Girdin Promotes Tumorigenesis and Chemoresistance in Lung Adenocarcinoma by Interacting with PKM2
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Lenti-sgRNA/Cre Virus
2.1.1. sgRNA Construction
2.1.2. Lentivirus Package
2.1.3. RT-PCR Measurements of Lentivirus Titer
2.2. Ethics Statement, Mice, and Tumor Initiation
2.3. Histology and Immunohistochemistry
2.4. Measurement of Glucose Assumption, Lactate, and ATP Production
2.5. PKM2 Activity Assay
2.6. Cell Culture, Transfection, Plasmids, and RNA Interference
2.7. Antibodies and Reagents
2.8. Data Analysis
3. Results
3.1. Girdin Knockout Severely Inhibits LUAD Tumorigenesis
3.2. Girdin Promotes LUAD Progression
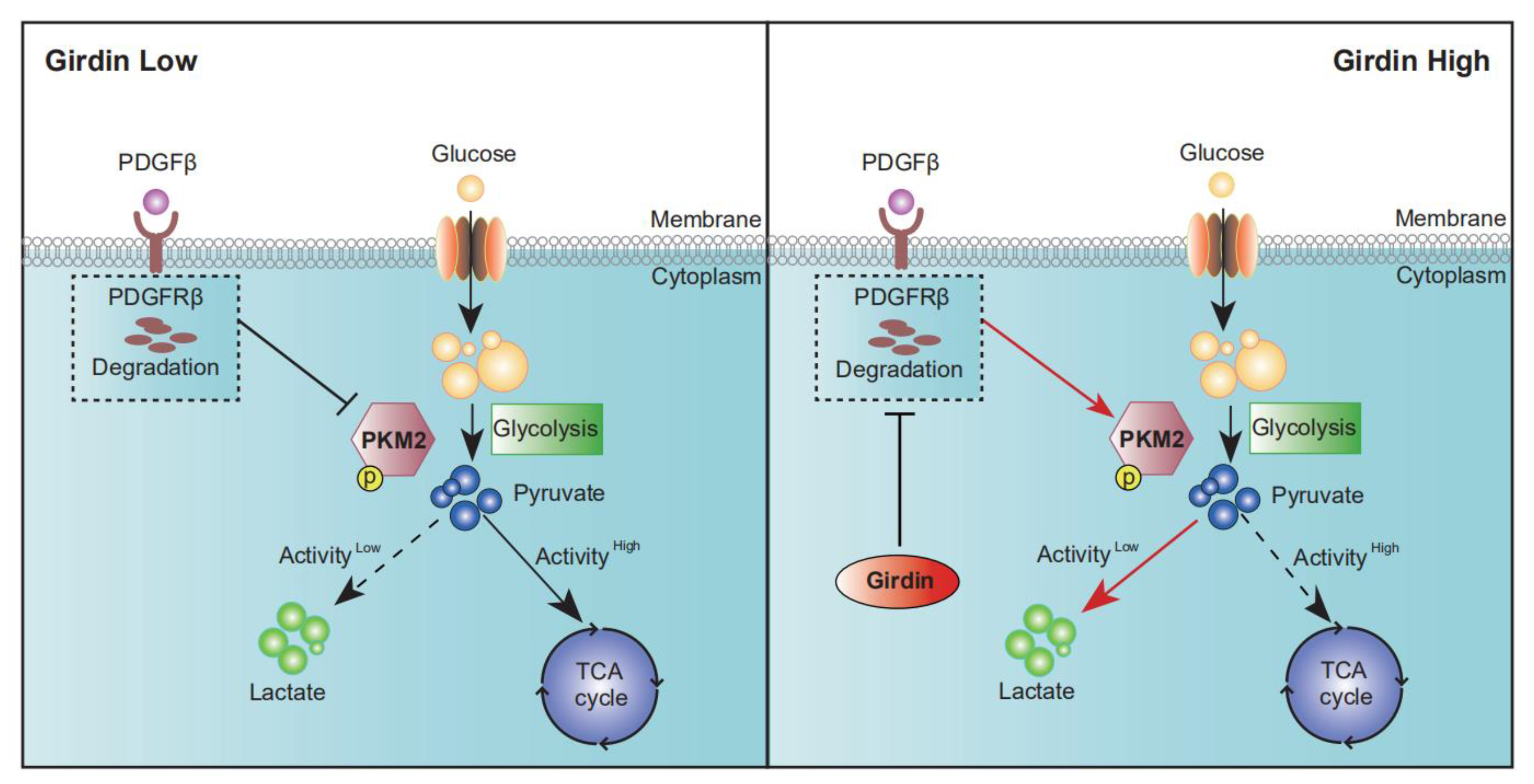
3.3. Girdin Enhances the Warburg Effect in LUAD
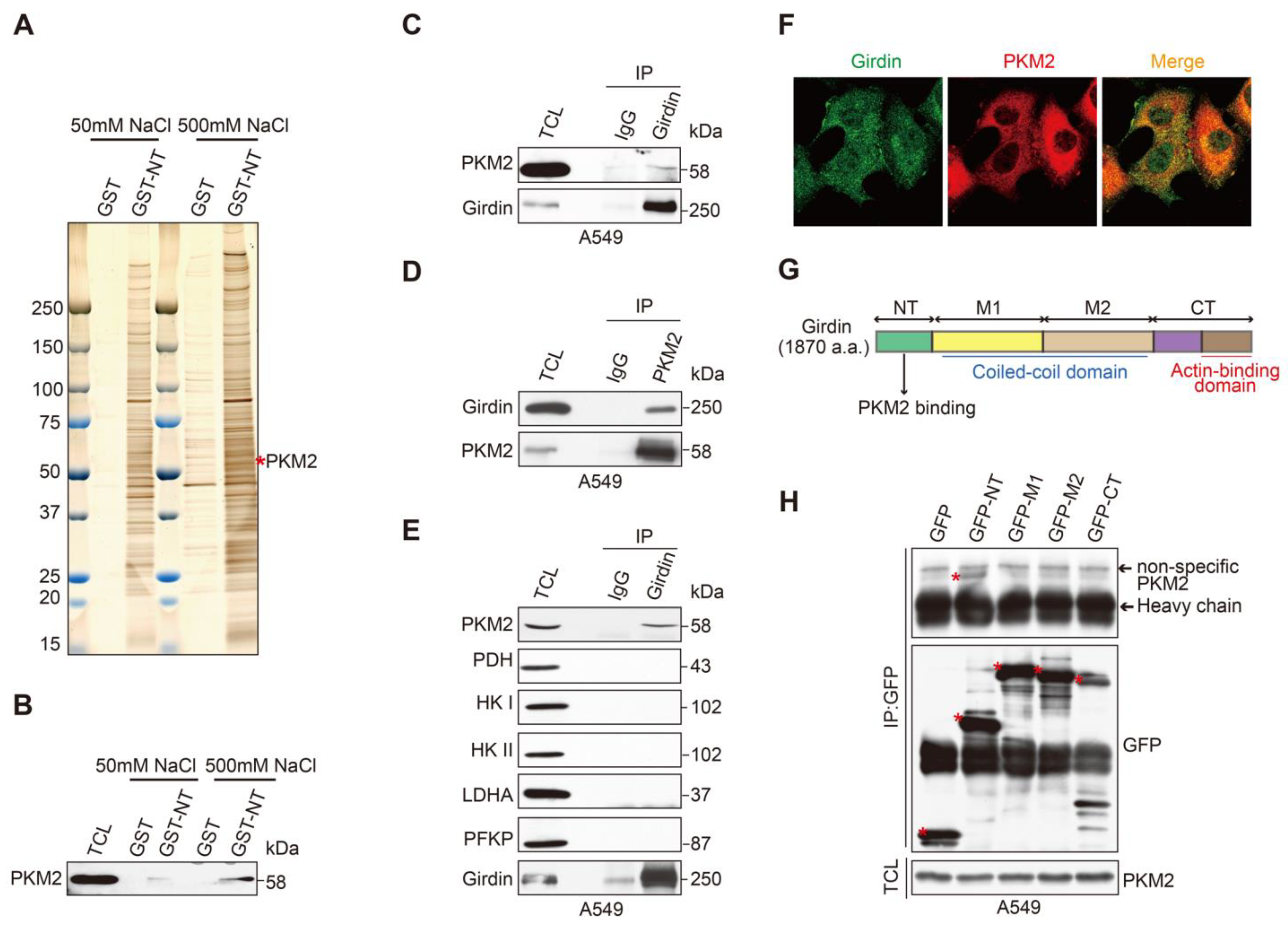
3.4. Identification of PKM2 as a Girdin-Interacting Protein
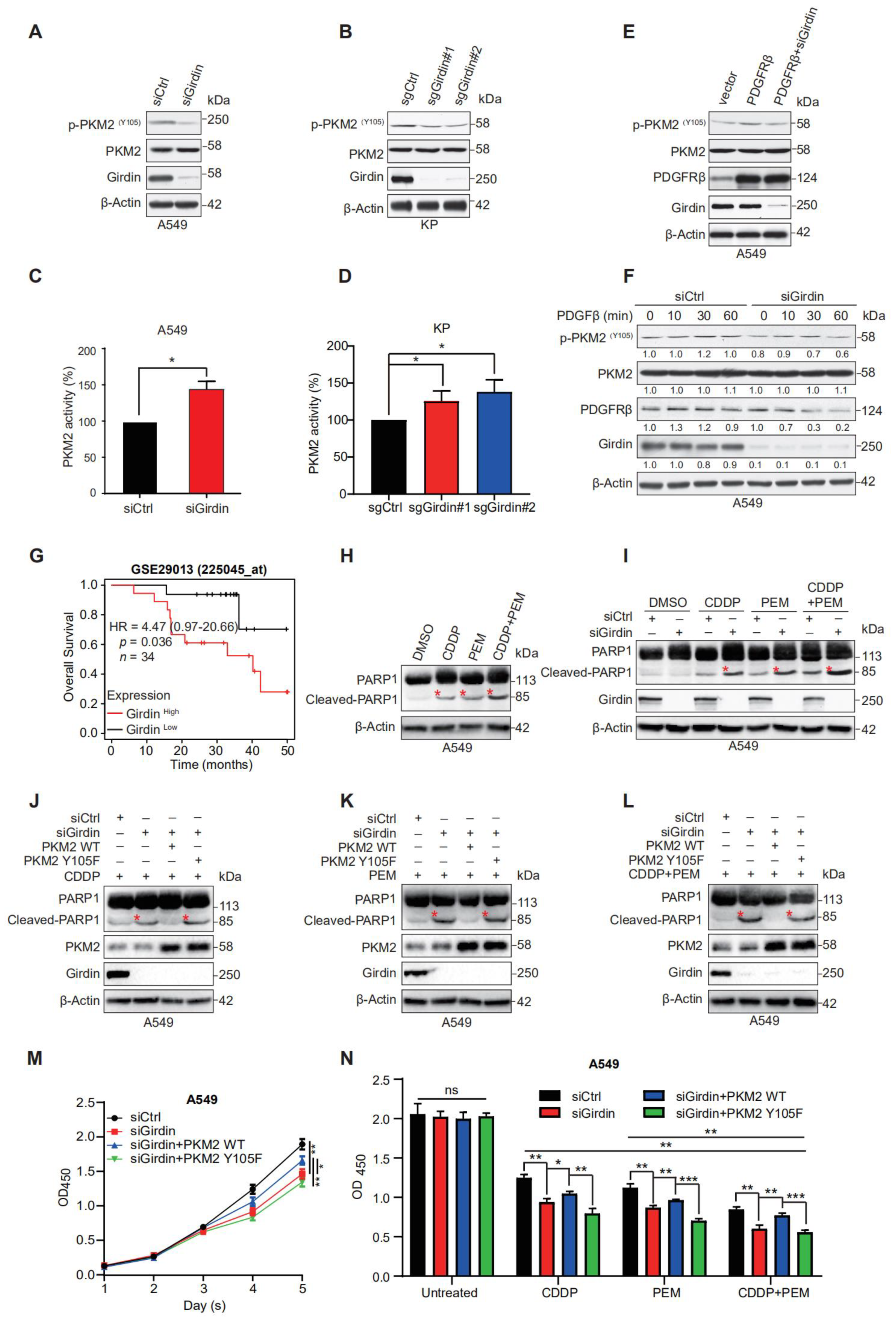
3.5. Girdin Leads to Chemoresistance by Regulating PKM2 Phosphorylation and Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Shulman, S. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2018, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, B.; Gottlieb, E. Rocking cell metabolism: Revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem. Sci. 2012, 37, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Kang, S. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal 2009, 2, ra73. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186. [Google Scholar] [CrossRef]
- Mazurek, S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 2011, 43, 969–980. [Google Scholar] [CrossRef]
- Jiang, P.; Enomoto, A. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 2008, 68, 1310–1318. [Google Scholar] [CrossRef]
- Natsume, A.; Kato, T. Girdin maintains the stemness of glioblastoma stem cells. Oncogene 2012, 31, 2715–2724. [Google Scholar] [CrossRef]
- Garcia-Marcos, M.; Jung, B.H. Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 2011, 25, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, A.; Murakami, H.; Asai, N.; Morone, N.; Watanabe, T.; Kawai, K.; Murakumo, Y.; Usukura, J.; Kaibuchi, K.; Takahashi, M. Akt PKB Regulates Actin Organization and Cell Motility via Girdin APE. Dev. Cell 2005, 9, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Anai, M.; Shojima, N. A novel protein kinase B (PKB)/AKT-binding protein enhances PKB kinase activity and regulates DNA synthesis. J. Biol. Chem. 2005, 280, 18525–18535. [Google Scholar]
- Le-Niculescu, H.; Niesman, I. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J. Biol. Chem. 2005, 280, 22012–22020. [Google Scholar] [CrossRef]
- Simpson, F.; Martin, S. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic 2005, 6, 442–458. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, Y.; Asai, N. Akt-Girdin signaling in cancer-associated fibroblasts contributes to tumor progression. Cancer Res. 2015, 75, 813–823. [Google Scholar] [CrossRef]
- Kitamura, T.; Asai, N. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat. Cell Biol. 2008, 10, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Enomoto, A. Regulation of cargo-selective endocytosis by dynamin 2 GTPase-activating protein girdin. EMBO J. 2014, 33, 2098–2112. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Enomoto, A. Girdin/GIV regulates collective cancer cell migration by controlling cell adhesion and cytoskeletal or-ganization. Cancer Sci. 2018, 109, 3643–3656. [Google Scholar] [CrossRef]
- Barbazan, J.; Dunkel, Y. Prognostic Impact of Modulators of G proteins in Circulating Tumor Cells from Patients with Metastatic Colorectal Cancer. Sci. Rep. 2016, 6, 22112. [Google Scholar] [CrossRef]
- Weng, L.; Han, Y.P. Negative regulation of amino acid signaling by MAPK-regulated 4F2hc/Girdin complex. PLoS Biol. 2018, 16, e2005090. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Feng, W. Girdin Knockdown Increases Gemcitabine Chemosensitivity to Pancreatic Cancer by Modulating Au-tophagy. Front. Oncol. 2021, 11, 618764. [Google Scholar] [CrossRef]
- Lizée, G.; Aerts, J.L.; Gonzales, M.I.; Chinnasamy, N.; Morgan, R.A.; Topalian, S.L. Real-time quantitative reverse transcriptase-polymerase chain reaction as a method for determining lentiviral vector titers and measuring transgene expression. Hum. Gene Ther. 2003, 14, 497–507. [Google Scholar] [CrossRef]
- DuPage, M.; Dooley, A.L. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009, 4, 1064–1072. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ramsey, M.R. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007, 448, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, F. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat. Commun. 2014, 5, 3261. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Hamblin, M.R. Regulation of Glycolysis by Non-coding RNAs in Cancer: Switching on the Warburg Effect. Mol. Ther. Oncolytics 2020, 19, 218–239. [Google Scholar] [CrossRef]
- Tamada, M.; Nagano, O. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, F.; Rumio, C. Glycolysis-induced drug resistance in tumors-A response to danger signals? Neoplasia 2021, 23, 234–245. [Google Scholar] [CrossRef]
- Wang, S.; Lei, Y. Girdin regulates the proliferation and apoptosis of pancreatic cancer cells via the PI3K/Akt signalling pathway. Oncol. Rep. 2018, 40, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Chen, X.S. Inhibition of AXL enhances chemosensitivity of human ovarian cancer cells to cisplatin via decreasing glycolysis. Acta Pharmacol. Sin. 2021, 42, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, M. Oncogenic Kinase-Induced PKM2 Tyrosine 105 Phosphorylation Converts Nononcogenic PKM2 to a Tumor Promoter and Induces Cancer Stem-like Cells. Cancer Res. 2018, 78, 2248–2261. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Schug, Z.T. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef]



| Primer | Sequences (5′ to 3′) |
|---|---|
| Lenti-sgTrp53/Cre-F | AGCTTTGTTTAAACGAGGGCCTATTTCCCATGAT |
| Lenti-sgTrp53/Cre-R | TTGGCGCGCCGCGAATTCAAAAAAGCACCG |
| Lenti-sgTrp53/Cre-sgGirdin-F | AGCTTTGTTTAAACGAATTGACGCGTATTGGGAT |
| Lenti-sgTrp53/Cre-sgGirdin-R | GCCAAGCTCGGCGCGCCATT |
| Primer | Sequences (5′ to 3′) |
|---|---|
| WPRE-F | TCAGCTCCTTTCCGGGACTT |
| WPRE-R | ATTGAGGGCCGAAGGGACGT |
| RagB-F | GCAAGACACCTTCATGGAAA |
| RagB-R | GCATGTCCTTTTCCAGTTCG |
| Primer | Sequences (5′ to 3′) |
|---|---|
| Kras-F | CTAGCCACCATGGCTTGAGT |
| Kras-R | TCCGAATTCAGTGACTACAGATG |
| LSL-F | CGGCATGGACGAGCTGTACAAG |
| LSL-R | TCAGCAAACACAGTGCACACCAC |
| WT-F | CCCAAAGTCGCTCTGAGTTGTTA |
| WT-R | TCGGGTGAGCATGTCTTTAATCT |
| Primer | Sequences (5′ to 3′) |
|---|---|
| siGirdin-F | AAGAAGGCTTAGGCAGGAATT |
| siGirdin-R | AATTCCTGCCTAAGCCTTCTT |
| sgGiridn#1-F | CACCGGTCATGAACTGCTCCAGAA |
| sgGiridn#1-R | AAACTTCTGGAGCAGTTCATGACC |
| sgGiridn#2-F | CACCGCTAAAGCCACATACTCATCC |
| sgGiridn#2-R | AAACGGATGAGTATGTGGCTTTAGC |
| Characteristic | Total | Girdin-Low (%) | Girdin-High (%) | p Value (χ2 Test) |
|---|---|---|---|---|
| Number | 57 | 28 (49.1) | 29 (50.9) | |
| Age (y) | ||||
| <55 | 27 | 12 (21.1) | 15 (26.3) | 0.5026 |
| ≥55 | 30 | 16 (28.1) | 14 (24.5) | |
| Gender | ||||
| Male | 32 | 13 (22.8) | 19 (33.3) | 0.1465 |
| Female | 25 | 15 (26.3) | 10 (17.6) | |
| Smoking | ||||
| Yes | 20 | 7 (12.3) | 13 (22.8) | 0.1168 |
| No | 37 | 21 (36.8) | 16 (28.1) | |
| Clinical stage | ||||
| I | 29 | 19 (33.3) | 10 (17.6) | 0.0118 |
| II~IV | 28 | 9 (15.8) | 19 (33.3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, F.; Yang, D.; Tang, F.; Lu, C.; He, X.; Chen, S.; Yang, Z.; Gong, S.; Sun, L.; Enomoto, A.; et al. Girdin Promotes Tumorigenesis and Chemoresistance in Lung Adenocarcinoma by Interacting with PKM2. Cancers 2022, 14, 5688. https://doi.org/10.3390/cancers14225688
Cao F, Yang D, Tang F, Lu C, He X, Chen S, Yang Z, Gong S, Sun L, Enomoto A, et al. Girdin Promotes Tumorigenesis and Chemoresistance in Lung Adenocarcinoma by Interacting with PKM2. Cancers. 2022; 14(22):5688. https://doi.org/10.3390/cancers14225688
Chicago/Turabian StyleCao, Fuyang, Desong Yang, Feiyu Tang, Can Lu, Xiang He, Songming Chen, Zhanghuan Yang, Siyuan Gong, Lunquan Sun, Atsushi Enomoto, and et al. 2022. "Girdin Promotes Tumorigenesis and Chemoresistance in Lung Adenocarcinoma by Interacting with PKM2" Cancers 14, no. 22: 5688. https://doi.org/10.3390/cancers14225688
APA StyleCao, F., Yang, D., Tang, F., Lu, C., He, X., Chen, S., Yang, Z., Gong, S., Sun, L., Enomoto, A., Takahashi, M., & Weng, L. (2022). Girdin Promotes Tumorigenesis and Chemoresistance in Lung Adenocarcinoma by Interacting with PKM2. Cancers, 14(22), 5688. https://doi.org/10.3390/cancers14225688








