Immunogenic Death of Hepatocellular Carcinoma Cells in Mice Expressing Caspase-Resistant ROCK1 Is Not Replicated by ROCK Inhibitors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Model
2.2. Isolation of Hepatocytes
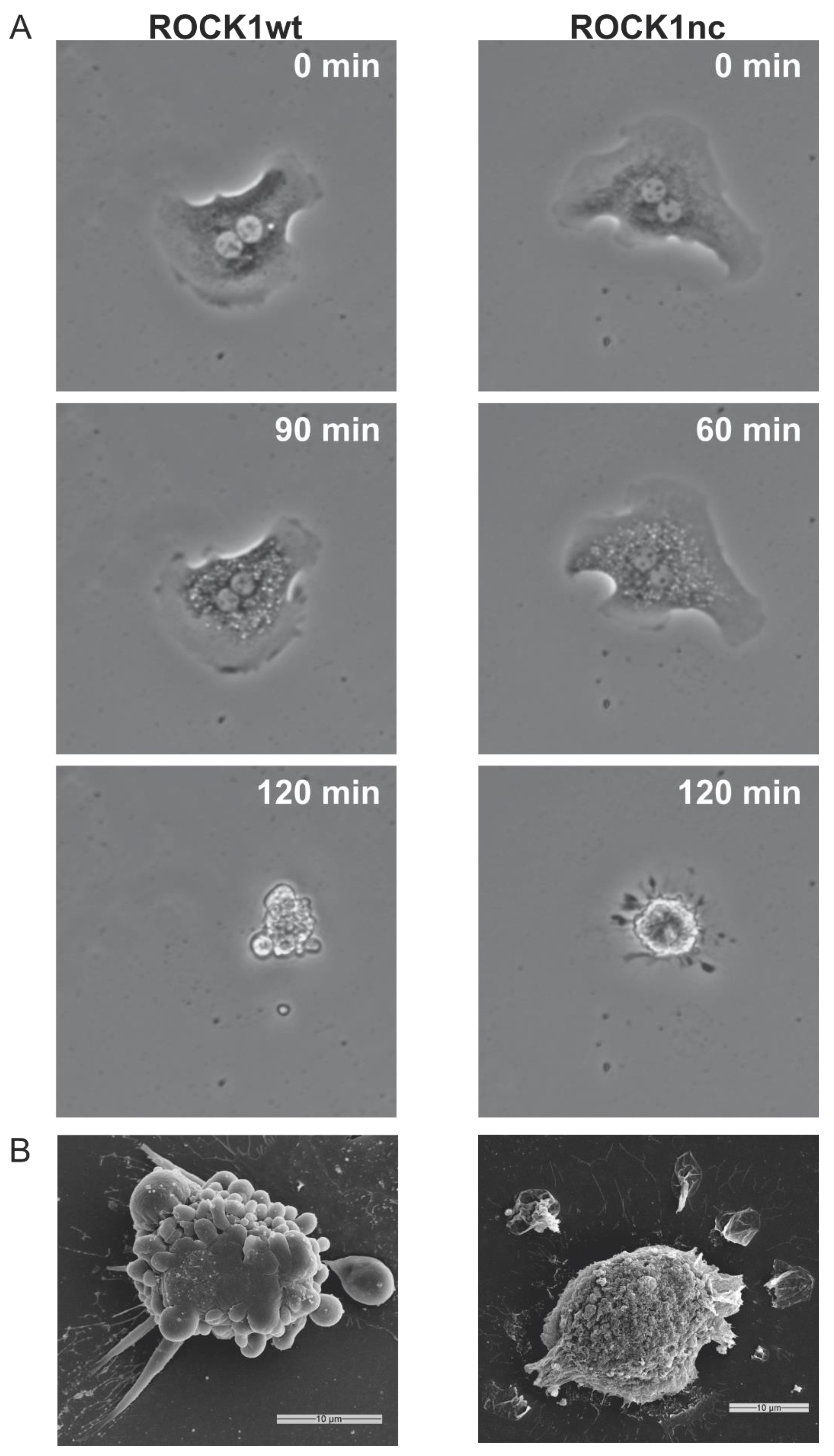
2.3. Time-Lapse Microscopy of Hepatocytes
2.4. Scanning Electron Microscopy (SEM)
2.5. Preparation and Administration of Substances
2.6. Tissue Collection and Fixation
2.7. Histology
2.8. RNA Isolation and Sequencing
2.9. Multiplex Cytokine Analysis
2.10. Immunohistochemistry
2.11. Immunofluorescence Staining of Frozen Sections
2.12. Hepatocellular Carcinoma Induction
2.13. Haematology
2.14. Liver Function
2.15. Flow Cytometry
- Neutrophil—Ly6G+ CD11b+
- Monocyte—Ly6G−, MHCII- SCClo, CD11b+ CD115+
- -
- Inflammatory Monocyte—Ly6Chi CD11c-
- -
- Resident monocyte—Ly6C− CD11c+
- Macrophage— Ly6G−, MHCII+ or SCChi, CD11b+ F480+
- -
- Recruited Macrophage—Clec4f−
- -
- Kupfer Cell—Clec4f+
- Dendritic Cell—Ly6G−, MHCII+ or SCChi, F480−, CD11c+
- -
- Classical DC—PDCA1−, Siglec H−
- -
- Plasmacytoid DC—PDCA1+, Siglec H+
- T Cell—CD3+, CD19−
- -
- αβ T Cell—TCRβ+, TCRγδ−
- o
- CD4 T Cell—CD4+, CD8−
- ▪
- Treg—FoxP3+
- o
- CD8 T Cell—CD4−, CD8+
- -
- γδ T Cell—TCRβ−, TCRγδ+
- B Cell—CD3−, CD19+, B220+
- Natural Killer—NK1.1+
- -
- NK Cell—NK1.1+, CD3−
- -
- NKT Cell—NK1.1+, CD3+
2.16. Statistical Analysis
3. Results
3.1. Caspase Cleavage of ROCK1 Affects Hepatocyte Morphology, Liver Gene Expression and Cytokine Release following Genotoxin Treatment
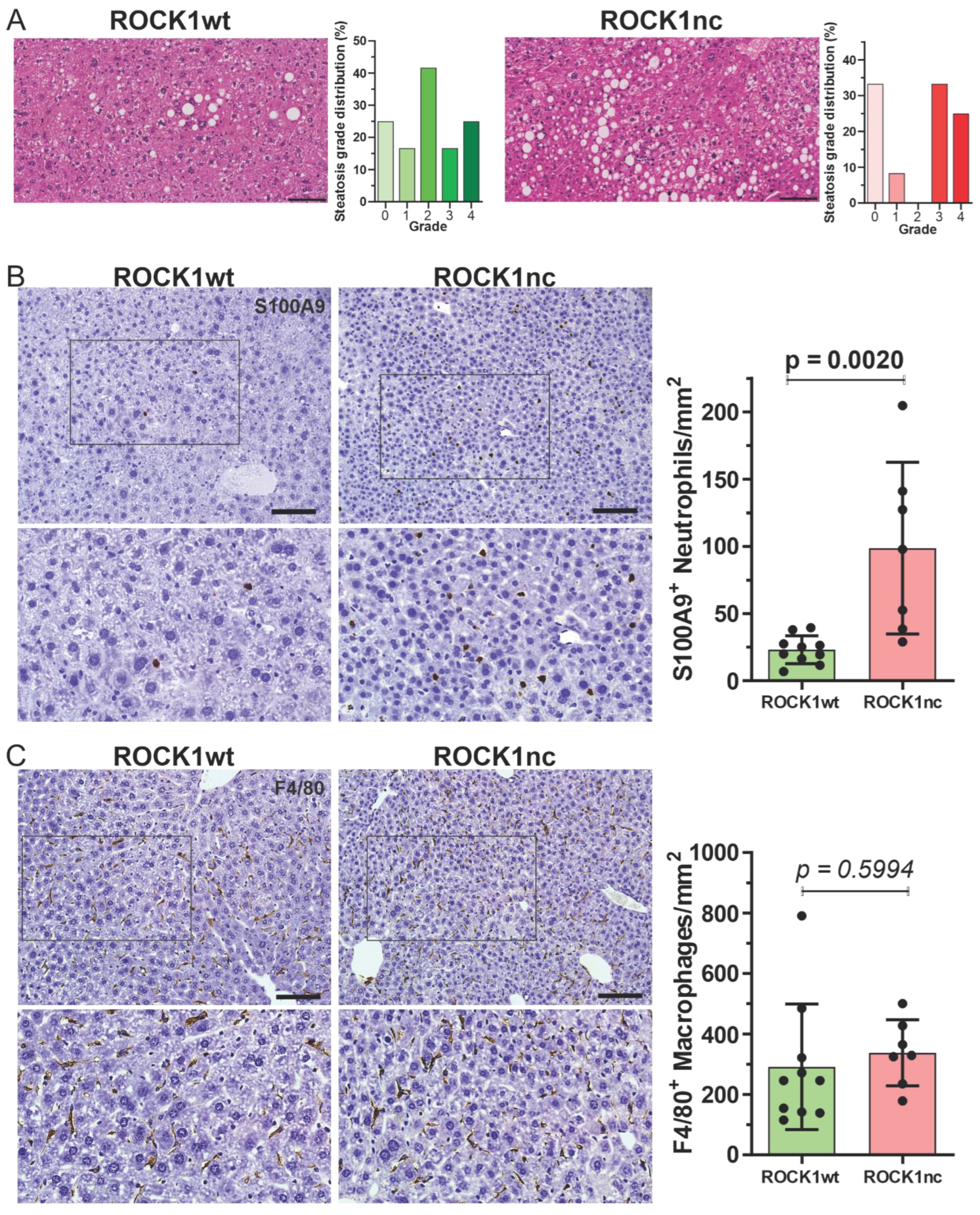
3.2. Hepatocellular Carcinomas from ROCK1nc Mice Show Increased Neutrophil Infiltration, CD3+, and CD8+ T Cell Recruitment Compared to ROCK1wt Mice
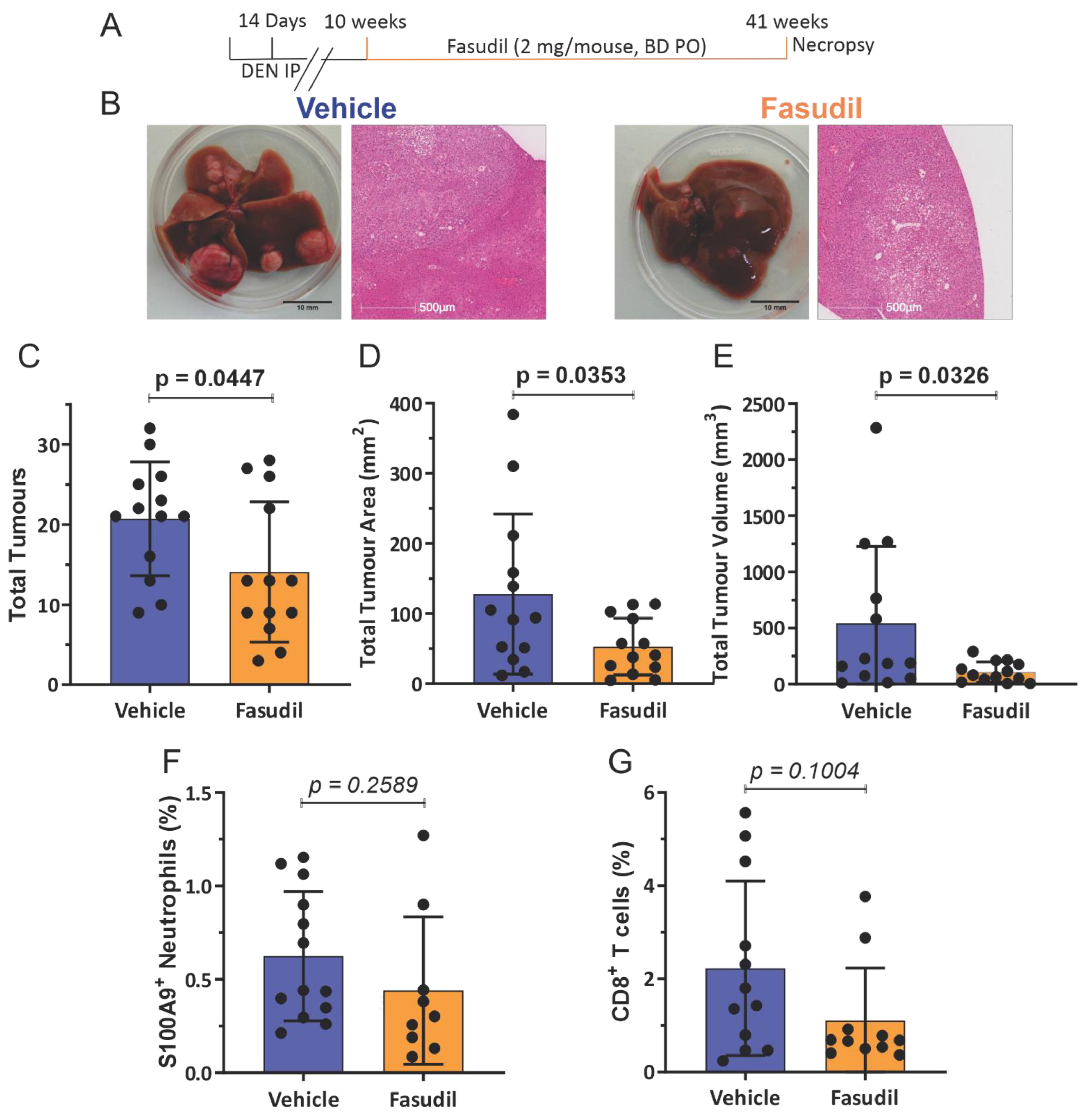
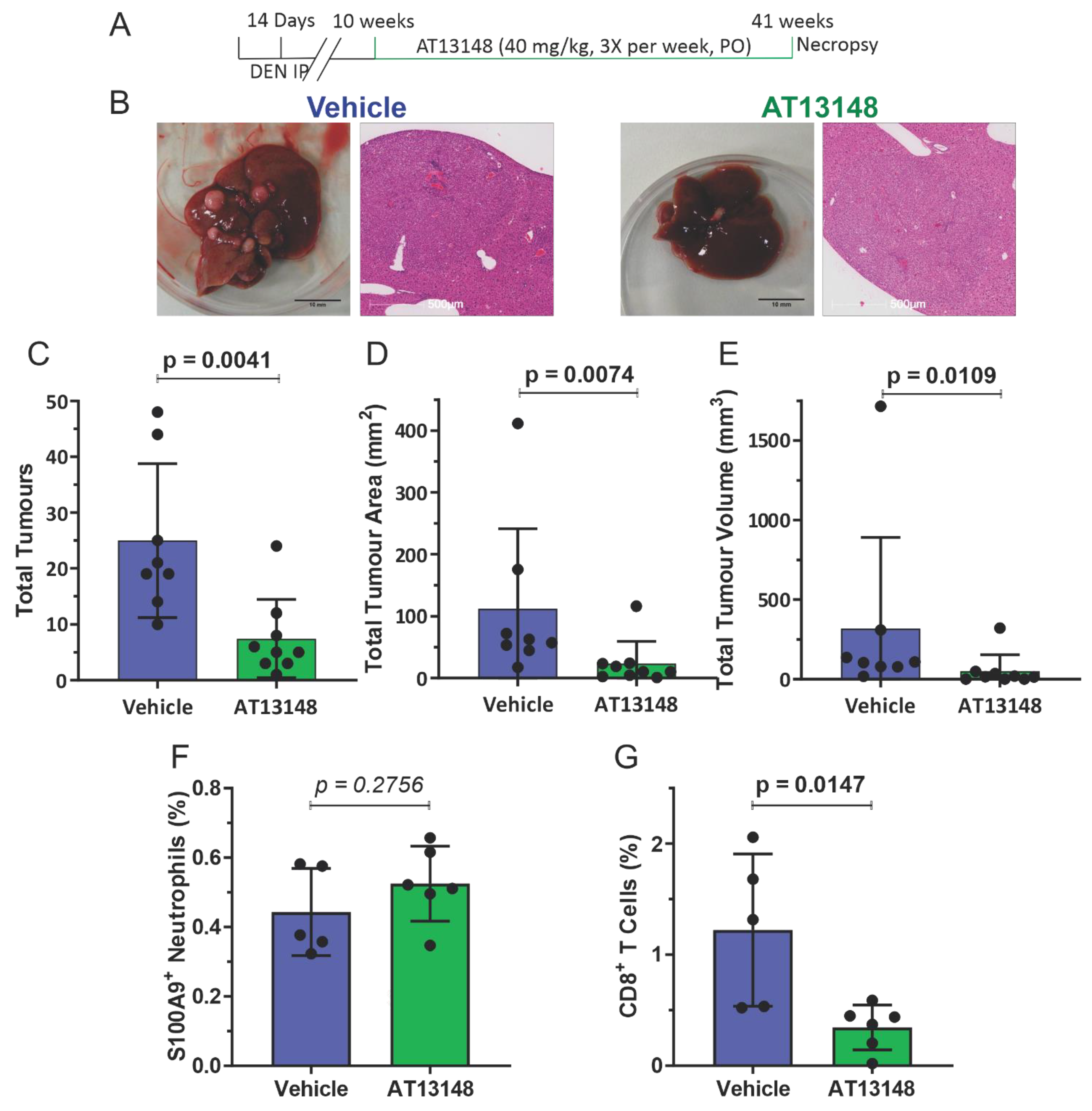
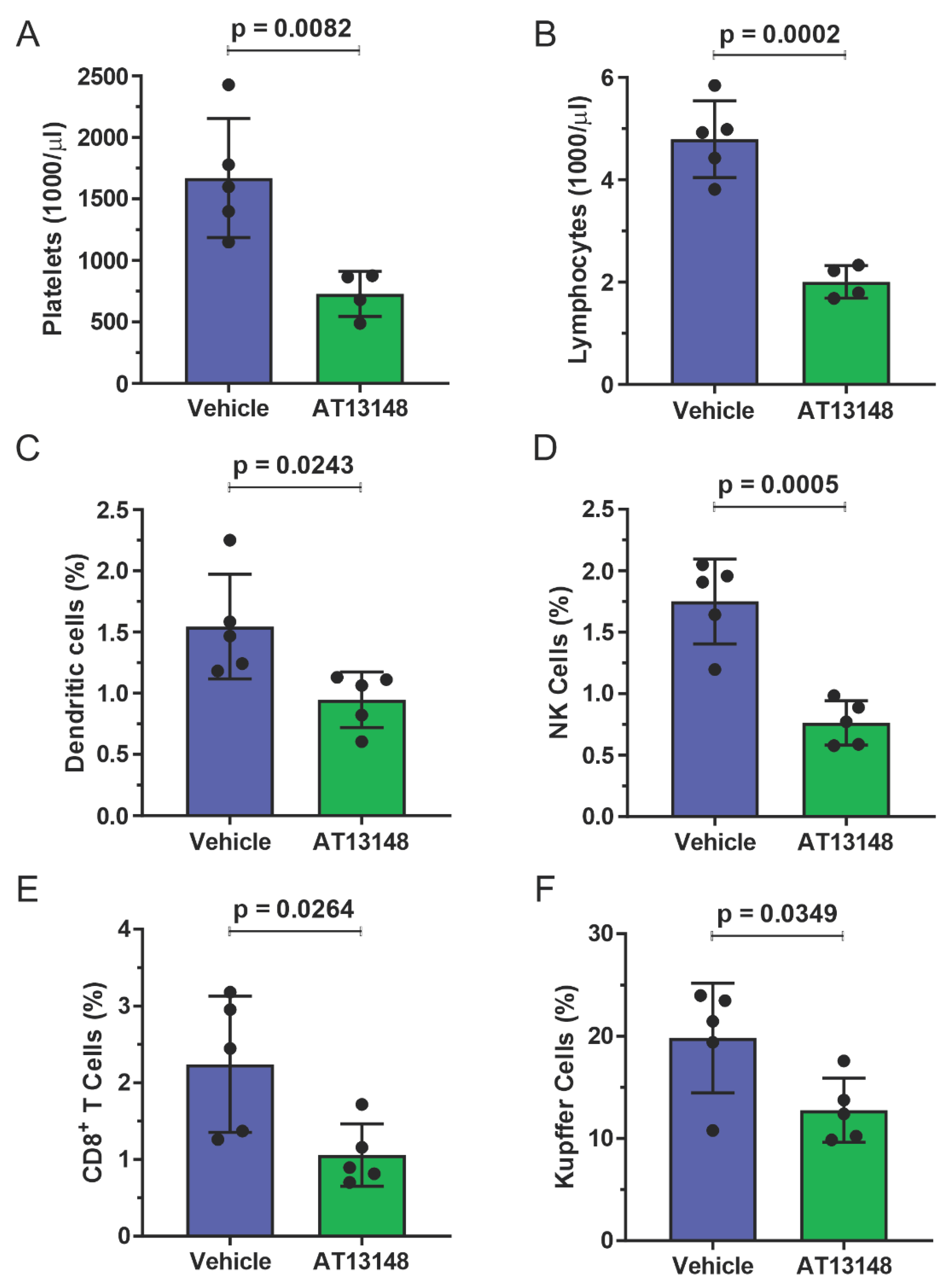
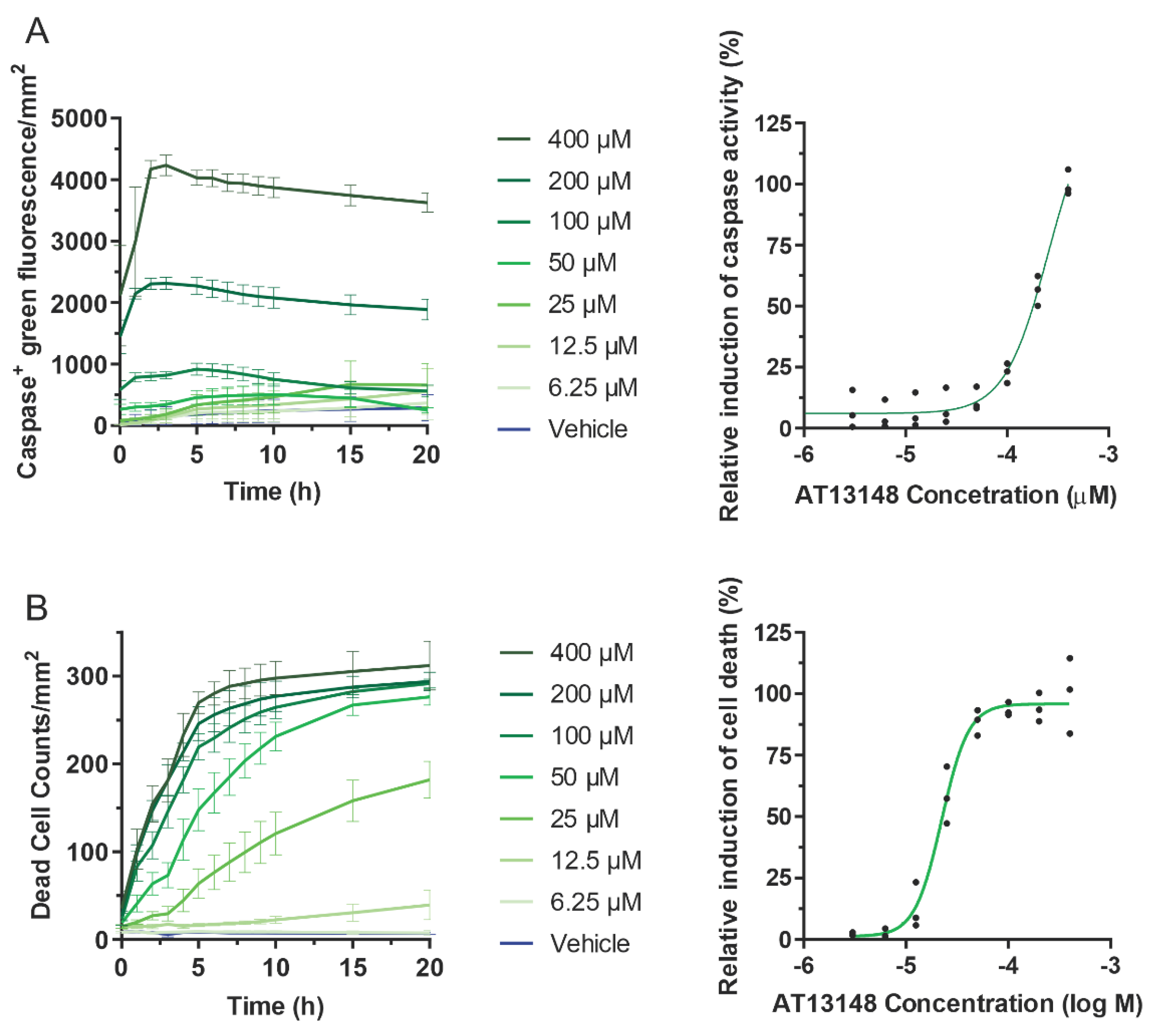
3.3. Pharmacological ROCK Inhibition Reduces DEN-Induced Liver Tumour Numbers without Increasing Immune Cell Recruitment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wickman, G.; Julian, L.; Olson, M.F. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012, 19, 735–742. [Google Scholar] [CrossRef]
- Elliott, M.R.; Ravichandran, K.S. Clearance of apoptotic cells: Implications in health and disease. J. Cell Biol. 2010, 189, 1059–1070. [Google Scholar] [CrossRef]
- Boada-Romero, E.; Martinez, J.; Heckmann, B.L.; Green, D.R. The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell Biol. 2020, 21, 398–414. [Google Scholar] [CrossRef]
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef]
- Kroemer, G.; Galassi, C.; Zitvogel, L.; Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 2022, 23, 487–500. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Galluzzi, L.; Zitvogel, L.; Kroemer, G. Immunological Mechanisms Underneath the Efficacy of Cancer Therapy. Cancer Immunol. Res. 2016, 4, 895–902. [Google Scholar] [CrossRef]
- Petroni, G.; Buqué, A.; Zitvogel, L.; Kroemer, G.; Galluzzi, L. Immunomodulation by targeted anticancer agents. Cancer Cell 2021, 39, 310–345. [Google Scholar] [CrossRef]
- Hossain, D.M.S.; Javaid, S.; Cai, M.; Zhang, C.; Sawant, A.; Hinton, M.; Sathe, M.; Grein, J.; Blumenschein, W.; Pinheiro, E.M.; et al. Dinaciclib induces immunogenic cell death and enhances anti-PD1–mediated tumor suppression. J. Clin. Investig. 2018, 128, 644–654. [Google Scholar] [CrossRef]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Martins, L.M.; Kaufmann, S.H. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999, 68, 383–424. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.L.; Sahai, E.A.; Yeo, M.; Bosch, M.; Dewar, A.; Olson, M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001, 3, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Sebbagh, M.; Renvoize, C.; Hamelin, J.; Riche, N.; Bertoglio, J.; Breard, J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 2001, 3, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.R.; Coleman, M.L.; Li, S.; Robertson, D.; Sullivan, T.; Stewart, C.L.; Olson, M.F. Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. J. Cell Biol. 2005, 168, 245–255. [Google Scholar] [CrossRef]
- Julian, L.; Naylor, G.; Wickman, G.R.; Rath, N.; Castino, G.; Stevenson, D.; Bryson, S.; Munro, J.; McGarry, L.; Mullin, M.; et al. Defective apoptotic cell contractility provokes sterile inflammation, leading to liver damage and tumour suppression. Elife 2021, 10, e61983. [Google Scholar] [CrossRef]
- Wickman, G.R.; Julian, L.; Mardilovich, K.; Schumacher, S.; Munro, J.; Rath, N.; Zander, S.A.L.; Mleczak, A.; Sumpton, D.; Morrice, N.; et al. Blebs produced by actin-myosin contraction during apoptosis release damage-associated molecular pattern proteins before secondary necrosis occurs. Cell Death Differ. 2013, 20, 1293–1305. [Google Scholar] [CrossRef]
- Tolba, R.; Kraus, T.; Liedtke, C.; Schwarz, M.; Weiskirchen, R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab. Anim. 2015, 49, 59–69. [Google Scholar] [CrossRef]
- Kang, J.S.; Wanibuchi, H.; Morimura, K.; Gonzalez, F.J.; Fukushima, S. Role of CYP2E1 in Diethylnitrosamine-Induced Hepatocarcinogenesis In vivo. Cancer Res. 2007, 67, 11141–11146. [Google Scholar] [CrossRef]
- Nagumo, H.; Sasaki, Y.; Ono, Y.; Okamoto, H.; Seto, M.; Takuwa, Y. Rho kinase inhibitor HA-1077 prevents Rho-mediated myosin phosphatase inhibition in smooth muscle cells. Am. J. Physiol. Physiol. 2000, 278, C57–C65. [Google Scholar] [CrossRef]
- Sadok, A.; McCarthy, A.; Caldwell, J.; Collins, I.; Garrett, M.D.; Yeo, M.; Hooper, S.; Sahai, E.; Kuemper, S.; Mardakheh, F.K.; et al. Rho Kinase Inhibitors Block Melanoma Cell Migration and Inhibit Metastasis. Cancer Res. 2015, 75, 2272–2284. [Google Scholar] [CrossRef]
- McLeod, R.; Kumar, R.; Papadatos-Pastos, D.; Mateo, J.; Brown, J.S.; Garces, A.H.I.; Ruddle, R.; Decordova, S.; Jueliger, S.; Ferraldeschi, R.; et al. First-in-Human Study of AT13148, a Dual ROCK-AKT Inhibitor in Patients with Solid Tumors. Clin. Cancer Res. 2020, 26, 4777–4784. [Google Scholar] [CrossRef] [PubMed]
- Rath, N.; Munro, J.; Cutiongco, M.F.; Jagiełło, A.; Gadegaard, N.; McGarry, L.; Unbekandt, M.; Michalopoulou, E.; Kamphorst, J.J.; Sumpton, D.; et al. Rho kinase inhibition by AT13148 blocks pancreatic ductal adenocarcinoma invasion and tumor growth. Cancer Res. 2018, 78, 3321–3336. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Rath, N.; Pajic, M.; Olson, M.F.; Timpson, P. Targeting ROCK activity to disrupt and prime pancreatic cancer for chemotherapy. Small GTPases 2020, 11, 45–52. [Google Scholar] [CrossRef]
- Rath, N.; Morton, J.P.; Julian, L.; Helbig, L.; Kadir, S.; McGhee, E.J.; Anderson, K.I.; Kalna, G.; Mullin, M.; Pinho, A.V.; et al. ROCK signaling promotes collagen remodeling to facilitate invasive pancreatic ductal adenocarcinoma tumor cell growth. EMBO Mol. Med. 2017, 9, 198–218. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.S.; Lopez, J.I.; McGhee, E.J.; Croft, D.R.; Strachan, D.; Timpson, P.; Munro, J.; Schröder, E.; Zhou, J.; Brunton, V.G.; et al. Actomyosin-Mediated Cellular Tension Drives Increased Tissue Stiffness and β-Catenin Activation to Induce Epidermal Hyperplasia and Tumor Growth. Cancer Cell 2011, 19, 776–791. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, Y.-R.A.; O’Koren, E.G.; Hotten, D.F.; Kan, M.J.; Kopin, D.; Nelson, E.R.; Que, L.; Gunn, M.D. A Protocol for the Comprehensive Flow Cytometric Analysis of Immune Cells in Normal and Inflamed Murine Non-Lymphoid Tissues. PLoS ONE 2016, 11, e0150606. [Google Scholar] [CrossRef]
- Bakiri, L.; Wagner, E.F. Mouse models for liver cancer. Mol. Oncol. 2013, 7, 206–223. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.M.; Iatropoulos, M.J.; Wang, C.X.; Ali, N.; Rivenson, A.; Peterson, L.A.; Schulz, C.; Gebhardt, R. Diethylnitrosamine exposure-responses for DNA damage, centrilobular cytotoxicity, cell proliferation and carcinogenesis in rat liver exhibit some non-linearities. Carcinogenesis 1996, 17, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.; Pyao, Y.; Jung, Y.; Lee, J.I.; Lee, W.K. A Modified Protocol of Diethylnitrosamine Administration in Mice to Model Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 5461. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; He, J.C.; Yang, Y.; Wang, J.M.; Qian, Y.W.; Yang, T.; Ji, L. The Prognostic Value of Tumor-infiltrating Lymphocytes in Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Sci. Rep. 2017, 7, 7525. [Google Scholar] [CrossRef] [PubMed]
- Mathai, A.M.; Kapadia, M.J.; Alexander, J.; Kernochan, L.E.; Swanson, P.E.; Yeh, M.M. Role of Foxp3-positive Tumor-infiltrating Lymphocytes in the Histologic Features and Clinical Outcomes of Hepatocellular Carcinoma. Am. J. Surg. Pathol. 2012, 36, 980–986. [Google Scholar] [CrossRef]
- Fu, J.; Xu, D.; Liu, Z.; Shi, M.; Zhao, P.; Fu, B.; Zhang, Z.; Yang, H.; Zhang, H.; Zhou, C.; et al. Increased Regulatory T Cells Correlate With CD8 T-Cell Impairment and Poor Survival in Hepatocellular Carcinoma Patients. Gastroenterology 2007, 132, 2328–2339. [Google Scholar] [CrossRef]
- Xu, X.; Tan, Y.; Qian, Y.; Xue, W.; Wang, Y.; Du, J.; Jin, L.; Ding, W. Clinicopathologic and prognostic significance of tumor-infiltrating CD8+ T cells in patients with hepatocellular carcinoma: A meta-analysis. Medicine 2019, 98, e13923. [Google Scholar] [CrossRef]
- Green, D.R.; Ferguson, T.; Zitvogel, L.; Kroemer, G. Immunogenic and tolerogenic cell death. Nat. Rev. Immunol. 2009, 9, 353–363. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Zitvogel, L.; Kroemer, G. The secret ally: Immunostimulation by anticancer drugs. Nat. Rev. Drug Discov. 2012, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, Y.; Li, B.; Li, C.; Guo, J.; You, J.; Hu, X.; Kuang, D.; Qi, S.; Liu, P.; et al. Targeting ROCK1/2 blocks cell division and induces mitotic catastrophe in hepatocellular carcinoma. Biochem. Pharmacol. 2021, 184, 114353. [Google Scholar] [CrossRef]
- Cassim, S.; Raymond, V.-A.; Lapierre, P.; Bilodeau, M. From in vivo to in vitro: Major metabolic alterations take place in hepatocytes during and following isolation. PLoS ONE 2017, 12, e0190366. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Kepp, O.; Zitvogel, L.; Kroemer, G. Clinical evidence that immunogenic cell death sensitizes to PD-1/PD-L1 blockade. Oncoimmunology 2019, 8, e1637188. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Zitvogel, L. Immune checkpoint inhibitors. J. Exp. Med. 2021, 218, e20201979. [Google Scholar] [CrossRef]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015, 28, 690–714. [Google Scholar] [CrossRef]
- Yu, S.; Wang, J.; Zheng, H.; Wang, R.; Johnson, N.; Li, T.; Li, P.; Lin, J.; Li, Y.; Yan, J.; et al. Pathogenesis from Inflammation to Cancer in NASH-Derived HCC. J. Hepatocell. Carcinoma 2022, 9, 855–867. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, D.; Guo, J.; Ren, Z.; Zhou, L.; Wang, S.; Xu, B.; Wang, R. Effect of Fasudil Hydrochloride, a Protein Kinase Inhibitor, on Cerebral Vasospasm and Delayed Cerebral Ischemic Symptoms After Aneurysmal Subarachnoid Hemorrhage; Results of a Randomized Trial of Fasudil Hydrochloride Versus Nimodipine. Neurol. Med. Chir. 2006, 46, 421–428. [Google Scholar] [CrossRef]
- Koch, J.C.; Kuttler, J.; Maass, F.; Lengenfeld, T.; Zielke, E.; Bähr, M.; Lingor, P. Compassionate Use of the ROCK Inhibitor Fasudil in Three Patients With Amyotrophic Lateral Sclerosis. Front. Neurol. 2020, 11, 173. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.C.; Tatenhorst, L.; Roser, A.-E.; Saal, K.-A.; Tönges, L.; Lingor, P. ROCK inhibition in models of neurodegeneration and its potential for clinical translation. Pharmacol. Ther. 2018, 189, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Fukumoto, Y.; Shimokawa, H. Rho-kinase: Important new therapeutic target in cardiovascular diseases. Am. J. Physiol.-Heart Circ. Physiol. 2011, 301, H287–H296. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Zheng, Y.; von Bornstadt, D.; Wei, Y.; Balcioglu, A.; Daneshmand, A.; Yalcin, N.; Yu, E.; Herisson, F.; Atalay, Y.B.; et al. Selective ROCK2 Inhibition In Focal Cerebral Ischemia. Ann. Clin. Transl. Neurol. 2014, 1, 2–14. [Google Scholar] [CrossRef]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef]
- Vennin, C.; Chin, V.T.; Warren, S.C.; Lucas, M.C.; Herrmann, D.; Magenau, A.; Melenec, P.; Walters, S.N.; del Monte-Nieto, G.; Conway, J.R.W.; et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl. Med. 2017, 9, eaai8504. [Google Scholar] [CrossRef]
- Rath, N.; Olson, M.F. Rho-associated kinases in tumorigenesis: Re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012, 13, 900–908. [Google Scholar] [CrossRef]


| Antigen (Clone) | Species | Supplier (Catalogue Number) | Antigen Retrieval Solution | Retrieval Method | Antibody Dilution; Time & Temperature |
|---|---|---|---|---|---|
| CD3 (SP7) | Rabbit | Vector (VP-RM01) | Citrate buffer pH 6 | WB; 98 °C; 25 min | 1:75; 35 min, RT |
| F4/80 (Cl:A3-1) | Rat | Abcam (ab6640) | Proteinase K | 10 min at RT | 1:400; 35 min; RT |
| Foxp3 (FJK-16s) | Rat | eBioscience (14-5773-80) | Tris-EDTA buffer pH 9 | WB; 98 °C; 20 min | 1:50; O/N; 4 °C |
| S100A9 (M-19) | Goat | Santa Cruz (sc-8115) | Citrate buffer pH 6 | WB; 98 °C; 25 min | 1:1000; 35 min; RT |
| CD8 (4SM15) | Rat | eBioscience (14-0808-82) | Tris-EDTA buffer pH 9 | WB; 98 °C; 20 min | 1:500; 35 min; RT |
| Lymphoid Panel | Myeloid Panel | ||||||
|---|---|---|---|---|---|---|---|
| Antigen (Clone) | Supplier | Dilu-tion | Fluoro-Phore | Antigen (Clone) | Supplier | Dilu-tion | Fluoro-Phore |
| B220 (RA3-6B2) | Biolegend | 1:200 | PerCPCy5.5 | CD11b (M1/70) | Biolegend | 1:200 | BV570 |
| CD3 (17A2) | Biolegend | 1:200 | FITC | CD11c (N418) | Biolegend | 1:200 | BV711 |
| CD4 (RN4-5) | eBio-science | 1:200 | SB600 | CD45 (30-F11) | Biolegend | 1:200 | BV510 |
| CD8 (53-6.7) | Biolegend | 1:200 | PECy7 | CD115 (AFS98) | Biolegend | 1:100 | BV421 |
| CD19 (6D5) | Biolegend | 1:200 | PE | Clec4f | Biolegend | 1:100 | Goat/AF594 |
| CD25 (PC61) | Biolegend | 1:100 | AF700 | F4/80 (BM8) | Biolegend | 1:100 | Biotin/PE |
| CD45 (30-F11) | Biolegend | 1:200 | BV510 | Ly6C (HK1.4) | Biolegend | 1:400 | PECy7 |
| FoxP3 (MF14) | Biolegend | 1:100 | AF647 | Ly6G (1A8) | Biolegend | 1:200 | AF700 |
| NK1.1 (PK136) | Biolegend | 1:200 | BV711 | MHCII (M5/114.15.2) | Biolegend | 1:500 | FITC |
| TCRβ (A57-597) | Biolegend | 1:100 | BV421 | PDCA1 (927) | Biolegend | 1:100 | PerCPCy5.5 |
| TCRγδ (GL3) | Biolegend | 1:100 | Biotin/AF594 | Siglec H (551) | Biolegend | 1:100 | APC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naylor, G.; Julian, L.; Watson-Bryce, S.; Mullin, M.; Nibbs, R.J.; Olson, M.F. Immunogenic Death of Hepatocellular Carcinoma Cells in Mice Expressing Caspase-Resistant ROCK1 Is Not Replicated by ROCK Inhibitors. Cancers 2022, 14, 5943. https://doi.org/10.3390/cancers14235943
Naylor G, Julian L, Watson-Bryce S, Mullin M, Nibbs RJ, Olson MF. Immunogenic Death of Hepatocellular Carcinoma Cells in Mice Expressing Caspase-Resistant ROCK1 Is Not Replicated by ROCK Inhibitors. Cancers. 2022; 14(23):5943. https://doi.org/10.3390/cancers14235943
Chicago/Turabian StyleNaylor, Gregory, Linda Julian, Steven Watson-Bryce, Margaret Mullin, Robert J. Nibbs, and Michael F. Olson. 2022. "Immunogenic Death of Hepatocellular Carcinoma Cells in Mice Expressing Caspase-Resistant ROCK1 Is Not Replicated by ROCK Inhibitors" Cancers 14, no. 23: 5943. https://doi.org/10.3390/cancers14235943
APA StyleNaylor, G., Julian, L., Watson-Bryce, S., Mullin, M., Nibbs, R. J., & Olson, M. F. (2022). Immunogenic Death of Hepatocellular Carcinoma Cells in Mice Expressing Caspase-Resistant ROCK1 Is Not Replicated by ROCK Inhibitors. Cancers, 14(23), 5943. https://doi.org/10.3390/cancers14235943







