Comprehensive Analysis of Cuproptosis-Related Genes in Prognosis and Immune Infiltration of Hepatocellular Carcinoma Based on Bulk and Single-Cell RNA Sequencing Data
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Acquisition
2.2. Identification of Prognostic CRGs and Selection of Differentially Expressed Genes Related to Cuproptosis (CRGs-DEGs)
2.3. Development and Validation of the Prediction Model
2.4. Functional Enrichment Analysis and Immune Infiltration Score
2.5. Single-Cell RNA seq (scRNA-seq) Analysis
2.6. Cell Culturing
2.7. Multiplex Immunofluorescence Staining
2.8. Real-Time Polymerase Chain Reaction (PCR)
2.9. Statistical Analyses
3. Results
3.1. Prognostic and Differential Analysis of CRGs in the TCGA Cohort
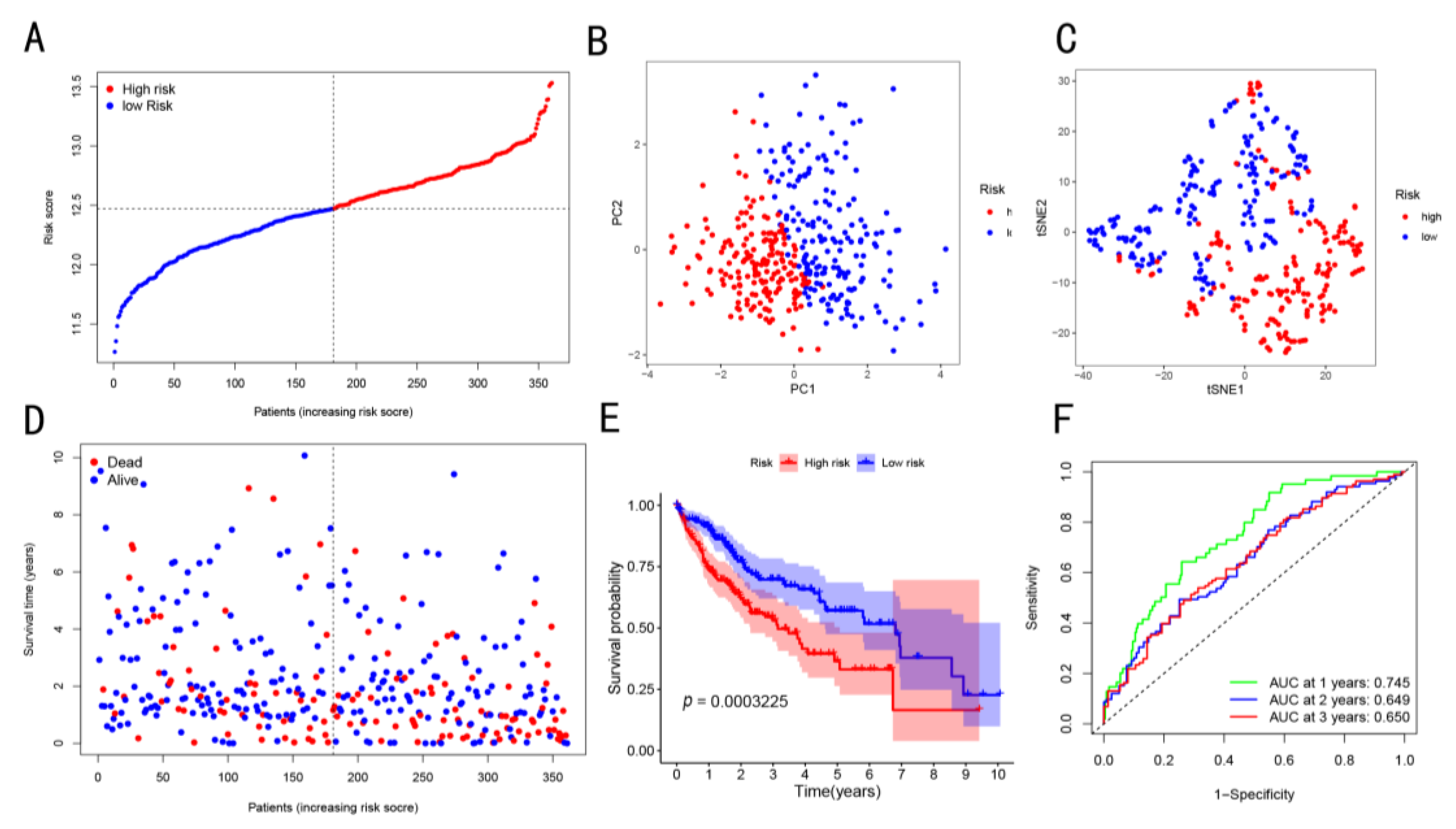
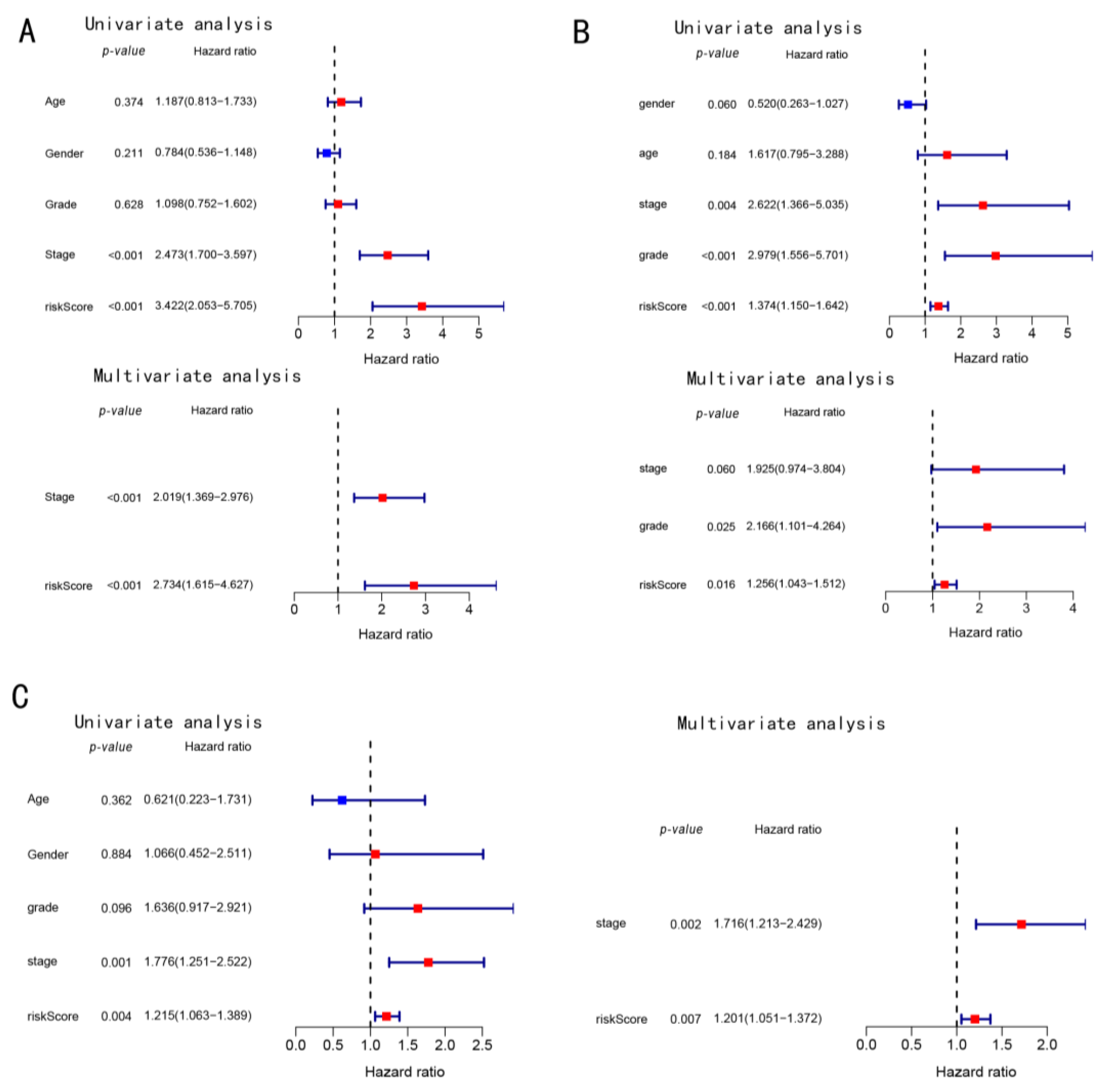
3.2. Construction and Exploration of Prognostic Models in the TCGA Cohort
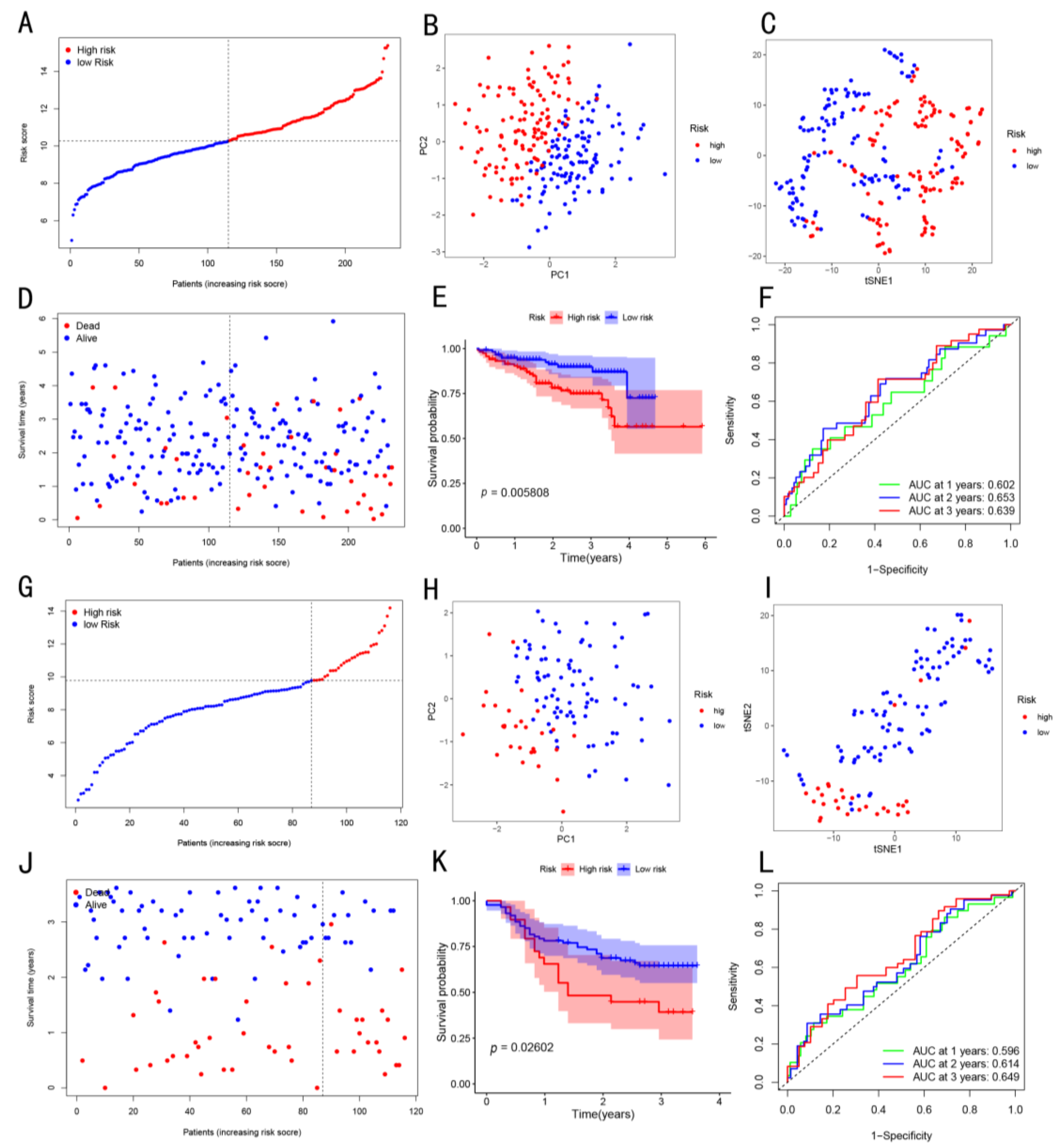
3.3. Prognostic Potential of the Constructed Four-Gene Signature
3.4. Functional Enrichment Analysis in Three Cohorts
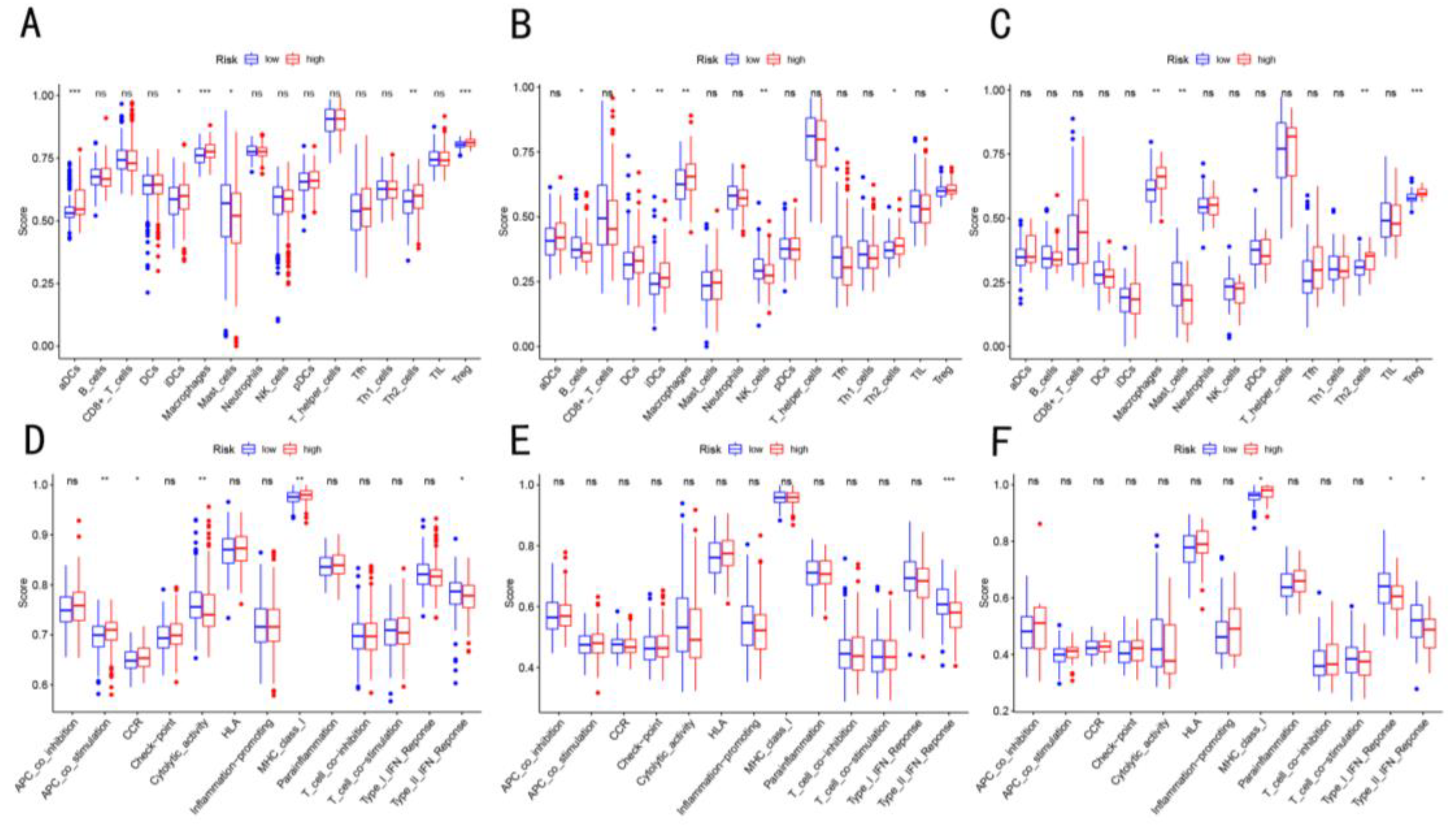
3.5. Enrichment Score of Immune Cells Infiltration in Three Cohorts
3.6. Immunosuppressive T cell Related to Cuproptosis Based on scRNA-seq
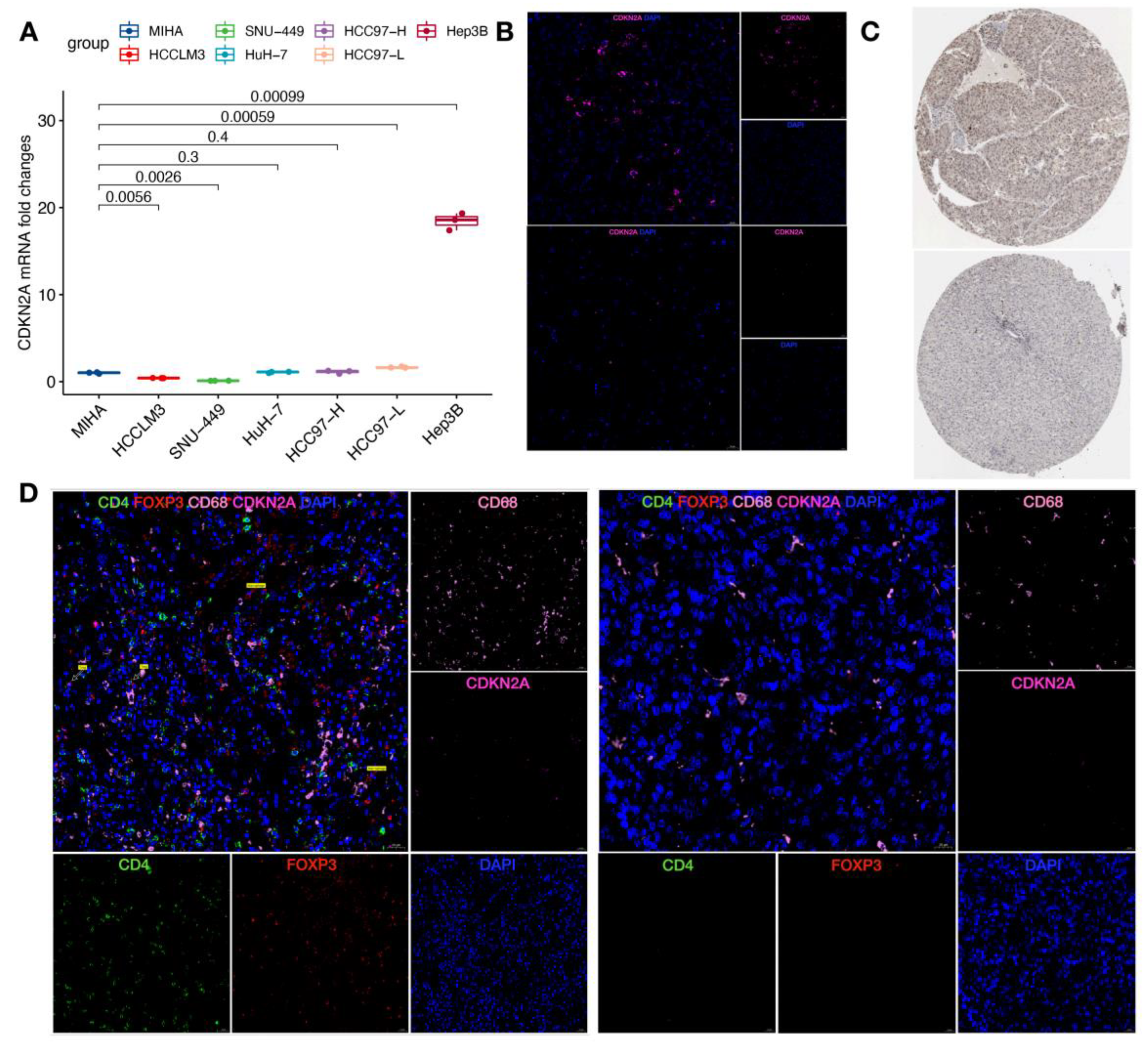
3.7. Validation of Cuproptosis-Related Genes and Immune Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 7. [Google Scholar] [CrossRef]
- Blockhuys, S.; Celauro, E.; Hildesjö, C.; Feizi, A.; Stål, O.; Fierro-González, J.C.; Wittung-Stafshede, P. Defining the human copper proteome and analysis of its expression variation in cancers. Metallomics 2017, 9, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Dai, L.-J.; Wu, S.-Y.; Xiao, Y.; Ma, D.; Jiang, Y.-Z.; Shao, Z.-M. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021, 14, 98. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Liang, J.-Y.; Wang, D.-S.; Lin, H.-C.; Chen, X.-X.; Yang, H.; Zheng, Y.; Li, Y.-H. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int. J. Biol. Sci. 2020, 16, 2430–2441. [Google Scholar] [CrossRef]
- Deng, M.; Sun, S.; Zhao, R.; Guan, R.; Zhang, Z.; Li, S.; Wei, W.; Guo, R. The pyroptosis-related gene signature predicts prognosis and indicates immune activity in hepatocellular carcinoma. Mol. Med. 2022, 28, 16. [Google Scholar] [CrossRef]
- Bian, Z.; Fan, R.; Xie, L. A Novel Cuproptosis-Related Prognostic Gene Signature and Validation of Differential Expression in Clear Cell Renal Cell Carcinoma. Genes 2022, 13, 851. [Google Scholar] [CrossRef] [PubMed]
- Shelton, P.; Jaiswal, A.K. The transcription factor NF-E2-related factor 2 (Nrf2): A protooncogene? FASEB J. 2013, 27, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.; Rojo, A.I.; Arias, E.; Díaz-Carretero, A.; Cuervo, A.M.; Cuadrado, A. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy 2018, 14, 1310–1322. [Google Scholar] [CrossRef] [PubMed]
- Lu, V.M.; O’Connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neurooncol. 2020, 148, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.-C.; Llovet, J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015, 149, 1226–1239.e4. [Google Scholar] [CrossRef] [PubMed]
- Rumping, L.; Tessadori, F.; Pouwels, P.J.W.; Vringer, E.; Wijnen, J.P.; Bhogal, A.A.; Savelberg, S.M.C.; Duran, K.J.; Bakkers, M.J.G.; Ramos, R.J.J.; et al. GLS hyperactivity causes glutamate excess, infantile cataract and profound developmental delay. Hum. Mol. Genet. 2019, 28, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cao, Y.; Meng, G.; Qian, L.; Xu, T.; Yan, C.; Luo, O.; Wang, S.; Wei, J.; Ding, Y.; et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. eBioMedicine 2019, 39, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gan, G.; Wang, X.; Xu, T.; Xie, W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy 2019, 15, 1258–1279. [Google Scholar] [CrossRef] [PubMed]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Oura, K.; Morishita, A.; Tani, J.; Masaki, T. Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: A Review. Int. J. Mol. Sci. 2021, 22, 5801. [Google Scholar] [CrossRef]
- Zhang, Q.; He, Y.; Luo, N.; Patel, S.J.; Han, Y.; Gao, R.; Modak, M.; Carotta, S.; Haslinger, C.; Kind, D.; et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019, 179, 829–845.e20. [Google Scholar] [CrossRef]
- Zhou, S.-L.; Zhou, Z.-J.; Hu, Z.-Q.; Huang, X.-W.; Wang, Z.; Chen, E.-B.; Fan, J.; Cao, Y.; Dai, Z.; Zhou, J. Tumor-Associated Neutrophils Recruit Macrophages and T-Regulatory Cells to Promote Progression of Hepatocellular Carcinoma and Resistance to Sorafenib. Gastroenterology 2016, 150, 1646–1658.e17. [Google Scholar] [CrossRef]
- Fu, J.; Xu, D.; Liu, Z.; Shi, M.; Zhao, P.; Fu, B.; Zhang, Z.; Yang, H.; Zhang, H.; Zhou, C.; et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007, 132, 2328–2339. [Google Scholar] [CrossRef]
- Takahashi, H.; Kawaguchi, T.; Yan, L.; Peng, X.; Qi, Q.; Morris, L.G.T.; Chan, T.A.; Tsung, A.; Otsuji, E.; Takabe, K. Immune Cytolytic Activity for Comprehensive Understanding of Immune Landscape in Hepatocellular Carcinoma. Cancers 2020, 12, 1221. [Google Scholar] [CrossRef]
- Li, H.; van der Leun, A.M.; Yofe, I.; Lubling, Y.; Gelbard-Solodkin, D.; van Akkooi, A.C.J.; van den Braber, M.; Rozeman, E.A.; Haanen, J.B.A.G.; Blank, C.U.; et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019, 176, 775–789.e18. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, J.; Yin, S.; Shi, J.; Zheng, M.; He, C.; Meng, H.; Han, Y.; Han, J.; Guo, J.; et al. The expression of cuproptosis-related genes in hepatocellular carcinoma and their relationships with prognosis. Front. Oncol. 2022, 12, 992468. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Li, W.; Tu, J.; Cao, Z.; Li, J.; Cao, H.; Liang, J.; Liang, Y.; Yu, Q.; Li, G. Identification of cuproptosis-related subtypes, cuproptosis-related gene prognostic index in hepatocellular carcinoma. Front. Immunol. 2022, 13, 989156. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, S.; Li, Z.; Qin, W.; Tong, Q.; Liu, C.; Wang, Z.; Liu, Z.; Xu, X. Comprehensive multiomics analysis of cuproptosis-related gene characteristics in hepatocellular carcinoma. Front. Genet. 2022, 13, 942387. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Wang, M.; Meng, Y.; Chen, D.; Xu, Y.; Jiang, X.; Xiong, Z. Prognostic Implication of a Cuproptosis-Related miRNA Signature in Hepatocellular Carcinoma. J. Healthc. Eng. 2022, 2022, 4694323. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, Y.; Xia, Y.; Liu, Y.; Gan, J. Cuproptosis-related lncRNAs predict the prognosis and immune response in hepatocellular carcinoma. Clin. Exp. Med. 2022, 20, 275. [Google Scholar] [CrossRef]
- Luo, L.; Hu, X.; Huang, A.; Liu, X.; Wang, L.; Du, T.; Liu, L.; Li, M. A Noval Established Cuproptosis-Associated LncRNA Signature for Prognosis Prediction in Primary Hepatic Carcinoma. Evid. Based Complement. Alternat. Med. 2022, 2022, 2075638. [Google Scholar]




| TCGA | ICGC (LIRI-JP) | In-House RNA-seq | |
|---|---|---|---|
| Number of patients | 361 | 231 | 116 |
| Age (median, range) | 61 (16–90) | 69 (31–89) | 49.5 (36–76) |
| Gender | |||
| Female | 117 | 61 | 15 |
| Male | 244 | 170 | 101 |
| Grade | |||
| Grade 1–2 | 224 | 53 | 15 |
| Grade 3–4 | 132 | 159 | 101 |
| unknown | 5 | 19 | 0 |
| AFP | |||
| ≤200 | 196 | NA | 63 |
| >200 | 75 | NA | 53 |
| unknown | 90 | NA | 0 |
| Stage | |||
| Stage I | 167 | 36 | 58 |
| Stage II | 82 | 105 | 32 |
| Stage III | 84 | 71 | 26 |
| Stage IV | 4 | 19 | 0 |
| unknown | 24 | 0 | 0 |
| vascular_tumor | |||
| Macro | 16 | NA | 23 |
| Micro | 89 | NA | 49 |
| None | 200 | NA | 44 |
| unknown | 56 | NA | NA |
| Survival status | |||
| OS days | 588 (0–3675) | 780 (10–2160) | 945 (0–1320) |
| Cuproptosis-Related Genes | CRGs-DEGs | Prognosis Value | Intersect Genes | Signature Model |
|---|---|---|---|---|
| NFE2L2 | √ | √ | √ | |
| NLRP3 | √ | |||
| ATP7B | ||||
| ATP7A | √ | √ | √ | |
| SLC31A1 | √ | |||
| FDX1 | √ | |||
| LIAS | √ | |||
| LIPT1 | √ | √ | √ | √ |
| LIPT2 | √ | √ | √ | |
| DLD | ||||
| DLAT | √ | √ | √ | √ |
| PDHA1 | √ | √ | √ | |
| PDHB | √ | |||
| MTF1 | √ | √ | √ | |
| GLS | √ | √ | √ | √ |
| CDKN2A | √ | √ | √ | √ |
| DBT | √ | |||
| GCSH | √ | |||
| DLST |
| Characteristics | TCGA-LIHC Cohort | ICGC-LIRP-JP Cohort | Internal RNA-seq Cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| High Risk | Low Risk | p-Value | High Risk | Low Risk | p-Value | High Risk | Low Risk | p-Value | |
| Gender | 0.76348 | 0.43206 | 0.63174 | ||||||
| Female | 57 | 60 | 28 | 33 | 5 | 10 | |||
| Male | 123 | 121 | 88 | 82 | 24 | 77 | |||
| Age | 0.72431 | 0.46656 | 0.20261 | ||||||
| ≤65 year | 113 | 111 | 42 | 47 | 28 | 70 | |||
| >65 year | 66 | 70 | 74 | 68 | 1 | 12 | |||
| unknown | 1 | 0 | 0 | 0 | 0 | 0 | |||
| Grade | 0.00845 | 0.01173 | - | ||||||
| G1 and G2 | 100 | 124 | 69 | 84 | 29 | 86 | |||
| G3 and G4 | 78 | 54 | 38 | 21 | 0 | 0 | |||
| unknown | 2 | 3 | 9 | 10 | 0 | 1 | |||
| AFP | 0.97195 | - | 0.45128 | ||||||
| ≤200 | 91 | 105 | - | - | 14 | 49 | |||
| >200 | 35 | 40 | - | - | 15 | 38 | |||
| unknown | 54 | 36 | - | - | 0 | 0 | |||
| vascular_tumor | 0.73062 | - | 0.18493 | ||||||
| Yes | 51 | 54 | - | - | 21 | 51 | |||
| no | 93 | 107 | - | - | 8 | 36 | |||
| unknown | 36 | 20 | - | - | - | - | |||
| Stage | 0.00136 | 0.11724 | 0.44054 | ||||||
| I and II | 109 | 140 | 65 | 76 | 24 | 66 | |||
| III and IV | 56 | 32 | 51 | 39 | 5 | 21 | |||
| unknown | 15 | 9 | 0 | 0 | 0 | 0 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Guo, Y.; Wu, Z.; Huang, J.; Xiang, B. Comprehensive Analysis of Cuproptosis-Related Genes in Prognosis and Immune Infiltration of Hepatocellular Carcinoma Based on Bulk and Single-Cell RNA Sequencing Data. Cancers 2022, 14, 5713. https://doi.org/10.3390/cancers14225713
Yang C, Guo Y, Wu Z, Huang J, Xiang B. Comprehensive Analysis of Cuproptosis-Related Genes in Prognosis and Immune Infiltration of Hepatocellular Carcinoma Based on Bulk and Single-Cell RNA Sequencing Data. Cancers. 2022; 14(22):5713. https://doi.org/10.3390/cancers14225713
Chicago/Turabian StyleYang, Chenglei, Yanlin Guo, Zongze Wu, Juntao Huang, and Bangde Xiang. 2022. "Comprehensive Analysis of Cuproptosis-Related Genes in Prognosis and Immune Infiltration of Hepatocellular Carcinoma Based on Bulk and Single-Cell RNA Sequencing Data" Cancers 14, no. 22: 5713. https://doi.org/10.3390/cancers14225713
APA StyleYang, C., Guo, Y., Wu, Z., Huang, J., & Xiang, B. (2022). Comprehensive Analysis of Cuproptosis-Related Genes in Prognosis and Immune Infiltration of Hepatocellular Carcinoma Based on Bulk and Single-Cell RNA Sequencing Data. Cancers, 14(22), 5713. https://doi.org/10.3390/cancers14225713





