The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFβ Pathway
Abstract
Simple Summary
Abstract
1. p53 Protein Family
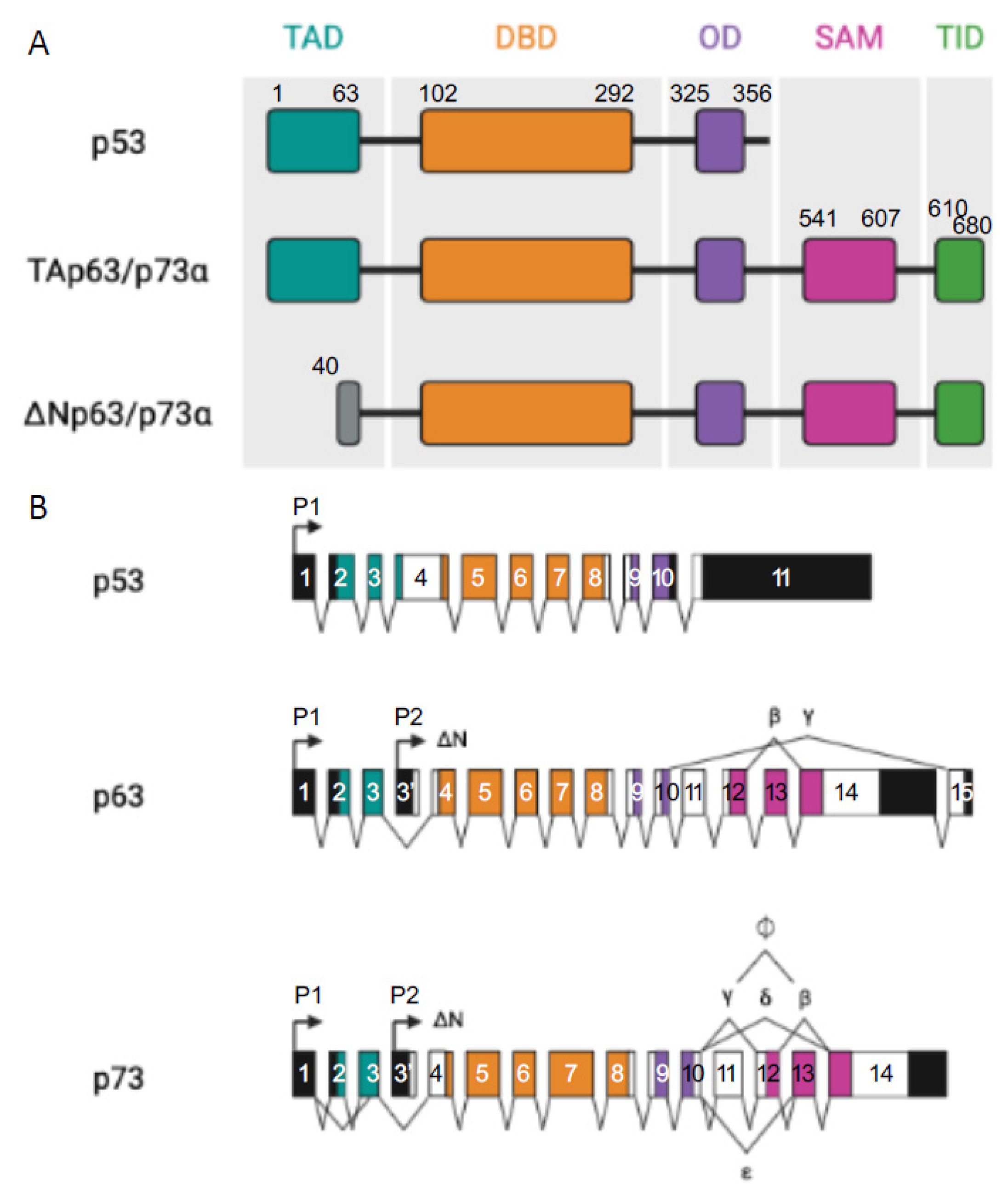
p53 Protein Family: Structure and Functions
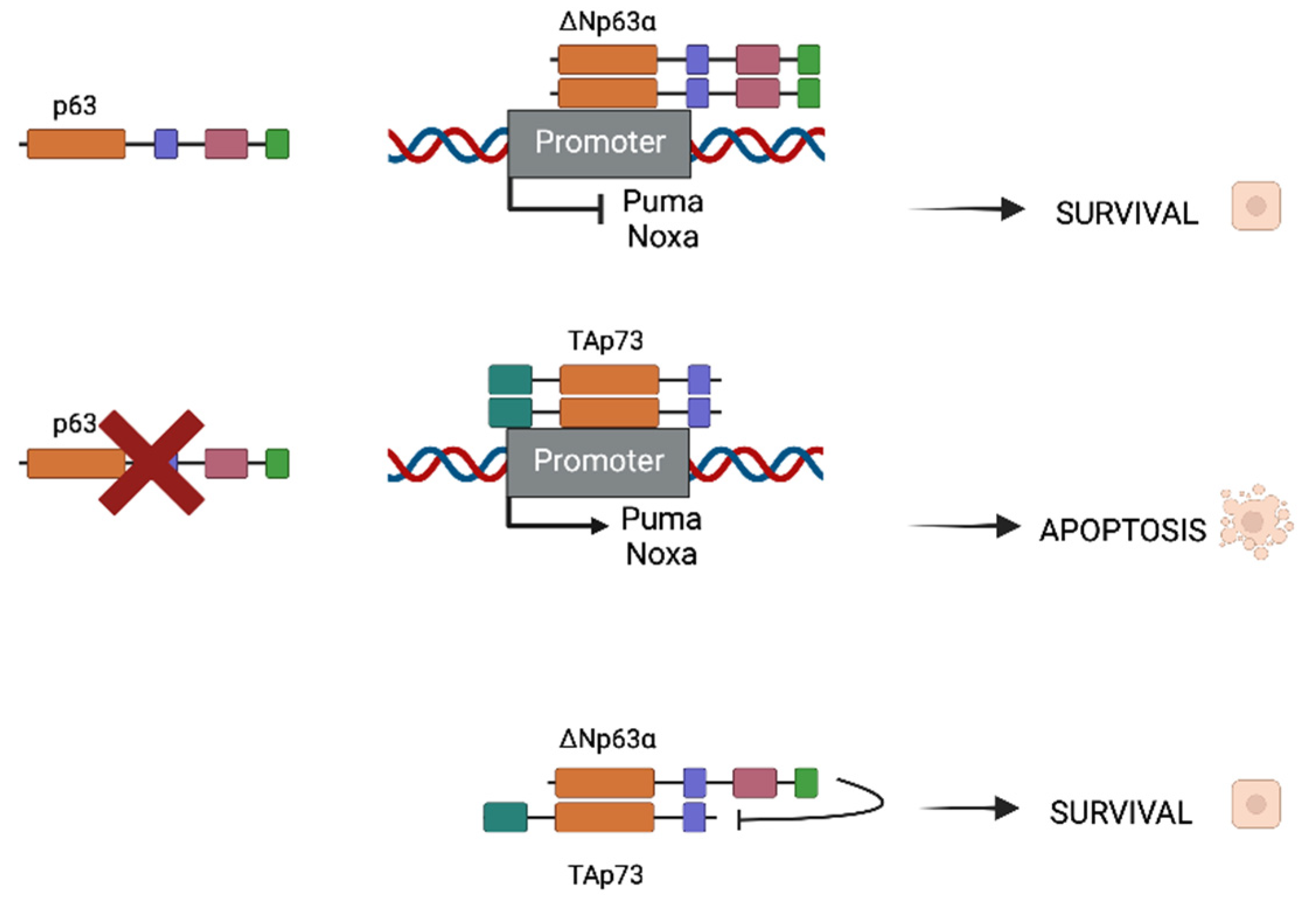
2. p63 and p73 Interactions in the Head and Neck Squamous Cell Carcinoma Model (HNSCC)
3. p53 Protein Family in Primary Tumors and Metastatic Dissemination
p53 Protein Family and microRNA-Regulated Metastasis
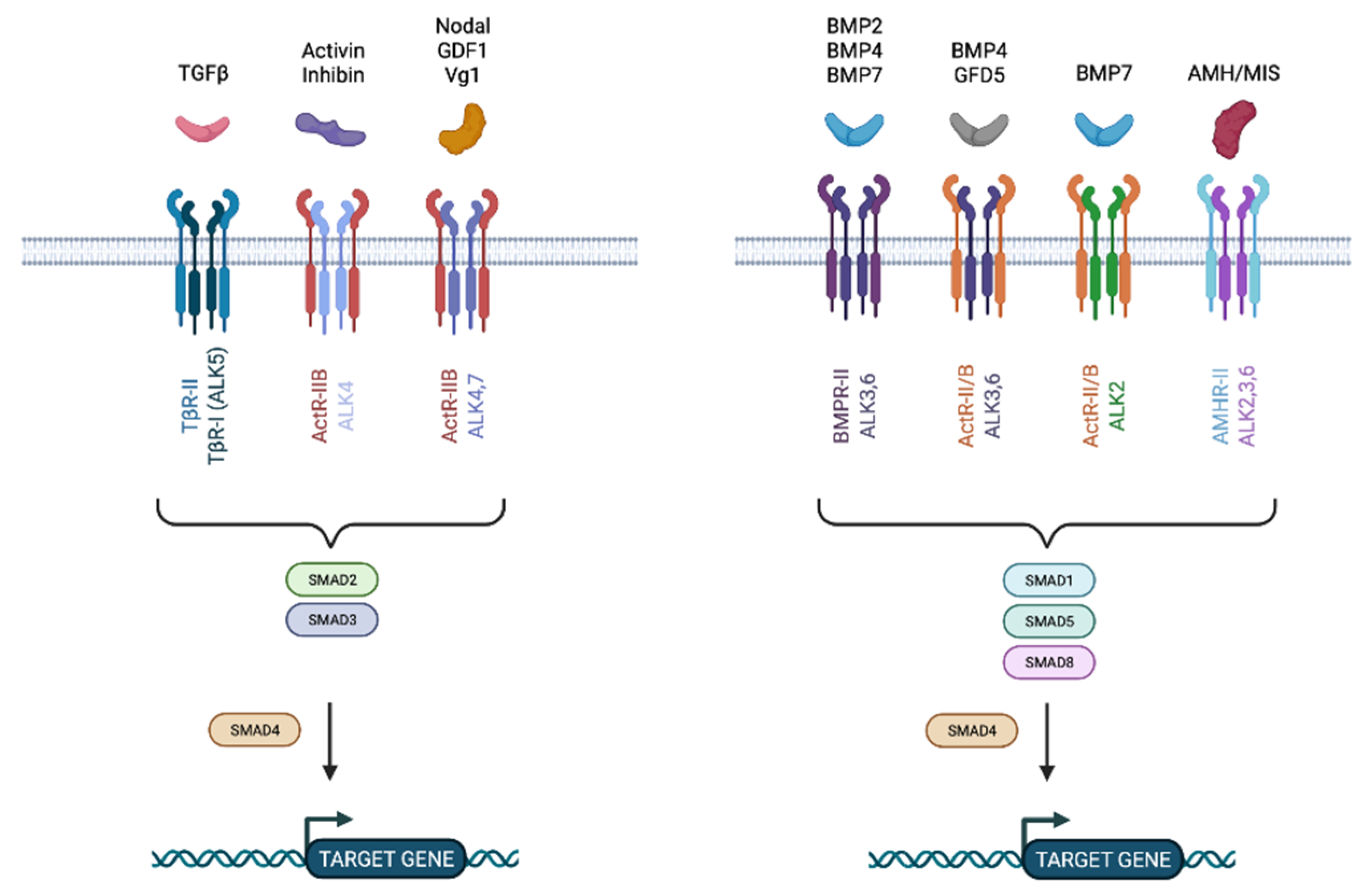
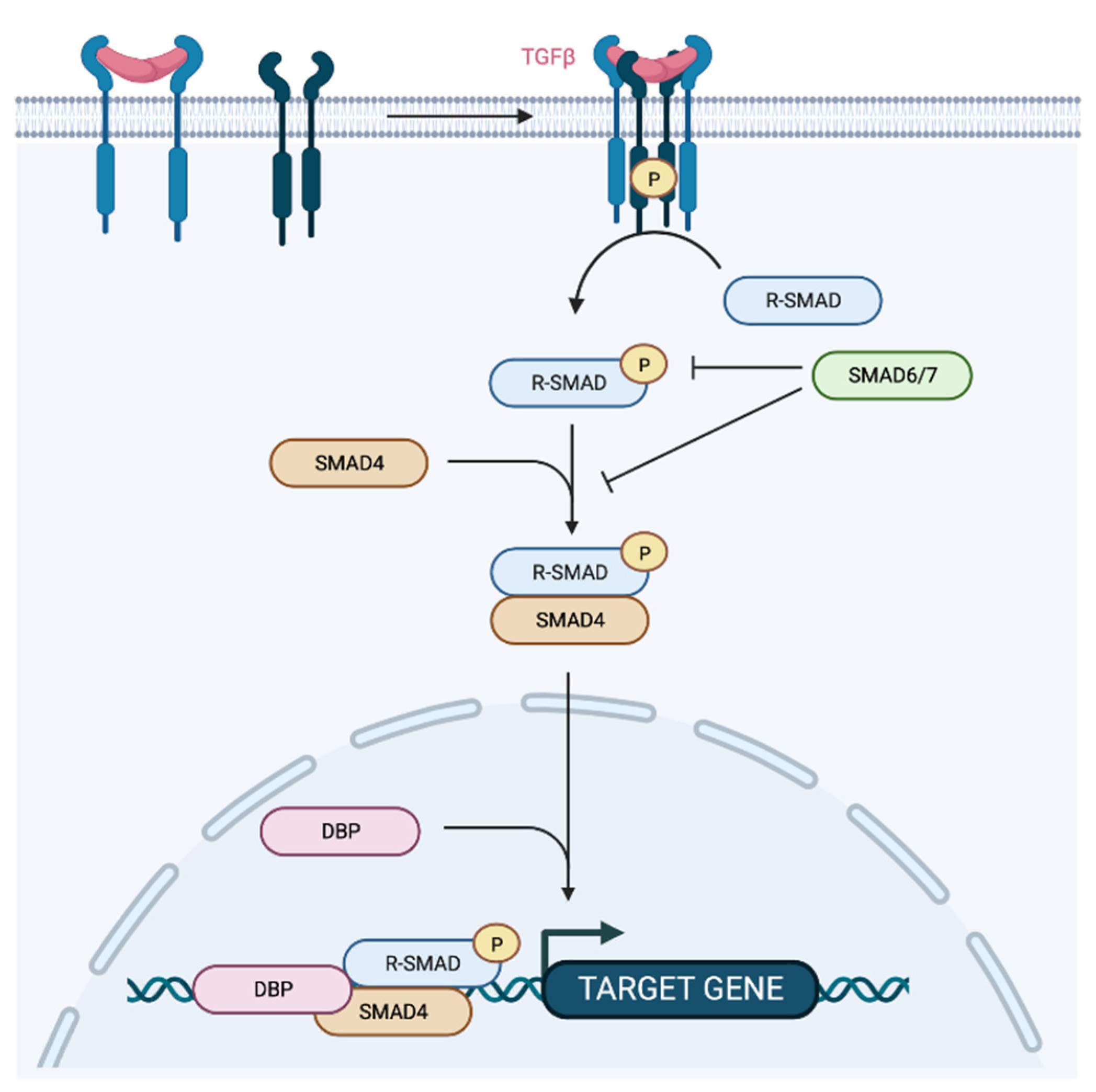
4. The TGFβ Pathway
4.1. TGFβ Pathway in Cancer
4.1.1. TGFβ as a Tumor Suppressor
4.1.2. TGFβ as a Tumor and Metastasis Promoter
5. Role of p53 Protein Family in TGFβ Pathway and Metastasis
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Levrero, M.; De Laurenzi, V.; Costanzo, A.; Gong, J.; Wang, J.Y.; Melino, G. The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J. Cell Sci. 2000, 113 Pt 10, 1661–1670. [Google Scholar] [CrossRef]
- Mills, A.A.; Zheng, B.; Wang, X.J.; Vogel, H.; Roop, D.R.; Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef]
- Pozniak, C.D.; Radinovic, S.; Yang, A.; McKeon, F.; Kaplan, D.R.; Miller, F.D. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000, 289, 304–306. [Google Scholar] [CrossRef]
- Yang, A.; McKeon, F. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 2000, 1, 199–207. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Moll, U.M.; Slade, N. p63 and p73: Roles in development and tumor formation. Mol. Cancer Res. MCR 2004, 2, 371–386. [Google Scholar] [CrossRef]
- Deyoung, M.P.; Ellisen, L.W. p63 and p73 in human cancer: Defining the network. Oncogene 2007, 26, 5169–5183. [Google Scholar] [CrossRef]
- Leong, C.O.; Vidnovic, N.; DeYoung, M.P.; Sgroi, D.; Ellisen, L.W. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J. Clin. Investig. 2007, 117, 1370–1380. [Google Scholar] [CrossRef]
- Chi, S.W.; Ayed, A.; Arrowsmith, C.H. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999, 18, 4438–4445. [Google Scholar] [CrossRef]
- Serber, Z.; Lai, H.C.; Yang, A.; Ou, H.D.; Sigal, M.S.; Kelly, A.E.; Darimont, B.D.; Duijf, P.H.; Van Bokhoven, H.; McKeon, F.; et al. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 2002, 22, 8601–8611. [Google Scholar] [CrossRef]
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dotsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 1998, 2, 305–316. [Google Scholar] [CrossRef]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef]
- Gaiddon, C.; Lokshin, M.; Ahn, J.; Zhang, T.; Prives, C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 2001, 21, 1874–1887. [Google Scholar] [CrossRef]
- Melino, G.; De Laurenzi, V.; Vousden, K.H. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2002, 2, 605–615. [Google Scholar] [CrossRef]
- Dohn, M.; Zhang, S.; Chen, X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 2001, 20, 3193–3205. [Google Scholar] [CrossRef]
- Gong, J.G.; Costanzo, A.; Yang, H.Q.; Melino, G.; Kaelin, W.G., Jr.; Levrero, M.; Wang, J.Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999, 399, 806–809. [Google Scholar] [CrossRef]
- Suh, Y.; Amelio, I.; Guerrero Urbano, T.; Tavassoli, M. Clinical update on cancer: Molecular oncology of head and neck cancer. Cell Death Dis. 2014, 5, e1018. [Google Scholar] [CrossRef]
- Safdari, Y.; Khalili, M.; Farajnia, S.; Asgharzadeh, M.; Yazdani, Y.; Sadeghi, M. Recent advances in head and neck squamous cell carcinoma—A review. Clin. Biochem. 2014, 47, 1195–1202. [Google Scholar] [CrossRef]
- Parkin, D.M.; Pisani, P.; Ferlay, J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 1999, 80, 827–841. [Google Scholar] [CrossRef]
- Vokes, E.E.; Weichselbaum, R.R.; Lippman, S.M.; Hong, W.K. Head and neck cancer. N. Engl. J. Med. 1993, 328, 184–194. [Google Scholar] [CrossRef]
- DeYoung, M.P.; Johannessen, C.M.; Leong, C.O.; Faquin, W.; Rocco, J.W.; Ellisen, L.W. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006, 66, 9362–9368. [Google Scholar] [CrossRef]
- Hibi, K.; Trink, B.; Patturajan, M.; Westra, W.H.; Caballero, O.L.; Hill, D.E.; Ratovitski, E.A.; Jen, J.; Sidransky, D. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2000, 97, 5462–5467. [Google Scholar] [CrossRef]
- Bjorkqvist, A.M.; Husgafvel-Pursiainen, K.; Anttila, S.; Karjalainen, A.; Tammilehto, L.; Mattson, K.; Vainio, H.; Knuutila, S. DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosom. Cancer 1998, 22, 79–82. [Google Scholar] [CrossRef]
- Rocco, J.W.; Leong, C.O.; Kuperwasser, N.; DeYoung, M.P.; Ellisen, L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006, 9, 45–56. [Google Scholar] [CrossRef]
- Sniezek, J.C.; Matheny, K.E.; Westfall, M.D.; Pietenpol, J.A. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope 2004, 114, 2063–2072. [Google Scholar] [CrossRef]
- Rodriguez Calleja, L.; Jacques, C.; Lamoureux, F.; Baud’huin, M.; Tellez Gabriel, M.; Quillard, T.; Sahay, D.; Perrot, P.; Amiaud, J.; Charrier, C.; et al. DeltaNp63alpha Silences a miRNA Program to Aberrantly Initiate a Wound-Healing Program That Promotes TGFbeta-Induced Metastasis. Cancer Res. 2016, 76, 3236–3251. [Google Scholar] [CrossRef]
- Kakuki, T.; Kurose, M.; Takano, K.; Kondoh, A.; Obata, K.; Nomura, K.; Miyata, R.; Kaneko, Y.; Konno, T.; Takahashi, S.; et al. Dysregulation of junctional adhesion molecule-A via p63/GATA-3 in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 33887–33900. [Google Scholar] [CrossRef]
- Dong, J.; Li, J.; Li, Y.; Ma, Z.; Yu, Y.; Wang, C.Y. Transcriptional super-enhancers control cancer stemness and metastasis genes in squamous cell carcinoma. Nat. Commun. 2021, 12, 3974. [Google Scholar] [CrossRef]
- Tan, E.H.; Morton, J.P.; Timpson, P.; Tucci, P.; Melino, G.; Flores, E.R.; Sansom, O.J.; Vousden, K.H.; Muller, P.A. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene 2014, 33, 3325–3333. [Google Scholar] [CrossRef]
- Muller, P.A.; Caswell, P.T.; Doyle, B.; Iwanicki, M.P.; Tan, E.H.; Karim, S.; Lukashchuk, N.; Gillespie, D.A.; Ludwig, R.L.; Gosselin, P.; et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009, 139, 1327–1341. [Google Scholar] [CrossRef]
- Ory, B.; Ramsey, M.R.; Wilson, C.; Vadysirisack, D.D.; Forster, N.; Rocco, J.W.; Rothenberg, S.M.; Ellisen, L.W. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J. Clin. Investig. 2011, 121, 809–820. [Google Scholar] [CrossRef]
- Cai, B.H.; Hsu, Y.C.; Yeh, F.Y.; Lin, Y.R.; Lu, R.Y.; Yu, S.J.; Shaw, J.F.; Wu, M.H.; Tsai, Y.Z.; Lin, Y.C.; et al. P63 and P73 Activation in Cancers with p53 Mutation. Biomedicines 2022, 10, 1490. [Google Scholar] [CrossRef]
- Petrenko, O.; Zaika, A.; Moll, U.M. deltaNp73 facilitates cell immortalization and cooperates with oncogenic Ras in cellular transformation in vivo. Mol. Cell. Biol. 2003, 23, 5540–5555. [Google Scholar] [CrossRef][Green Version]
- Rufini, A.; Agostini, M.; Grespi, F.; Tomasini, R.; Sayan, B.S.; Niklison-Chirou, M.V.; Conforti, F.; Velletri, T.; Mastino, A.; Mak, T.W.; et al. p73 in Cancer. Genes Cancer 2011, 2, 491–502. [Google Scholar] [CrossRef]
- Su, X.; Chakravarti, D.; Cho, M.S.; Liu, L.; Gi, Y.J.; Lin, Y.L.; Leung, M.L.; El-Naggar, A.; Creighton, C.J.; Suraokar, M.B.; et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010, 467, 986–990. [Google Scholar] [CrossRef]
- Cho, M.S.; Chan, I.L.; Flores, E.R. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle 2010, 9, 2434–2441. [Google Scholar] [CrossRef]
- Danilov, A.V.; Neupane, D.; Nagaraja, A.S.; Feofanova, E.V.; Humphries, L.A.; Direnzo, J.; Korc, M. DeltaNp63alpha-Mediated Induction of Epidermal Growth Factor Receptor Promotes Pancreatic Cancer Cell Growth and Chemoresistance. PLoS ONE 2011, 6, e26815. [Google Scholar] [CrossRef]
- Lindsay, J.; McDade, S.S.; Pickard, A.; McCloskey, K.D.; McCance, D.J. Role of DeltaNp63gamma in epithelial to mesenchymal transition. J. Biol. Chem. 2011, 286, 3915–3924. [Google Scholar] [CrossRef]
- Oh, J.E.; Kim, R.H.; Shin, K.H.; Park, N.H.; Kang, M.K. DeltaNp63alpha protein triggers epithelial-mesenchymal transition and confers stem cell properties in normal human keratinocytes. J. Biol. Chem. 2011, 286, 38757–38767. [Google Scholar] [CrossRef]
- Melino, G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011, 18, 1487–1499. [Google Scholar] [CrossRef]
- Tucci, P.; Agostini, M.; Grespi, F.; Markert, E.K.; Terrinoni, A.; Vousden, K.H.; Muller, P.A.; Dotsch, V.; Kehrloesser, S.; Sayan, B.S.; et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 15312–15317. [Google Scholar] [CrossRef]
- Liang, S.; Tang, X.; Ye, T.; Xiang, W. HER2 induces cell scattering and invasion through Np63alpha and E-cadherin. Biochem. Cell Biol. 2022, 100, 403–412. [Google Scholar] [CrossRef]
- Georges, S.; Calleja, L.R.; Jacques, C.; Lavaud, M.; Moukengue, B.; Lecanda, F.; Quillard, T.; Gabriel, M.T.; Cartron, P.F.; Baud’huin, M.; et al. Loss of miR-198 and -206 during primary tumor progression enables metastatic dissemination in human osteosarcoma. Oncotarget 2018, 9, 35726–35741. [Google Scholar] [CrossRef][Green Version]
- Wang, C.; Wang, Y.; Fu, Z.; Huang, W.; Yu, Z.; Wang, J.; Zheng, K.; Zhang, S.; Li, S.; Chen, J. MiR-29b-3p Inhibits Migration and Invasion of Papillary Thyroid Carcinoma by Downregulating COL1A1 and COL5A1. Front. Oncol. 2022, 12, 837581. [Google Scholar] [CrossRef]
- Zhao, E.; Li, X.; You, B.; Wang, J.; Hou, W.; Wu, Q. Identification of a Five-miRNA Signature for Diagnosis of Kidney Renal Clear Cell Carcinoma. Front. Genet 2022, 13, 857411. [Google Scholar] [CrossRef]
- Hermeking, H. p53 enters the microRNA world. Cancer Cell 2007, 12, 414–418. [Google Scholar] [CrossRef]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef]
- Shi, L.; Jackstadt, R.; Siemens, H.; Li, H.; Kirchner, T.; Hermeking, H. p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2014, 74, 532–542. [Google Scholar] [CrossRef]
- Ren, D.; Wang, M.; Guo, W.; Zhao, X.; Tu, X.; Huang, S.; Zou, X.; Peng, X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR145. Int. J. Oncol. 2013, 42, 1473–1481. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, C.; Wu, R.; Hu, W. Tumor suppressor p53 meets microRNAs. J. Mol. Cell Biol. 2011, 3, 44–50. [Google Scholar] [CrossRef]
- Stacy, A.J.; Craig, M.P.; Sakaram, S.; Kadakia, M. DeltaNp63alpha and microRNAs: Leveraging the epithelial-mesenchymal transition. Oncotarget 2017, 8, 2114–2129. [Google Scholar] [CrossRef]
- Lin, C.; Li, X.; Zhang, Y.; Guo, Y.; Zhou, J.; Gao, K.; Dai, J.; Hu, G.; Lv, L.; Du, J.; et al. The microRNA feedback regulation of p63 in cancer progression. Oncotarget 2015, 6, 8434–8453. [Google Scholar] [CrossRef]
- Boominathan, L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010, 29, 613–639. [Google Scholar] [CrossRef]
- Bui, N.H.B.; Napoli, M.; Davis, A.J.; Abbas, H.A.; Rajapakshe, K.; Coarfa, C.; Flores, E.R. Spatiotemporal Regulation of DeltaNp63 by TGFbeta-Regulated miRNAs Is Essential for Cancer Metastasis. Cancer Res 2020, 80, 2833–2847. [Google Scholar] [CrossRef]
- Franzen, P.; ten Dijke, P.; Ichijo, H.; Yamashita, H.; Schulz, P.; Heldin, C.H.; Miyazono, K. Cloning of a TGF beta type I receptor that forms a heteromeric complex with the TGF beta type II receptor. Cell 1993, 75, 681–692. [Google Scholar] [CrossRef]
- Cheifetz, S.; Hernandez, H.; Laiho, M.; ten Dijke, P.; Iwata, K.K.; Massague, J. Distinct transforming growth factor-beta (TGF-beta) receptor subsets as determinants of cellular responsiveness to three TGF-beta isoforms. J. Biol. Chem. 1990, 265, 20533–20538. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Pierce, D.F., Jr.; Gorska, A.E.; Chytil, A.; Meise, K.S.; Page, D.L.; Coffey, R.J., Jr.; Moses, H.L. Mammary tumor suppression by transforming growth factor beta 1 transgene expression. Proc. Natl. Acad. Sci. USA 1995, 92, 4254–4258. [Google Scholar] [CrossRef]
- Kim, B.G.; Li, C.; Qiao, W.; Mamura, M.; Kasprzak, B.; Anver, M.; Wolfraim, L.; Hong, S.; Mushinski, E.; Potter, M.; et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature 2006, 441, 1015–1019. [Google Scholar] [CrossRef]
- Becker, C.; Fantini, M.C.; Schramm, C.; Lehr, H.A.; Wirtz, S.; Nikolaev, A.; Burg, J.; Strand, S.; Kiesslich, R.; Huber, S.; et al. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity 2004, 21, 491–501. [Google Scholar] [CrossRef]
- Munger, K.; Pietenpol, J.A.; Pittelkow, M.R.; Holt, J.T.; Moses, H.L. Transforming growth factor beta 1 regulation of c-myc expression, pRB phosphorylation, and cell cycle progression in keratinocytes. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1992, 3, 291–298. [Google Scholar]
- Warner, B.J.; Blain, S.W.; Seoane, J.; Massague, J. Myc downregulation by transforming growth factor beta required for activation of the p15(Ink4b) G(1) arrest pathway. Mol. Cell. Biol. 1999, 19, 5913–5922. [Google Scholar] [CrossRef]
- Derynck, R.; Akhurst, R.J.; Balmain, A. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 2001, 29, 117–129. [Google Scholar] [CrossRef]
- Hahn, S.A.; Schutte, M.; Hoque, A.T.; Moskaluk, C.A.; da Costa, L.T.; Rozenblum, E.; Weinstein, C.L.; Fischer, A.; Yeo, C.J.; Hruban, R.H.; et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996, 271, 350–353. [Google Scholar] [CrossRef]
- Eppert, K.; Scherer, S.W.; Ozcelik, H.; Pirone, R.; Hoodless, P.; Kim, H.; Tsui, L.C.; Bapat, B.; Gallinger, S.; Andrulis, I.L.; et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 1996, 86, 543–552. [Google Scholar] [CrossRef]
- Bottinger, E.P.; Jakubczak, J.L.; Haines, D.C.; Bagnall, K.; Wakefield, L.M. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor beta receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res. 1997, 57, 5564–5570. [Google Scholar]
- Turco, A.; Coppa, A.; Aloe, S.; Baccheschi, G.; Morrone, S.; Zupi, G.; Colletta, G. Overexpression of transforming growth factor beta-type II receptor reduces tumorigenicity and metastastic potential of K-ras-transformed thyroid cells. Int. J. Cancer 1999, 80, 85–91. [Google Scholar] [CrossRef]
- Tang, B.; de Castro, K.; Barnes, H.E.; Parks, W.T.; Stewart, L.; Bottinger, E.P.; Danielpour, D.; Wakefield, L.M. Loss of responsiveness to transforming growth factor beta induces malignant transformation of nontumorigenic rat prostate epithelial cells. Cancer Res. 1999, 59, 4834–4842. [Google Scholar]
- Rahimi, R.A.; Leof, E.B. TGF-beta signaling: A tale of two responses. J. Cell Biochem. 2007, 102, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Parvani, J.G.; Schiemann, W.P. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J. Mammary Gland Biol. Neoplasia 2010, 15, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Juarez, P.; Guise, T.A. TGF-beta in cancer and bone: Implications for treatment of bone metastases. Bone 2011, 48, 23–29. [Google Scholar] [CrossRef]
- Oft, M.; Peli, J.; Rudaz, C.; Schwarz, H.; Beug, H.; Reichmann, E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996, 10, 2462–2477. [Google Scholar] [CrossRef] [PubMed]
- Piek, E.; Moustakas, A.; Kurisaki, A.; Heldin, C.H.; ten Dijke, P. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 1999, 112 Pt 24, 4557–4568. [Google Scholar] [CrossRef] [PubMed]
- Siegel, P.M.; Shu, W.; Cardiff, R.D.; Muller, W.J.; Massague, J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. USA 2003, 100, 8430–8435. [Google Scholar] [CrossRef] [PubMed]
- Padua, D.; Zhang, X.H.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massague, J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.J. Manipulating the environment of cancer cells in bone: A novel therapeutic approach. J. Clin. Investig. 2002, 110, 1399–1401. [Google Scholar] [CrossRef]
- Kakonen, S.M.; Selander, K.S.; Chirgwin, J.M.; Yin, J.J.; Burns, S.; Rankin, W.A.; Grubbs, B.G.; Dallas, M.; Cui, Y.; Guise, T.A. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 2002, 277, 24571–24578. [Google Scholar] [CrossRef]
- Kang, Y.; He, W.; Tulley, S.; Gupta, G.P.; Serganova, I.; Chen, C.R.; Manova-Todorova, K.; Blasberg, R.; Gerald, W.L.; Massague, J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 13909–13914. [Google Scholar] [CrossRef]
- Morice, S.; Danieau, G.; Tesfaye, R.; Mullard, M.; Brion, R.; Dupuy, M.; Ory, B.; Brounais-Le Royer, B.; Corre, I.; Redini, F.; et al. Involvement of the TGF-beta Signaling Pathway in the Development of YAP-Driven Osteosarcoma Lung Metastasis. Front. Oncol. 2021, 11, 765711. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Chen, J.; Wang, D.; Li, Q.; Yang, X.; Wang, J.; Pan, Z.; Xiao, Z.J.; Yi, Y. TGF-beta1 Facilitates TAp63alpha Protein Lysosomal Degradation to Promote Pancreatic Cancer Cell Migration. Biology 2021, 10, 597. [Google Scholar] [CrossRef]
- Niu, M.; He, Y.; Xu, J.; Ding, L.; He, T.; Yi, Y.; Fu, M.; Guo, R.; Li, F.; Chen, H.; et al. Noncanonical TGF-beta signaling leads to FBXO3-mediated degradation of DeltaNp63alpha promoting breast cancer metastasis and poor clinical prognosis. PLoS Biol. 2021, 19, e3001113. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.K.; Nigri, J.; Lac, S.; Leca, J.; Bressy, C.; Berthezene, P.; Bartholin, L.; Chan, P.; Calvo, E.; Iovanna, J.L.; et al. TAp73 loss favors Smad-independent TGF-beta signaling that drives EMT in pancreatic ductal adenocarcinoma. Cell Death Differ. 2016, 23, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, Y.; Lee, W.L.; Goh, M.X.; Ito, Y. Role of TAp73alpha in induction of apoptosis by transforming growth factor-beta in gastric cancer cells. FEBS Lett. 2008, 582, 2663–2667. [Google Scholar] [CrossRef]
- Niemantsverdriet, M.; Nagle, P.; Chiu, R.K.; Langendijk, J.A.; Kampinga, H.H.; Coppes, R.P. DeltaNp73 enhances promoter activity of TGF-beta induced genes. PLoS ONE 2012, 7, e50815. [Google Scholar] [CrossRef][Green Version]
- Yang, Z.; Zhang, Y.; Wang, L. A feedback inhibition between miRNA-127 and TGFbeta/c-Jun cascade in HCC cell migration via MMP13. PLoS ONE 2013, 8, e65256. [Google Scholar] [CrossRef]
- Diaz-Lopez, A.; Moreno-Bueno, G.; Cano, A. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag. Res. 2014, 6, 205–216. [Google Scholar] [CrossRef]
- Adorno, M.; Cordenonsi, M.; Montagner, M.; Dupont, S.; Wong, C.; Hann, B.; Solari, A.; Bobisse, S.; Rondina, M.B.; Guzzardo, V.; et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 2009, 137, 87–98. [Google Scholar] [CrossRef]
- Vasilaki, E.; Morikawa, M.; Koinuma, D.; Mizutani, A.; Hirano, Y.; Ehata, S.; Sundqvist, A.; Kawasaki, N.; Cedervall, J.; Olsson, A.K.; et al. Ras and TGF-beta signaling enhance cancer progression by promoting the DeltaNp63 transcriptional program. Sci. Signal 2016, 9, ra84. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez Calleja, L.; Lavaud, M.; Tesfaye, R.; Brounais-Le-Royer, B.; Baud’huin, M.; Georges, S.; Lamoureux, F.; Verrecchia, F.; Ory, B. The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFβ Pathway. Cancers 2022, 14, 5948. https://doi.org/10.3390/cancers14235948
Rodriguez Calleja L, Lavaud M, Tesfaye R, Brounais-Le-Royer B, Baud’huin M, Georges S, Lamoureux F, Verrecchia F, Ory B. The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFβ Pathway. Cancers. 2022; 14(23):5948. https://doi.org/10.3390/cancers14235948
Chicago/Turabian StyleRodriguez Calleja, Lidia, Melanie Lavaud, Robel Tesfaye, Bénédicte Brounais-Le-Royer, Marc Baud’huin, Steven Georges, François Lamoureux, Franck Verrecchia, and Benjamin Ory. 2022. "The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFβ Pathway" Cancers 14, no. 23: 5948. https://doi.org/10.3390/cancers14235948
APA StyleRodriguez Calleja, L., Lavaud, M., Tesfaye, R., Brounais-Le-Royer, B., Baud’huin, M., Georges, S., Lamoureux, F., Verrecchia, F., & Ory, B. (2022). The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFβ Pathway. Cancers, 14(23), 5948. https://doi.org/10.3390/cancers14235948







