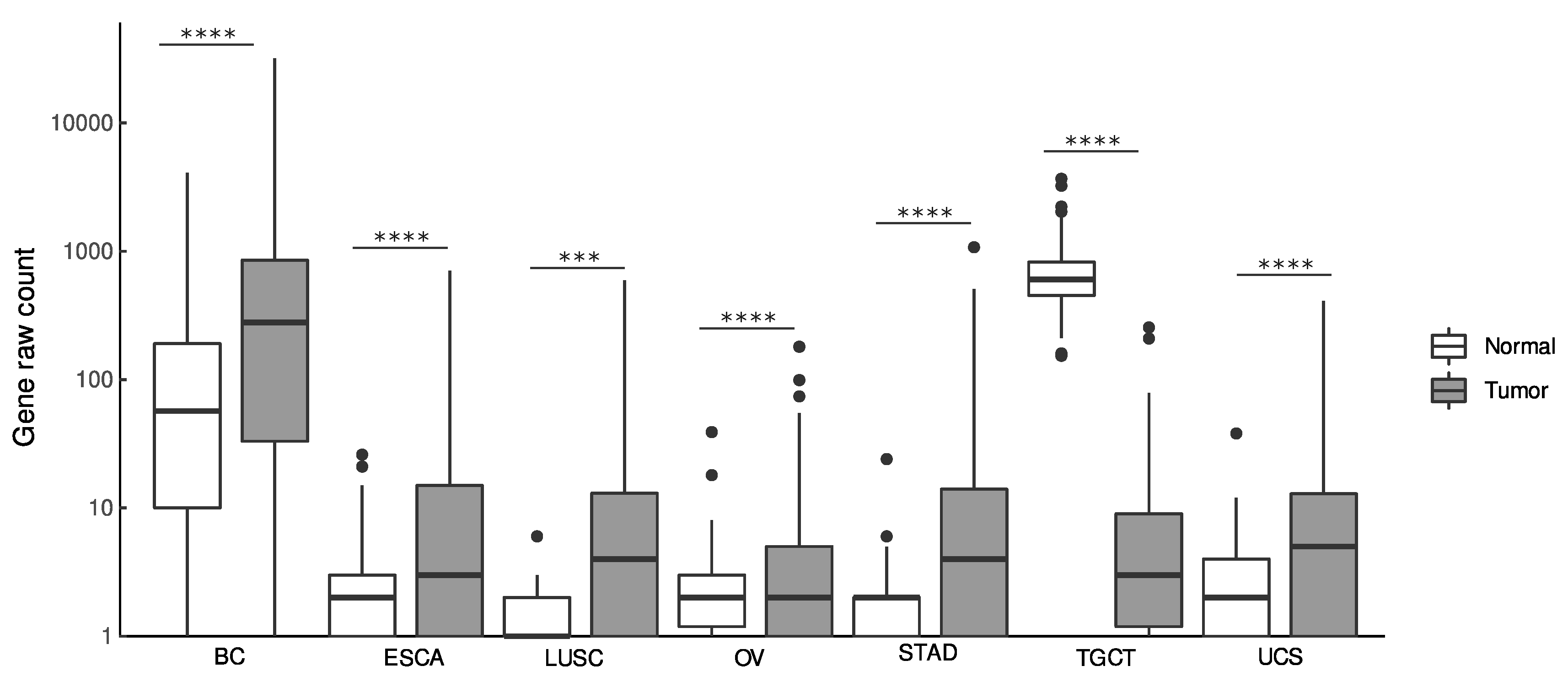
3.1. Diagnostic Value of LINC01087
3.2. Prognostic Value of LINC01087 in Cancer
3.3. Impact of LINC01087 on Cancer-Related Pathways
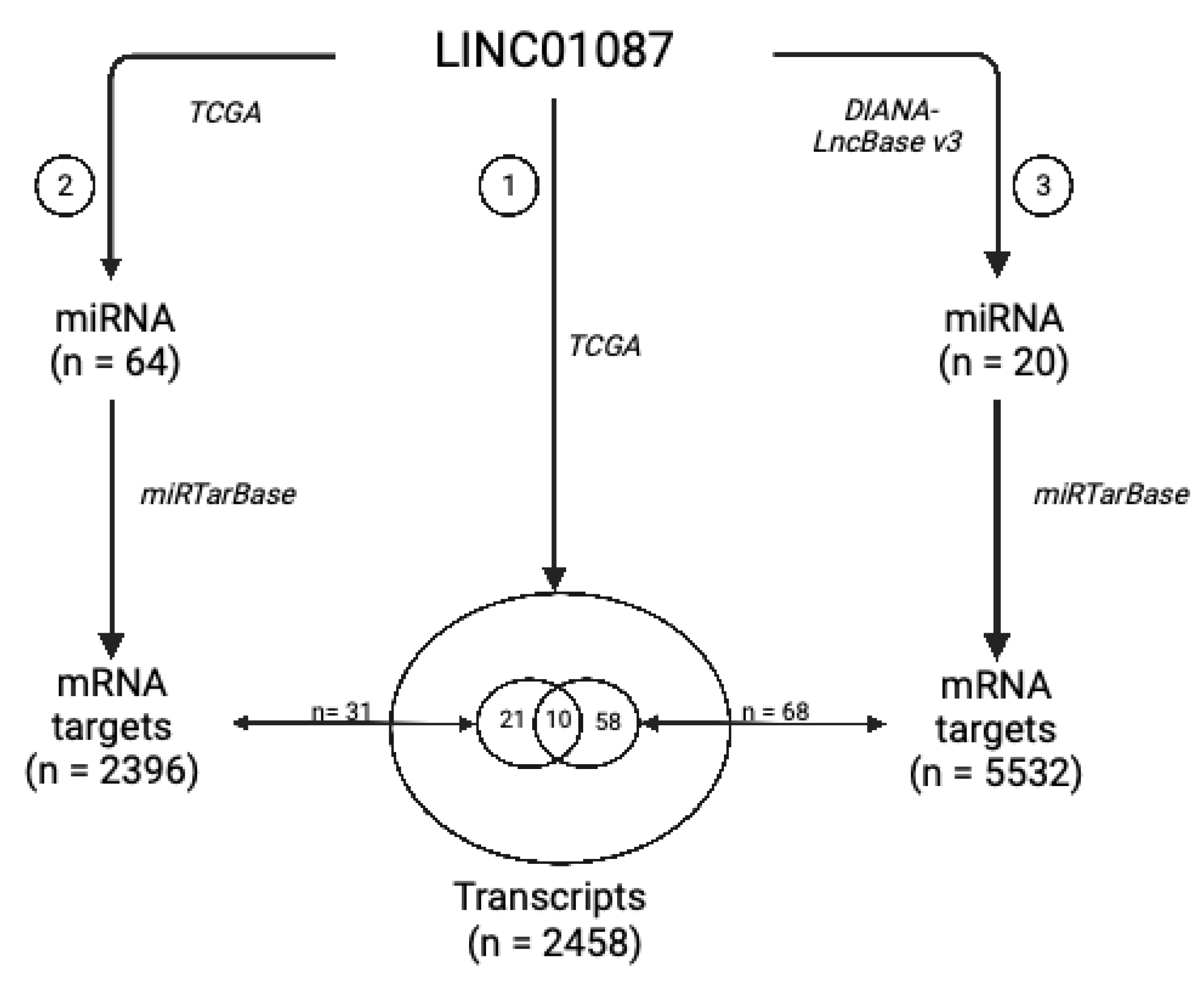
3.4. LINC01087 Functions as Competing Endogenous RNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Hayes, D.F. Cancer Biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Precone, V.; Del Monaco, V.; Esposito, M.V.; De Palma, F.D.E.; Ruocco, A.; Salvatore, F.; D’Argenio, V. Cracking the Code of Human Diseases Using Next-Generation Sequencing: Applications, Challenges, and Perspectives. Biomed Res. Int. 2015, 2015, 161648. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef]
- Srivastava, A.; Creek, D.J. Discovery and Validation of Clinical Biomarkers of Cancer: A Review Combining Metabolomics and Proteomics. Proteomics 2019, 19, e1700448. [Google Scholar] [CrossRef]
- Arechederra, M.; Recalde, M.; Gárate-Rascón, M.; Fernández-Barrena, M.G.; Ávila, M.A.; Berasain, C. Epigenetic Biomarkers for the Diagnosis and Treatment of Liver Disease. Cancers 2021, 13, 1265. [Google Scholar] [CrossRef]
- Parrella, P. The Value of Epigenetic Biomarkers in Breast Cancer. Biomark. Med. 2018, 12, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Reddy, D.; Gupta, S. Global Histone Post-Translational Modifications and Cancer: Biomarkers for Diagnosis, Prognosis and Treatment? World J. Biol. Chem. 2015, 6, 333–345. [Google Scholar] [CrossRef]
- Lu, H.; Liu, Y.; Wang, J.; Fu, S.; Wang, L.; Huang, C.; Li, J.; Xie, L.; Wang, D.; Li, D.; et al. Detection of Ovarian Cancer Using Plasma Cell-Free DNA Methylomes. Clin. Epigenetics 2022, 14, 74. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-Coding RNA Networks in Cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- von Deimling, A.; Korshunov, A.; Hartmann, C. The next Generation of Glioma Biomarkers: MGMT Methylation, BRAF Fusions and IDH1 Mutations. Brain Pathol. 2011, 21, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Palanca-Ballester, C.; Hervas, D.; Villalba, M.; Valdes-Sanchez, T.; Garcia, D.; Alcoriza-Balaguer, M.I.; Benet, M.; Martinez-Tomas, R.; Briones-Gomez, A.; Galbis-Caravajal, J.; et al. Translation of a Tissue Epigenetic Signature to Circulating Free DNA Suggests BCAT1 as a Potential Noninvasive Diagnostic Biomarker for Lung Cancer. Clin. Epigenetics 2022, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Clifton, S.; Locke, W.; Luu, P.-L.; Du, Q.; Lam, D.; Armstrong, N.J.; Kumar, B.; Deng, N.; Harvey, K.; et al. Identification of DNA Methylation Biomarkers with Potential to Predict Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin. Epigenetics 2021, 13, 226. [Google Scholar] [CrossRef]
- De Palma, F.D.E.; Luglio, G.; Tropeano, F.P.; Pagano, G.; D’Armiento, M.; Kroemer, G.; Maiuri, M.C.; De Palma, G.D. The Role of Micro-RNAs and Circulating Tumor Markers as Predictors of Response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Int. J. Mol. Sci. 2020, 21, 7040. [Google Scholar] [CrossRef]
- Sarkar, D.; Diermeier, S.D. Circular RNAs: Potential Applications as Therapeutic Targets and Biomarkers in Breast Cancer. Noncoding RNA 2021, 7, 2. [Google Scholar] [CrossRef]
- De Palma, F.D.E.; Raia, V.; Kroemer, G.; Maiuri, M.C. The Multifaceted Roles of MicroRNAs in Cystic Fibrosis. Diagnostics 2020, 10, 1102. [Google Scholar] [CrossRef]
- De Palma, F.D.E.; D’Argenio, V.; Pol, J.; Kroemer, G.; Maiuri, M.C.; Salvatore, F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers 2019, 11, 1017. [Google Scholar] [CrossRef]
- De Palma, F.D.E.; Salvatore, F.; Pol, J.G.; Kroemer, G.; Maiuri, M.C. Circular RNAs as Potential Biomarkers in Breast Cancer. Biomedicines 2022, 10, 725. [Google Scholar] [CrossRef]
- Cabús, L.; Lagarde, J.; Curado, J.; Lizano, E.; Pérez-Boza, J. Current Challenges and Best Practices for Cell-Free Long RNA Biomarker Discovery. Biomark. Res. 2022, 10, 62. [Google Scholar] [CrossRef]
- Di Cecilia, S.; Zhang, F.; Sancho, A.; Li, S.D.; Aguilo, F.; Sun, Y.; Rengasamy, M.; Zhang, W.; Vecchio, L.D.; Salvatore, F.; et al. RBM5-AS1 Is Critical for Self-Renewal of Colon Cancer Stem-like Cells. Cancer Res. 2016, 76, 5615–5627. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a Complete Map of the Human Long Non-Coding RNA Transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long Non-Coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Hou, Z.; Zhao, W.; Zhou, J.; Shen, L.; Zhan, P.; Xu, C.; Chang, C.; Bi, H.; Zou, J.; Yao, X.; et al. A Long Noncoding RNA Sox2ot Regulates Lung Cancer Cell Proliferation and Is a Prognostic Indicator of Poor Survival. Int. J. Biochem. Cell Biol. 2014, 53, 380–388. [Google Scholar] [CrossRef]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-Associated Long Non-Coding RNA Drives Gastric Cancer Development and Promotes Peritoneal Metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef]
- Deng, H.-Y.; Wang, Y.-C.; Ni, P.-Z.; Lin, Y.-D.; Chen, L.-Q. Long Noncoding RNAs Are Novel Potential Prognostic Biomarkers for Esophageal Squamous Cell Carcinoma: An Overview. J. Thorac. Dis. 2016, 8, E653–E659. [Google Scholar] [CrossRef]
- Lu, C.; Wei, D.; Zhang, Y.; Wang, P.; Zhang, W. Long Non-Coding RNAs as Potential Diagnostic and Prognostic Biomarkers in Breast Cancer: Progress and Prospects. Front. Oncol. 2021, 11, 710538. [Google Scholar] [CrossRef] [PubMed]
- De Palma, F.D.E.; Del Monaco, V.; Pol, J.G.; Kremer, M.; D’Argenio, V.; Stoll, G.; Montanaro, D.; Uszczyńska-Ratajczak, B.; Klein, C.C.; Vlasova, A.; et al. The Abundance of the Long Intergenic Non-Coding RNA 01087 Differentiates between Luminal and Triple-Negative Breast Cancers and Predicts Patient Outcome. Pharmacol. Res. 2020, 161, 105249. [Google Scholar] [CrossRef] [PubMed]
- Flippot, R.; Malouf, G.G.; Su, X.; Mouawad, R.; Spano, J.-P.; Khayat, D. Cancer Subtypes Classification Using Long Non-Coding RNA. Oncotarget 2016, 7, 54082–54093. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qian, J.; Wang, Y.; Zhang, J.; You, Y. Long Noncoding RNA Profiles Reveal Three Molecular Subtypes in Glioma. CNS Neurosci. Ther. 2014, 20, 339–343. [Google Scholar] [CrossRef]
- Su, X.; Malouf, G.G.; Chen, Y.; Zhang, J.; Yao, H.; Valero, V.; Weinstein, J.N.; Spano, J.-P.; Meric-Bernstam, F.; Khayat, D.; et al. Comprehensive Analysis of Long Non-Coding RNAs in Human Breast Cancer Clinical Subtypes. Oncotarget 2014, 5, 9864–9876. [Google Scholar] [CrossRef]
- Tian, Y.-H.; Jia, L.-W.; Liu, Z.-F.; Chen, Y.-H. LINC01087 Inhibits Glioma Cell Proliferation and Migration, and Increases Cell Apoptosis via MiR-384/Bcl-2 Axis. Aging 2021, 13, 20808–20819. [Google Scholar] [CrossRef]
- Chen, W.; Wang, F.; Zhang, J.; Li, C.; Hong, L. LINC01087 Indicates a Poor Prognosis of Glioma Patients with Preoperative MRI. Funct. Integr. Genom. 2022, 22, 55–64. [Google Scholar] [CrossRef]
- Yin, Y.; Huang, J.; Shi, H.; Huang, Y.; Huang, Z.; Song, M.; Yin, L. LINC01087 Promotes the Proliferation, Migration, and Invasion of Thyroid Cancer Cells by Upregulating PPM1E. J. Oncol. 2022, 2022, 7787378. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, F.; Qiang, G.; Zhang, Z.; Ma, Q.; Hao, Y.; Xing, H.; Liang, C. Novel LncRNA Panel as for Prognosis in Esophageal Squamous Cell Carcinoma Based on CeRNA Network Mechanism. Comput. Math. Methods Med. 2021, 2021, 8020879. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 3 June 2021).
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, Statistics and Computing, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 978-0-387-95457-8. [Google Scholar]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing Experimentally Supported MiRNA Targets on Non-Coding Transcripts. Nucleic Acids Res. 2020, 48, D101–D110. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. MiRTarBase 2020: Updates to the Experimentally Validated MicroRNA–Target Interaction Database. Nucleic Acids Res. 2019, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed]
- Calmon, M.F.; Jeschke, J.; Zhang, W.; Dhir, M.; Siebenkäs, C.; Herrera, A.; Tsai, H.-C.; O’Hagan, H.M.; Pappou, E.P.; Hooker, C.M.; et al. Epigenetic Silencing of Neurofilament Genes Promotes an Aggressive Phenotype in Breast Cancer. Epigenetics 2015, 10, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Johnson, R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell 2019, 73, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kuscu, C.; Banach, A.; Zhang, Q.; Pulkoski-Gross, A.; Kim, D.; Liu, J.; Roth, E.; Li, E.; Shroyer, K.R.; et al. MiR-181a-5p Inhibits Cancer Cell Migration and Angiogenesis via Downregulation of Matrix Metalloproteinase-14. Cancer Res. 2015, 75, 2674–2685. [Google Scholar] [CrossRef]
- Ardizzone, A.; Calabrese, G.; Campolo, M.; Filippone, A.; Giuffrida, D.; Esposito, F.; Colarossi, C.; Cuzzocrea, S.; Esposito, E.; Paterniti, I. Role of MiRNA-19a in Cancer Diagnosis and Poor Prognosis. Int. J. Mol. Sci. 2021, 22, 4697. [Google Scholar] [CrossRef] [PubMed]
- Melling, N.; Harutyunyan, L.; Hube-Magg, C.; Kluth, M.; Simon, R.; Lebok, P.; Minner, S.; Tsourlakis, M.C.; Koop, C.; Graefen, M.; et al. High-Level HOOK3 Expression Is an Independent Predictor of Poor Prognosis Associated with Genomic Instability in Prostate Cancer. PLoS ONE 2015, 10, e0134614. [Google Scholar] [CrossRef]
- Huang, W.; Li, B.-R.; Feng, H. PLAG1 Silencing Promotes Cell Chemosensitivity in Ovarian Cancer via the IGF2 Signaling Pathway. Int. J. Mol. Med. 2020, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jaberi-Douraki, M.; Wallace, N.A. Predicting the Prognostic Value of POLI Expression in Different Cancers via a Machine Learning Approach. Int. J. Mol. Sci. 2022, 23, 8571. [Google Scholar] [CrossRef] [PubMed]
- Katabi, N.; Xu, B.; Jungbluth, A.A.; Zhang, L.; Shao, S.Y.; Lane, J.; Ghossein, R.; Antonescu, C.R. PLAG1 Immunohistochemistry Is a Sensitive Marker for Pleomorphic Adenoma: A Comparative Study with PLAG1 Genetic Abnormalities. Histopathology 2018, 72, 285. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Meng, J.; Gong, W.; Zhang, Z.; Gan, X.; Wang, J.; Wu, Z.; Liu, B.; Qu, L.; Wang, L. Elevated SNRPA1, as a Promising Predictor Reflecting Severe Clinical Outcome via Effecting Tumor Immunity for CcRCC, Is Related to Cell Invasion, Metastasis, and Sunitinib Sensitivity. Front. Immunol. 2022, 13, 842096. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Feng, X.; Cui, Y.; Wang, L.; Gan, J.; Zhao, Y.; Meng, Q. Expression Characteristic, Immune Signature, and Prognosis Value of EFNA Family Identified by Multi-Omics Integrative Analysis in Pan-Cancer. BMC Cancer 2022, 22, 871. [Google Scholar] [CrossRef]
- Ebron, J.S.; Shankar, E.; Singh, J.; Sikand, K.; Weyman, C.M.; Gupta, S.; Lindner, D.J.; Liu, X.; Campbell, M.J.; Shukla, G.C. MiR-644a Disrupts Oncogenic Transformation and Warburg Effect by Direct Modulation of Multiple Genes of Tumor-Promoting Pathways. Cancer Res. 2019, 79, 1844–1856. [Google Scholar] [CrossRef]
- Heverhagen, A.E.; Legrand, N.; Wagner, V.; Fendrich, V.; Bartsch, D.K.; Slater, E.P. Overexpression of MicroRNA MiR-7-5p Is a Potential Biomarker in Neuroendocrine Neoplasms of the Small Intestine. Neuroendocrinology 2018, 106, 312–317. [Google Scholar] [CrossRef]
- Long, M.; Zhan, M.; Xu, S.; Yang, R.; Chen, W.; Zhang, S.; Shi, Y.; He, Q.; Mohan, M.; Liu, Q.; et al. MiR-92b-3p Acts as a Tumor Suppressor by Targeting Gabra3 in Pancreatic Cancer. Mol. Cancer 2017, 16, 167. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, X.; Zhou, X.; Wu, B.; Zhu, D.; Jia, W.; Chu, J.; Wang, J.; Wu, J.; Kong, L. MiR-3174 Promotes Proliferation and Inhibits Apoptosis by Targeting FOXO1 in Hepatocellular Carcinoma. Biochem. Biophys. Res. Commun. 2020, 526, 889–897. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Lu, Q.; Fei, X.; Lu, C.; Li, C.; Chen, H. MiR-34a-5p Inhibits Proliferation, Migration, Invasion and Epithelial-Mesenchymal Transition in Esophageal Squamous Cell Carcinoma by Targeting LEF1 and Inactivation of the Hippo-YAP1/TAZ Signaling Pathway. J. Cancer 2020, 11, 3072–3081. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, H.; Feng, X.; Li, H.; Qiu, C.; Yi, X.; Tang, H.; Zhang, J. Long Non-Coding RNA FENDRR Acts as a MiR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol. Ther. Nucleic Acids 2019, 17, 516–529. [Google Scholar] [CrossRef]
- Zheng, Y.P.; Wu, L.; Gao, J.; Wang, Y. Tumor Suppressive Role of MiR-569 in Lung Cancer. Oncol. Lett. 2018, 15, 4087–4092. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hong, S.; Liu, Z. LncRNA LINC00641 Predicts Prognosis and Inhibits Bladder Cancer Progression through MiR-197-3p/KLF10/PTEN/PI3K/AKT Cascade. Biochem. Biophys. Res. Commun. 2018, 503, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.-Y.; Shan, T.-D.; Pan, X.-T.; Tian, Z.-B.; Liu, X.-S.; Liu, F.-G.; Sun, X.-G.; Xue, H.-G.; Li, X.-H.; Han, Y.; et al. The LncRNA ZEB1-AS1 Sponges MiR-181a-5p to Promote Colorectal Cancer Cell Proliferation by Regulating Wnt/β-Catenin Signaling. Cell Cycle 2018, 17, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Hao, Y.; Ren, H.; Dang, Z.; Xu, H.; Xue, X.; Gao, Y. MiR-1305 Inhibits The Progression Of Non-Small Cell Lung Cancer By Regulating MDM2. Cancer Manag. Res. 2019, 11, 9529–9540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wan, H.; Zhang, X. LncRNA LHFPL3-AS1 Contributes to Tumorigenesis of Melanoma Stem Cells via the MiR-181a-5p/BCL2 Pathway. Cell Death Dis. 2020, 11, 950. [Google Scholar] [CrossRef]
- She, J.-K.; Fu, D.-N.; Zhen, D.; Gong, G.-H.; Zhang, B. LINC01087 Is Highly Expressed in Breast Cancer and Regulates the Malignant Behavior of Cancer Cells Through MiR-335-5p/Rock1. OncoTargets Ther. 2020, 13, 9771–9783. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, Q.; Lu, Z.; Wang, L.; Ding, L.; Xia, L.; Zhang, H.; Wang, M.; Chen, Y.; Li, G. Role of the Nervous System in Cancers: A Review. Cell Death Discov. 2021, 7, 76. [Google Scholar] [CrossRef]
- Li, Y.-H.; Liu, Y.; Li, Y.-D.; Liu, Y.-H.; Li, F.; Ju, Q.; Xie, P.-L.; Li, G.-C. GABA Stimulates Human Hepatocellular Carcinoma Growth through Overexpressed GABAA Receptor Theta Subunit. World J. Gastroenterol. 2012, 18, 2704–2711. [Google Scholar] [CrossRef]
- Takehara, A.; Hosokawa, M.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Nakamura, Y.; Nakagawa, H. γ-Aminobutyric Acid (GABA) Stimulates Pancreatic Cancer Growth through Overexpressing GABAA Receptor π Subunit. Cancer Res. 2007, 67, 9704–9712. [Google Scholar] [CrossRef]
- Watanabe, M.; Maemura, K.; Kanbara, K.; Tamayama, T.; Hayasaki, H. GABA and GABA Receptors in the Central Nervous System and Other Organs. In International Review of Cytology, A Survey of Cell Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2002; Volume 213, pp. 1–47. [Google Scholar]
- Le, A.; Udupa, S.; Zhang, C. The Metabolic Interplay between Cancer and Other Diseases. Trends Cancer 2019, 5, 809–821. [Google Scholar] [CrossRef]
- Gumireddy, K.; Li, A.; Kossenkov, A.V.; Sakurai, M.; Yan, J.; Li, Y.; Xu, H.; Wang, J.; Zhang, P.J.; Zhang, L.; et al. The MRNA-Edited Form of GABRA3 Suppresses GABRA3-Mediated Akt Activation and Breast Cancer Metastasis. Nat. Commun. 2016, 7, 10715. [Google Scholar] [CrossRef] [PubMed]
- Keihani, S.; Kluever, V.; Mandad, S.; Bansal, V.; Rahman, R.; Fritsch, E.; Gomes, L.C.; Gärtner, A.; Kügler, S.; Urlaub, H.; et al. The Long Noncoding RNA NeuroLNC Regulates Presynaptic Activity by Interacting with the Neurodegeneration-Associated Protein TDP-43. Sci. Adv. 2019, 5, eaay2670. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Wu, P.; Zhong, Z.; He, Z.; Li, W. Long Non-Coding RNA Gm37494 Alleviates Osteoarthritis Chondrocyte Injury via the MicroRNA-181a-5p/GABRA1 Axis. J. Orthop. Surg. Res. 2022, 17, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-X.; Chen, Z.-H.; Xu, Y.; Chen, J.-W.; Weng, H.-W.; Yun, M.; Zheng, Z.-S.; Chen, C.; Wu, B.-L.; Li, E.-M.; et al. Downregulation of MicroRNA-644a Promotes Esophageal Squamous Cell Carcinoma Aggressiveness and Stem Cell–like Phenotype via Dysregulation of PITX2. Clin. Cancer Res. 2017, 23, 298–310. [Google Scholar] [CrossRef]
- Liang, W.; Liao, Y.; Li, Z.; Wang, Y.; Zheng, S.; Xu, X.; Ran, F.; Tang, B.; Wang, Z. MicroRNA-644a Promotes Apoptosis of Hepatocellular Carcinoma Cells by Downregulating the Expression of Heat Shock Factor 1. Cell Commun. Signal. 2018, 16, 30. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, Y.-S.L.; Matthen, M.; Yoon, A.; Schwartz, G.K.; Bala, S.; Taylor, A.M.; Momen-Heravi, F. Down-Regulation of the Tumor Suppressor MiR-34a Contributes to Head and Neck Cancer by up-Regulating the MET Oncogene and Modulating Tumor Immune Evasion. J. Exp. Clin. Cancer Res. 2021, 40, 70. [Google Scholar] [CrossRef]
- Xiao, H. MiR-7-5p Suppresses Tumor Metastasis of Non-Small Cell Lung Cancer by Targeting NOVA2. Cell Mol. Biol. Lett. 2019, 24, 60. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, T.; Du, Y.; Hu, X.; Xia, W. LncRNA LUCAT1/MiR-181a-5p Axis Promotes Proliferation and Invasion of Breast Cancer via Targeting KLF6 and KLF15. BMC Mol. Cell Biol. 2020, 21, 69. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ryu, D.; Lim, S.W.; Ryu, K.J.; Choi, M.E.; Yoon, S.E.; Kim, K.; Park, C.; Kim, S.J. Exosomal MiR-1305 in the Oncogenic Activity of Hypoxic Multiple Myeloma Cells: A Biomarker for Predicting Prognosis. J. Cancer 2021, 12, 2825–2834. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Zhang, G.; Luo, C.; Zhang, S.; Luo, R.; Deng, B. Hsa-MiR-7-5p Suppresses Proliferation, Migration and Promotes Apoptosis in Hepatocellular Carcinoma Cell Lines by Inhibiting SPC24 Expression. Biochem. Biophys. Res. Commun. 2021, 561, 80–87. [Google Scholar] [CrossRef]
- Pan, Y.; Jin, K.; Xie, X.; Wang, K.; Zhang, H. MicroRNA-19a-3p Inhibits the Cellular Proliferation and Invasion of Non-small Cell Lung Cancer by Downregulating UBAP2L. Exp. Ther. Med. 2020, 20, 2252–2261. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.-M.; Yu, X.-N.; Liu, T.-T.; Zhu, H.-R.; Shi, X.; Bilegsaikhan, E.; Guo, H.-Y.; Song, G.-Q.; Weng, S.-Q.; Huang, X.-X.; et al. MicroRNA-19a-3p Promotes Tumor Metastasis and Chemoresistance through the PTEN/Akt Pathway in Hepatocellular Carcinoma. Biomed. Pharmacother. 2018, 105, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Ye, L.; Han, Z.; Liu, Y.; Yang, Y.; Peng, Z.; Fan, J. MiR-19b-3p Promotes Colon Cancer Proliferation and Oxaliplatin-Based Chemoresistance by Targeting SMAD4: Validation by Bioinformatics and Experimental Analyses. J. Exp. Clin. Cancer Res. 2017, 36, 131. [Google Scholar] [CrossRef]
- Wei, Y.; Guo, S.; Tang, J.; Wen, J.; Wang, H.; Hu, X.; Gu, Q. MicroRNA-19b-3p Suppresses Gastric Cancer Development by Negatively Regulating Neuropilin-1. Cancer Cell Int. 2020, 20, 193. [Google Scholar] [CrossRef]
- Chaluvally-Raghavan, P.; Zhang, F.; Pradeep, S.; Hamilton, M.P.; Zhao, X.; Rupaimoole, R.; Moss, T.; Lu, Y.; Yu, S.; Pecot, C.V.; et al. Copy Number Gain of Hsa-MiR-569 at 3q26.2 Leads to Loss of TP53INP1 and Aggressiveness of Epithelial Cancers. Cancer Cell 2014, 26, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, Y.; Hao, M.; Zhu, R. LINC00665 Functions as a Competitive Endogenous RNA to Regulate AGTR1 Expression by Sponging MiR-34a-5p in Glioma. Oncol. Rep. 2021, 45, 1202–1212. [Google Scholar] [CrossRef]
- Shao, T.; Hu, Y.; Tang, W.; Shen, H.; Yu, Z.; Gu, J. The Long Noncoding RNA HOTAIR Serves as a MicroRNA-34a-5p Sponge to Reduce Nucleus Pulposus Cell Apoptosis via a NOTCH1-Mediated Mechanism. Gene 2019, 715, 144029. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Wang, M.; Li, X.; Cui, F. The Long Noncoding RNA NEAT1 Targets MiR-34a-5p and Drives Nasopharyngeal Carcinoma Progression via Wnt/β-Catenin Signaling. Yonsei Med. J. 2019, 60, 336–345. [Google Scholar] [CrossRef]
- Gao, Y.; Luo, X.; Zhang, J. LincRNA-ROR Is Activated by H3K27 Acetylation and Induces EMT in Retinoblastoma by Acting as a Sponge of MiR-32 to Activate the Notch Signaling Pathway. Cancer Gene Ther. 2021, 28, 42–54. [Google Scholar] [CrossRef]
- Li, Q.-Y.; Shen, J.-Q.; Li, J.-H.; Dai, D.-F.; Saeed, M.; Li, C.-X. LINC00958/MiR-3174/PHF6 Axis Is Responsible for Triggering Proliferation, Migration and Invasion of Endometrial Cancer. Eur. Rev. Med. Pharm. Sci. 2021, 25, 6853–6861. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhao, Z.; Shang, J.; Li, G.; Zhang, R. LncRNA NEAT1 Promotes Glioma Cancer Progression via Regulation of MiR-98-5p/BZW1. Biosci. Rep. 2021, 41, BSR20200767. [Google Scholar] [CrossRef] [PubMed]
- Cilek, E.E.; Ozturk, H.; Dedeoglu, B.G. Construction of MiRNA-MiRNA Networks Revealing the Complexity of MiRNA-Mediated Mechanisms in Trastuzumab Treated Breast Cancer Cell Lines. PLoS ONE 2017, 12, e0185558. [Google Scholar] [CrossRef] [PubMed]
- Enokida, H.; Yoshino, H.; Matsushita, R.; Nakagawa, M. The Role of MicroRNAs in Bladder Cancer. Investig. Clin. Urol. 2016, 57, S60. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, K.; Hanazawa, T.; Fukumoto, I.; Kikkawa, N.; Okamoto, Y.; Seki, N. The MicroRNA Signatures: Aberrantly Expressed MicroRNAs in Head and Neck Squamous Cell Carcinoma. J. Hum. Genet. 2017, 62, 3–13. [Google Scholar] [CrossRef]
- Shams, R.; Saberi, S.; Zali, M.; Sadeghi, A.; Ghafouri-Fard, S.; Aghdaei, H.A. Identification of Potential MicroRNA Panels for Pancreatic Cancer Diagnosis Using Microarray Datasets and Bioinformatics Methods. Sci. Rep. 2020, 10, 7559. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, Y.; He, X.; Zheng, Z.; Xue, D. Functional Implication of Exosomal MiR-217 and MiR-23b-3p in the Progression of Prostate Cancer. OncoTargets Ther. 2020, 13, 11595–11606. [Google Scholar] [CrossRef]
- Lin, C.-W.; Yang, W.-E.; Lee, W.-J.; Hua, K.-T.; Hsieh, F.-K.; Hsiao, M.; Chen, C.-C.; Chow, J.-M.; Chen, M.-K.; Yang, S.-F.; et al. Lipocalin 2 Prevents Oral Cancer Metastasis through Carbonic Anhydrase IX Inhibition and Is Associated with Favourable Prognosis. Carcinogenesis 2016, 37, 712–722. [Google Scholar] [CrossRef]
- He, S.; Zeng, S.; Zhou, Z.-W.; He, Z.-X.; Zhou, S.-F. Hsa-MicroRNA-181a Is a Regulator of a Number of Cancer Genes and a Biomarker for Endometrial Carcinoma in Patients: A Bioinformatic and Clinical Study and the Therapeutic Implication. Drug Des. Dev. Ther. 2015, 9, 1103–1175. [Google Scholar] [CrossRef][Green Version]
- Sakiyama, T.; Kohno, T.; Mimaki, S.; Ohta, T.; Yanagitani, N.; Sobue, T.; Kunitoh, H.; Saito, R.; Shimizu, K.; Hirama, C.; et al. Association of Amino Acid Substitution Polymorphisms in DNA Repair Genes TP53, POLI, REV1 and LIG4 with Lung Cancer Risk. Int. J. Cancer 2005, 114, 730–737. [Google Scholar] [CrossRef]
- Hoff, A.M.; Kraggerud, S.M.; Alagaratnam, S.; Berg, K.C.G.; Johannessen, B.; Høland, M.; Nilsen, G.; Lingjærde, O.C.; Andrews, P.W.; Lothe, R.A.; et al. Frequent Copy Number Gains of SLC2A3 and ETV1 in Testicular Embryonal Carcinomas. Endocr. Relat. Cancer 2020, 27, 457–468. [Google Scholar] [CrossRef]
- Ziegler, G.C.; Almos, P.; McNeill, R.V.; Jansch, C.; Lesch, K.-P. Cellular Effects and Clinical Implications of SLC2A3 Copy Number Variation. J. Cell. Physiol. 2020, 235, 9021–9036. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; He, Z.; Qin, C.; Deng, X.; Bai, L.; Li, G.; Shi, J. SLC2A3 Promotes Macrophage Infiltration by Glycolysis Reprogramming in Gastric Cancer. Cancer Cell Int. 2020, 20, 503. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liang, J.; Duan, J.; Chen, L.; Li, H.; Zhen, T.; Zhang, F.; Dong, Y.; Shi, H.; Han, A. A Prognosis Marker SLC2A3 Correlates With EMT and Immune Signature in Colorectal Cancer. Front. Oncol. 2021, 11, 638099. [Google Scholar] [CrossRef]
- Chu, M.; Zheng, K.; Li, X.; Luo, Z.; Yang, X.; Wei, C. Comprehensive Analysis of the Role of SLC2A3 on Prognosis and Immune Infiltration in Head and Neck Squamous Cell Carcinoma. Anal. Cell. Pathol. 2022, 2022, e2371057. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hong, J.; Han, H.; Park, J.; Kim, D.; Park, H.; Ko, M.; Koh, Y.; Shin, D.-Y.; Yoon, S.-S. Decreased Vitamin C Uptake Mediated by SLC2A3 Promotes Leukaemia Progression and Impedes TET2 Restoration. Br. J. Cancer 2020, 122, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.-W.; Lu, Q.; Wang, L.-X.; Zhao, W.-Y.; Cao, Y.-Q.; Li, Y.-N.; Han, G.-S.; Liu, J.-M.; Yue, Z.-J. Decreased MiR-106a Inhibits Glioma Cell Glucose Uptake and Proliferation by Targeting SLC2A3 in GBM. BMC Cancer 2013, 13, 478. [Google Scholar] [CrossRef]
- Li, J.; Fu, F.; Wan, X.; Huang, S.; Wu, D.; Li, Y. Up-Regulated MiR-29c Inhibits Cell Proliferation and Glycolysis by Inhibiting SLC2A3 Expression in Prostate Cancer. Gene 2018, 665, 26–34. [Google Scholar] [CrossRef]
- Chen, D.; Wang, H.; Chen, J.; Li, Z.; Li, S.; Hu, Z.; Huang, S.; Zhao, Y.; He, X. MicroRNA-129-5p Regulates Glycolysis and Cell Proliferation by Targeting the Glucose Transporter SLC2A3 in Gastric Cancer Cells. Front Pharm. 2018, 9, 502. [Google Scholar] [CrossRef]
- Kania, A.; Klein, R. Mechanisms of Ephrin–Eph Signalling in Development, Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2016, 17, 240–256. [Google Scholar] [CrossRef]
- Wang, T.-H.; Chang, J.-L.; Ho, J.-Y.; Wu, H.-C.; Chen, T.-C. EphrinA5 Suppresses Colon Cancer Development by Negatively Regulating Epidermal Growth Factor Receptor Stability. FEBS J. 2012, 279, 251–263. [Google Scholar] [CrossRef]
- Herath, N.I.; Spanevello, M.D.; Sabesan, S.; Newton, T.; Cummings, M.; Duffy, S.; Lincoln, D.; Boyle, G.; Parsons, P.G.; Boyd, A.W. Over-Expression of Eph and Ephrin Genes in Advanced Ovarian Cancer: Ephrin Gene Expression Correlates with Shortened Survival. BMC Cancer 2006, 6, 144. [Google Scholar] [CrossRef] [PubMed]
- Giaginis, C.; Tsourouflis, G.; Zizi-Serbetzoglou, A.; Kouraklis, G.; Chatzopoulou, E.; Dimakopoulou, K.; Theocharis, S.E. Clinical Significance of Ephrin (Eph)-A1, -A2, -A4, -A5 and -A7 Receptors in Pancreatic Ductal Adenocarcinoma. Pathol. Oncol. Res. 2010, 16, 267–276. [Google Scholar] [CrossRef]
- Li, J.-J.; Liu, D.-P.; Liu, G.-T.; Xie, D. EphrinA5 Acts as a Tumor Suppressor in Glioma by Negative Regulation of Epidermal Growth Factor Receptor. Oncogene 2009, 28, 1759–1768. [Google Scholar] [CrossRef]
- Feng, J.; Guo, J.; Zhao, P.; Shen, J.; Chai, B.; Wang, J. MTOR Up-Regulation of SNRPA1 Contributes to Hepatocellular Carcinoma Development. Biosci. Rep. 2020, 40, BSR20193815. [Google Scholar] [CrossRef] [PubMed]
- Prognostic Value of Survival-Associated Splicing Factor SNRPA1 Overexpression and Its Potential Mechanism in Liver Cancer. Available online: https://www.fortunejournals.com/articles/prognostic-value-of-survivalassociated-splicing-factor-snrpa1-overexpression-and-its-potential-mechanism-in-liver-cancer.html (accessed on 1 August 2022).
- Zeng, Q.; Lei, F.; Chang, Y.; Gao, Z.; Wang, Y.; Gao, Q.; Niu, P.; Li, Q. An Oncogenic Gene, SNRPA1, Regulates PIK3R1, VEGFC, MKI67, CDK1 and Other Genes in Colorectal Cancer. Biomed Pharm. 2019, 117, 109076. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hao, Q.; Wang, S.; Cao, M.; Huang, Y.; Weng, X.; Wang, J.; Zhang, Z.; He, X.; Lu, H.; et al. Inactivation of the Tumor Suppressor P53 by Long Noncoding RNA RMRP. Proc. Natl. Acad. Sci. USA 2021, 118, e2026813118. [Google Scholar] [CrossRef]
- Ding, B.; Yao, M.; Fan, W.; Lou, W. Whole-Transcriptome Analysis Reveals a Potential Hsa_circ_0001955/Hsa_circ_0000977-Mediated MiRNA-MRNA Regulatory Sub-Network in Colorectal Cancer. Aging 2020, 12, 5259–5279. [Google Scholar] [CrossRef]
- Siddappa, M.; Wani, S.A.; Long, M.D.; Leach, D.A.; Mathé, E.A.; Bevan, C.L.; Campbell, M.J. Identification of Transcription Factor Co-Regulators That Drive Prostate Cancer Progression. Sci. Rep. 2020, 10, 20332. [Google Scholar] [CrossRef]
- Estrada-Cuzcano, A.; Neveling, K.; Kohl, S.; Banin, E.; Rotenstreich, Y.; Sharon, D.; Falik-Zaccai, T.C.; Hipp, S.; Roepman, R.; Wissinger, B.; et al. Mutations in C8orf37, Encoding a Ciliary Protein, Are Associated with Autosomal-Recessive Retinal Dystrophies with Early Macular Involvement. Am. J. Hum. Genet. 2012, 90, 102–109. [Google Scholar] [CrossRef]
- Heon, E.; Kim, G.; Qin, S.; Garrison, J.E.; Tavares, E.; Vincent, A.; Nuangchamnong, N.; Scott, C.A.; Slusarski, D.C.; Sheffield, V.C. Mutations in C8ORF37 Cause Bardet Biedl Syndrome (BBS21). Hum. Mol. Genet. 2016, 25, 2283–2294. [Google Scholar] [CrossRef]

| Diagnosis | Prognosis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TCGA and GTEx | TCGA | KM Plotter | ||||||||
| Expression | Clinical Interest (ROC) | OS | ||||||||
| Type of Tumor | Abbreviation | N. of Samples (N, T) | p-Value | log2FC | p-Value | AUC | p-Value | HR (95%CI) | p-Value | HR (95%CI) |
| Adrenocortical carcinoma | ACC | 337 (258, 79) | NS | −0.17 | 0.03 | 0.6 | NS | 1.15 (0.53–2.5) | NA | NA |
| Bladder cancer | BLCA | 452 (40, 412) | NS | 2.5 | 0.03 | 0.6 | NS | 1.01 (0.75–1.36) | NS | 1 (0.74–1.37) |
| Breast cancer | BC | 1685 (572, 1113) | 2.32E-35 | 2.2 | 1.16E-35 | 0.7 | 0.01 | 0.65 (0.47–0.9) | 0.0063 | 0.64 (0.46–0.88) |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 328 (22, 306) | NS | 0.2 | NS | 0.5 | NS | 0.91 (0.57–1.44) | NS | 0.92 (0.56–1.51) |
| Cholangiocarcinoma | CHOL | 270 (235, 35) | NS | 2.5 | NS | 0.5 | NS | 1.24 (0.46–3.38) | NA | NA |
| Colon adenocarcinoma | COAD | 1300 (820, 480) | NS | 0.8 | 0.05 | 0.5 | NS | 0.97 (0.61–1.52) | NA | NA |
| Esophageal carcinoma | ESCA | 1619 (1456, 163) | 4.08E-35 | 3.7 | 2.04E-35 | 0.8 | NS | 1.50 (0.92–2.45) | NS | 1.7 (0.89–3.23) |
| Glioblastoma multiforme | GBM | 2816 (2647, 169) | NS | −0.1 | NS | 0.5 | NS | 1.09 (0.76–1.57) | NA | NA |
| Lower grade glioma | LGG | 3174 (2642, 532) | 2.43E-04 | −0.3 | 0.0001 | 0.5 | NS | 0.98 (0.63–1.52) | NA | NA |
| Head-neck squamous cell carcinoma | HNSC | 710 (206, 504) | 1.55E-11 | 0.1 | 7.77E-12 | 0.6 | NS | 0.92 (0.69–1.21) | NS | 0.8 (0.59–1.09) |
| Hepatocellular carcinoma | HCC | 650 (276, 374) | 4.28E-07 | 0.8 | 2.14E-07 | 0.6 | NS | 1.38 (0.96–1.98) | 0.037 | 1.47 (1.02–2.13) |
| Acute myeloid leukemia | AML | 1079 (929, 150) | 9.46E-08 | −1 | 4.73E-08 | 0.6 | NS | 1.26 (0.82–1.93) | NA | NA |
| Lung adenocarcinoma | LUAD | 1176 (637, 539) | 0.002 | 1.3 | 0.001 | 0.5 | NS | 0.99 (0.73–1.35) | NS | 1.05 (0.77–1.44) |
| squamous cell carcinoma | LUSC | 551 (49, 502) | 0.003 | 2.7 | 0.001 | 0.6 | NS | 0.98 (0.75–1.29) | NS | 124 (0.94–1.63) |
| Skin cutaneous melanoma | SKCM | 2282 (1810, 472) | 7.99E-23 | 0.5 | 3.99E-23 | 0.6 | NS | 0.84 (0.64–1.12) | NA | NA |
| Ovarian cancer | OV | 561 (180, 381) | 1.11E-14 | 1.5 | 5.56E-15 | 0.7 | NS | 0.97 (0.74–1.26) | NS | 0.95 (0.73–1.24) |
| Pancreatic adenocarcinoma | PAAD | 511 (332, 179) | 6.86E-20 | −1.1 | 3.43E-20 | 0.7 | NS | 0.69 (0.45–1.06) | NS | 0.84 (0.53–1.34) |
| Pheochromocytoma and Paraganglioma | PCPG | 445 (261,184) | 3.87E-07 | −0.7 | 1.93E-07 | 0.6 | NS | 3.3e-09 (0–Inf) | NS | 0 (0–Inf) |
| Prostate adenocarcinoma | PRAD | 798 (297, 501) | NS | 1.6 | NS | 0.5 | NS | 0.94 (0.25–3.50) | NA | NA |
| Rectum adenocarcinoma | READ | 956 (789,167) | 0.001 | 0.7 | 4.73E-04 | 0.6 | NS | 0.82 (0.37–1.82) | NS | 0.67 (0.29–1.53) |
| Chromophobe renal cell carcinoma | chRCC | 179 (114, 65) | NS | 0.1 | NS | 0.5 | NS | 0.45 (0.05–3.56) | NA | NA |
| Clear cell renal cell carcinoma | ccRCC | 702 (161, 541) | 1.25E-15 | 0.8 | 6.24E-16 | 0.7 | NS | 1.20 (0.89–1.63) | NS | 1.19 (0.88–1.61) |
| Papillary renal cell carcinoma | pRCC | 412 (121, 291) | 0.005 | 0.9 | 0.002 | 0.6 | 2.86E-04 | 2.85 (1.58–5.17) | 0.0025 | 2.46 (1.35–4.49) |
| Sarcoma | SARC | 265 (2, 263) | NS | 0.6 | NS | 0.6 | NS | 0.66 (0.43–1.02) | NS | 0.71 (0.46–1.12) |
| Stomach adenocarcinoma | STAD | 766 (391, 375) | 2.49E-50 | 4 | 1.25E-50 | 0.8 | NS | 1.02 (0.73–1.41) | NS | 1.05 (0.76–1.47) |
| Testicular germ cell tumors | TGCT | 517 (361, 156) | 6.59E-73 | −6.1 | 3.30E-73 | 1.0 | NS | 0.76 (0.07–8.38) | NS | 0.85 (0.08–9.44) |
| Thymoma | THYM | 121 (2, 119) | NS | 5.2 | 0.047 | 0.8 | NS | 0.91 (0.24–3.40) | NS | 1.1 (0.3–4.4) |
| Thyroid carcinoma | THCA | 1224 (712, 512) | 3.13E-10 | −0.6 | 1.56E-10 | 0.6 | NS | 1.84 (0.67–5.08) | NS | 1.05 (0.36–3.02) |
| Uterine corpus endometrial carcinoma | UCEC | 731 (177, 554) | NS | 1.1 | NS | 0.5 | NS | 1.48 (0.97–2.24) | 0.0088 | 1.73 (1.14–2.62) |
| Uterine carcinosarcoma | UCS | 199 (142, 57) | 0.0001 | 3.2 | 6.69E-05 | 0.6 | NS | 1.57 (0.80–3.08) | NA | NA |
| BC | |||||
|---|---|---|---|---|---|
| Clinicopathological Features | n. of Total Cases | LINC01087 Expression | p-Value | ||
| Low | High | ||||
| pT | T1-T2 | 929 | 453 | 476 | 0.04 |
| T3-T4 | 180 | 103 | 77 | ||
| Tumor size | ≤2 cm | 282 | 123 | 159 | 0.01 |
| >2 cm | 827 | 433 | 394 | ||
| TGCT | |||||
| Clinicopathological Features | n. of Total Cases | LINC01087 Expression | p-Value | ||
| Tumor size | ≤2 cm | 80 | 41 | 39 | 0.05 |
| >2 cm | 58 | 40 | 18 | ||
| lncRNA | 10 miRNA Targets Extracted from Analysis 2 | 18 miRNA Targets Extracted from Analysis 3 | 10 mRNA Targets Shared between Analyses 1, 2, and 3 |
|---|---|---|---|
| LINC01087 | hsa-miR-644a | hsa-miR-7-5p | C8orf37 |
| hsa-miR-1305 | hsa-miR-92a-3p | EFNA5 | |
| hsa-miR-4505 | hsa-miR-181a-5p | HOOK3 | |
| hsa-miR-3671 | hsa-miR-181a-5p, hsa-miR-181b-5p, hsa-miR-181d-5p | IQCG | |
| hsa-miR-7975 | hsa-miR-197-3p | MYOZ3 | |
| hsa-miR-4505 | hsa-miR-423-5p | PCP4L1 | |
| hsa-miR-569 | hsa-miR-181a-5p, hsa-miR-181b-5p, hsa-miR-98-5p | PLAG1 | |
| hsa-miR-4255 | hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-92a-3p | POLI | |
| hsa-miR-606 | hsa-miR-34a-5p, hsa-miR-98-5p | SLC2A3 | |
| hsa-miR-3174 | hsa-miR-34a-5p, hsa-miR-98-5p | SNRPA1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Palma, F.D.E.; Carbonnier, V.; Salvatore, F.; Kroemer, G.; Pol, J.G.; Maiuri, M.C. Systematic Investigation of the Diagnostic and Prognostic Impact of LINC01087 in Human Cancers. Cancers 2022, 14, 5980. https://doi.org/10.3390/cancers14235980
De Palma FDE, Carbonnier V, Salvatore F, Kroemer G, Pol JG, Maiuri MC. Systematic Investigation of the Diagnostic and Prognostic Impact of LINC01087 in Human Cancers. Cancers. 2022; 14(23):5980. https://doi.org/10.3390/cancers14235980
Chicago/Turabian StyleDe Palma, Fatima Domenica Elisa, Vincent Carbonnier, Francesco Salvatore, Guido Kroemer, Jonathan G. Pol, and Maria Chiara Maiuri. 2022. "Systematic Investigation of the Diagnostic and Prognostic Impact of LINC01087 in Human Cancers" Cancers 14, no. 23: 5980. https://doi.org/10.3390/cancers14235980
APA StyleDe Palma, F. D. E., Carbonnier, V., Salvatore, F., Kroemer, G., Pol, J. G., & Maiuri, M. C. (2022). Systematic Investigation of the Diagnostic and Prognostic Impact of LINC01087 in Human Cancers. Cancers, 14(23), 5980. https://doi.org/10.3390/cancers14235980