Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomic Analysis
2.2. CDCA5 Immunohistochemistry
2.3. Immunohistochemical Analysis
2.4. Evaluation of the Functional Activity of CDCA5 in BC Cell Lines
2.5. Transient (siRNA) Knockdowns (KD) of CDCA5
2.6. Phenotypic and Mechanistic Characterisation of CDCA5 Depletion
2.7. Statistical Analysis
3. Results
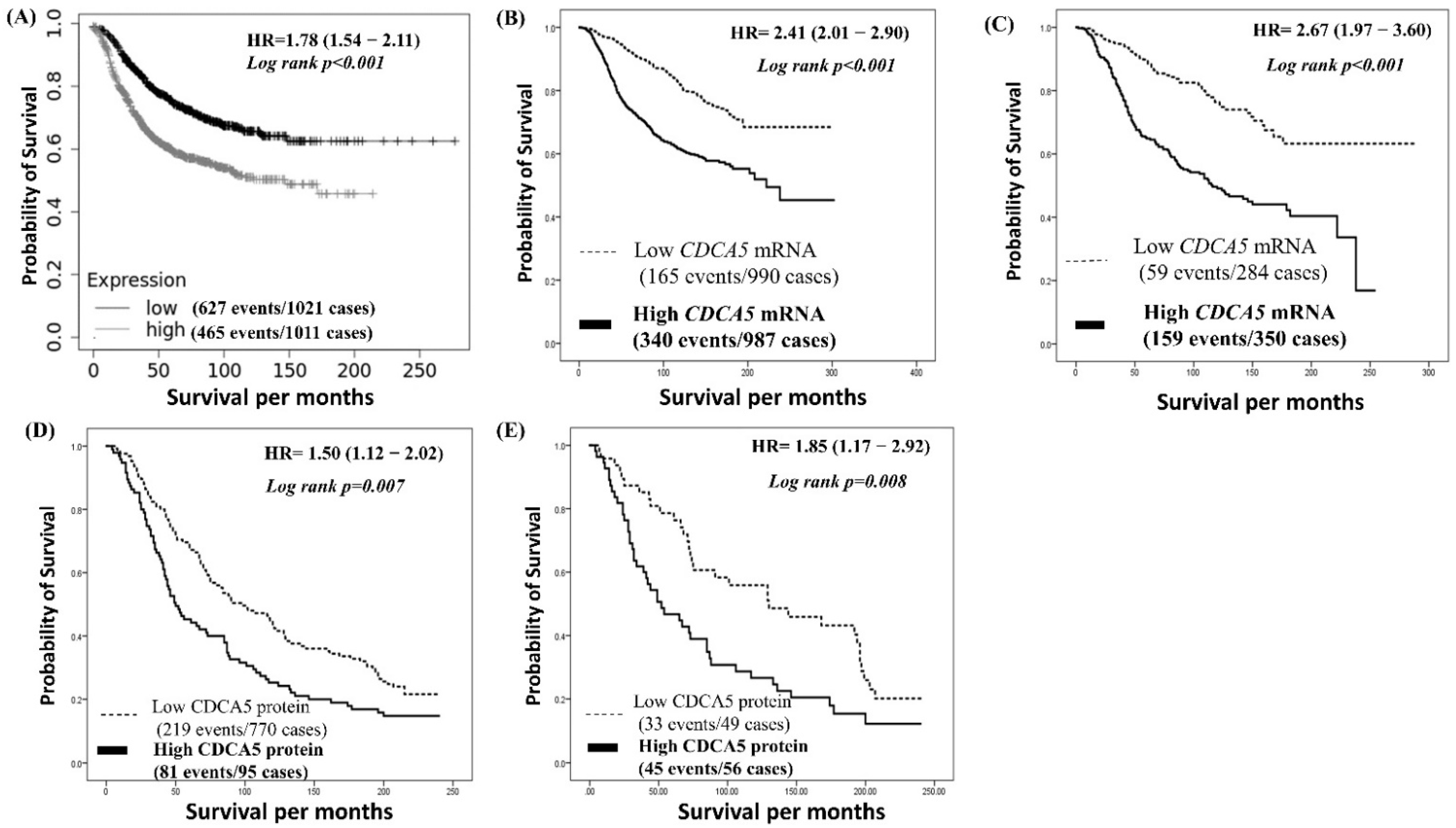
3.1. Significance of CDCA5 mRNA Expression in BC
3.2. CDCA5 Protein Expression
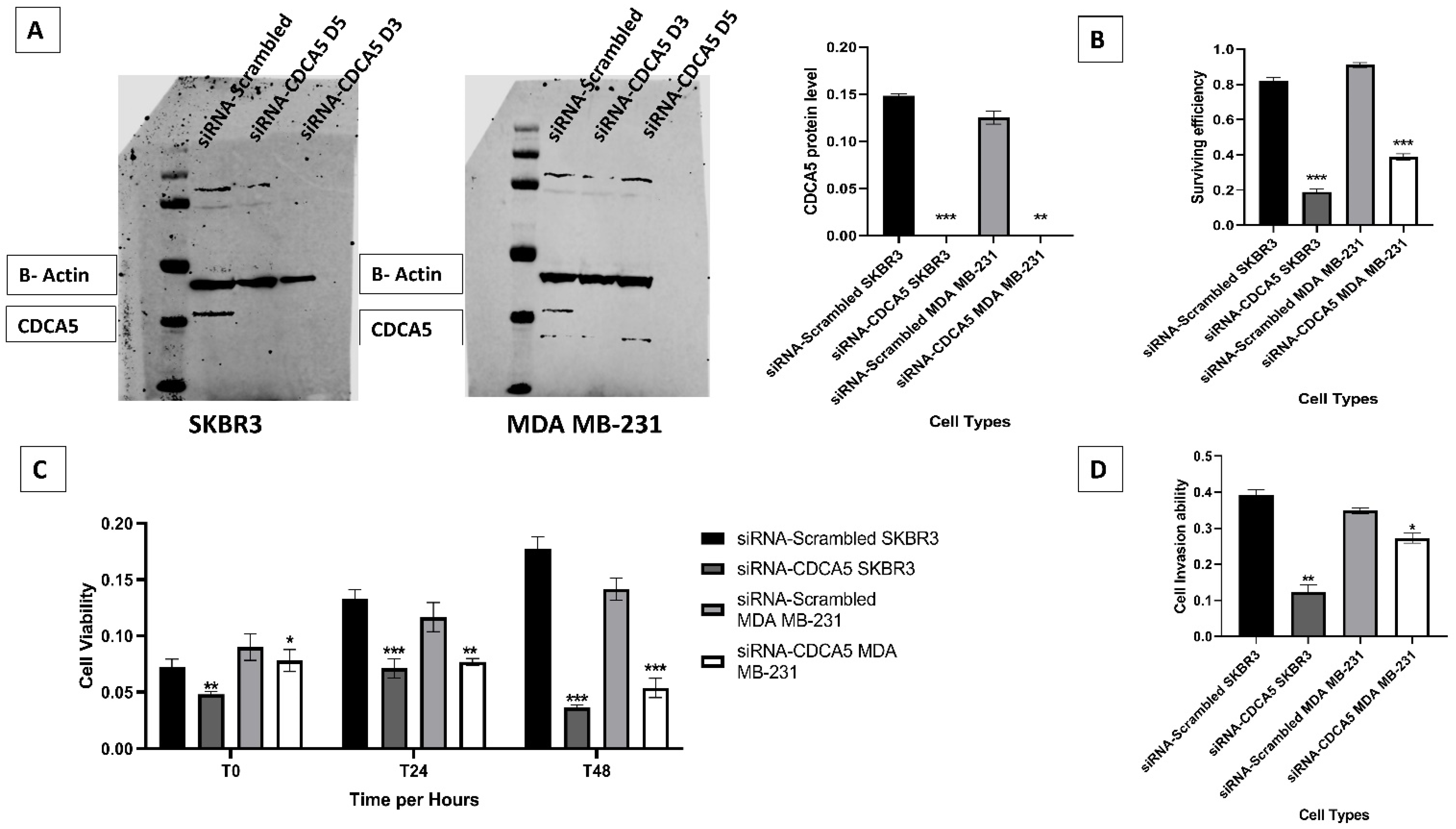
3.3. In Vitro Investigation of CDCA5
3.4. CDCA5 Promotes Cell Survival Efficiency, Proliferation and Invasion Ability
3.5. CDCA5 Increases Cell Migration and Cell Cycle Ability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aleskandarany, M.A.; Sonbul, S.N.; Mukherjee, A.; Rakha, E.A. Molecular Mechanisms Underlying Lymphovascular Invasion in Invasive Breast Cancer. Pathobiology 2015, 82, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Martin, S.; Lee, A.H.S.; Morgan, D.; Pharoah, P.D.P.; Hodi, Z.; Macmillan, D.; Ellis, I.O. The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer 2012, 118, 3670–3680. [Google Scholar] [CrossRef] [PubMed]
- Kariri, Y.A.; Aleskandarany, M.A.; Joseph, C.; Kurozumi, S.; Mohammed, O.J.; Toss, M.S.; Green, A.R.; Rakha, E.A. Molecular Complexity of Lymphovascular Invasion: The Role of Cell Migration in Breast Cancer as a Prototype. Pathobiology 2020, 87, 218–231. [Google Scholar] [CrossRef]
- Vijai, J.; Kirchhoff, T.; Schrader, K.A.; Brown, J.; Dutra-Clarke, A.V.; Manschreck, C.; Hansen, N.; Rau-Murthy, R.; Sarrel, K.; Przybylo, J.; et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet. 2013, 9, e1003220. [Google Scholar] [CrossRef] [PubMed]
- Johanneson, B.; Deutsch, K.; McIntosh, L.; Friedrichsen-Karyadi, D.M.; Janer, M.; Kwon, E.M.; Iwasaki, L.; Hood, L.; Ostrander, E.A.; Stanford, J.L. Suggestive genetic linkage to chromosome 11p11.2-q12.2 in hereditary prostate cancer families with primary kidney cancer. Prostate 2007, 67, 732–742. [Google Scholar] [CrossRef]
- Nishiyama, T.; Ladurner, R.; Schmitz, J.; Kreidl, E.; Schleiffer, A.; Bhaskara, V.; Bando, M.; Shirahige, K.; Hyman, A.A.; Mechtler, K.; et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 2010, 143, 737–749. [Google Scholar] [CrossRef]
- Hu, H.; Xiang, Y.; Zhang, X.Y.; Deng, Y.; Wan, F.J.; Huang, Y.; Liao, X.H.; Zhang, T.C. CDCA5 promotes the progression of breast cancer and serves as a potential prognostic biomarker. Oncol. Rep. 2022, 48, 1–11. [Google Scholar] [CrossRef]
- Borton, M.T.; Rashid, M.S.; Dreier, M.R.; Taylor, W.R. Multiple Levels of Regulation of Sororin by Cdk1 and Aurora B. J. Cell. Biochem. 2016, 117, 351–360. [Google Scholar] [CrossRef]
- Schmitz, J.; Watrin, E.; Lenart, P.; Mechtler, K.; Peters, J.M. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 2007, 17, 630–636. [Google Scholar] [CrossRef]
- Chen, T.; Huang, Z.; Tian, Y.; Wang, H.; Ouyang, P.; Chen, H.; Wu, L.; Lin, B.; He, R. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol. Rep. 2017, 38, 1822–1832. [Google Scholar] [CrossRef]
- Fu, G.; Xu, Z.; Chen, X.; Pan, H.; Wang, Y.; Jin, B. CDCA5 functions as a tumor promoter in bladder cancer by dysregulating mitochondria-mediated apoptosis, cell cycle regulation and PI3k/AKT/mTOR pathway activation. J. Cancer 2020, 11, 2408–2420. [Google Scholar] [CrossRef]
- Chen, H.; Chen, J.; Zhao, L.; Song, W.; Xuan, Z.; Chen, J.; Li, Z.; Song, G.; Hong, L.; Song, P.; et al. CDCA5, Transcribed by E2F1, Promotes Oncogenesis by Enhancing Cell Proliferation and Inhibiting Apoptosis via the AKT Pathway in Hepatocellular Carcinoma. J. Cancer 2019, 10, 1846–1854. [Google Scholar] [CrossRef]
- Shen, A.; Liu, L.; Chen, H.; Qi, F.; Huang, Y.; Lin, J.; Sferra, T.J.; Sankararaman, S.; Wei, L.; Chu, J.; et al. Cell division cycle associated 5 promotes colorectal cancer progression by activating the ERK signaling pathway. Oncogenesis 2019, 8, 19. [Google Scholar] [CrossRef]
- Tokuzen, N.; Nakashiro, K.; Tanaka, H.; Iwamoto, K.; Hamakawa, H. Therapeutic potential of targeting cell division cycle associated 5 for oral squamous cell carcinoma. Oncotarget 2016, 7, 2343–2353. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, S.; Joseph, C.; Sonbul, S.; Alsaeed, S.; Kariri, Y.; Aljohani, A.; Raafat, S.; Alsaleem, M.; Ogden, A.; Johnston, S.J.; et al. A key genomic subtype associated with lymphovascular invasion in invasive breast cancer. Br. J. Cancer 2019, 120, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. BioRxiv 2020, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Abd El-Rehim, D.M.; Ball, G.; Pinder, S.E.; Rakha, E.; Paish, C.; Robertson, J.F.; Macmillan, D.; Blamey, R.W.; Ellis, I.O. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer 2005, 116, 340–350. [Google Scholar] [CrossRef]
- McCarty, K.S., Jr.; McCarty, K.S., Sr. Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1984, 1, 297–308. [Google Scholar] [PubMed]
- Kariri, Y.A.; Joseph, C.; Kurozumi, S.; Toss, M.S.; Alsaleem, M.; Raafat, S.; Mongan, N.P.; Aleskandarany, M.A.; Green, A.R.; Rakha, E.A. Prognostic significance of KN motif and ankyrin repeat domains 1 (KANK1) in invasive breast cancer. Breast Cancer Res. Treat. 2019, 179, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Muftah, A.A.; Aleskandarany, M.A.; Al-Kaabi, M.M.; Sonbul, S.N.; Diez-Rodriguez, M.; Nolan, C.C.; Caldas, C.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Ki67 expression in invasive breast cancer: The use of tissue microarrays compared with whole tissue sections. Breast Cancer Res. Treat. 2017, 164, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Rolland, P.; Spendlove, I.; Madjd, Z.; Rakha, E.A.; Patel, P.; Ellis, I.O.; Durrant, L. The p53 positive Bcl-2 negative phenotype is an independent marker of prognosis in breast cancer. Int. J. Cancer 2007, 120, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Abd El Rehim, D.; Pinder, S.E.; Lewis, S.A.; Ellis, I.O. E-cadherin expression in invasive non-lobular carcinoma of the breast and its prognostic significance. Histopathology 2005, 46, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Kariri, Y.A.; Alsaleem, M.; Joseph, C.; Alsaeed, S.; Aljohani, A.; Shiino, S.; Mohammed, O.J.; Toss, M.S.; Green, A.R.; Rakha, E.A. The prognostic significance of interferon-stimulated gene 15 (ISG15) in invasive breast cancer. Breast Cancer Res. Treat. 2020, 185, 293–305. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Taube, S.E.; McShane, L.M.; Cavenagh, M.M.; Altman, D.G. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. J. Natl. Cancer Inst. 2018, 110, 803–811. [Google Scholar] [CrossRef]
- Wenzel, E.S.; Singh, A.T.K. Cell-cycle Checkpoints and Aneuploidy on the Path to Cancer. In Vivo 2018, 32, 1–5. [Google Scholar]
- Phan, N.N.; Wang, C.Y.; Li, K.L.; Chen, C.F.; Chiao, C.C.; Yu, H.G.; Huang, P.L.; Lin, Y.C. Distinct expression of CDCA3, CDCA5, and CDCA8 leads to shorter relapse free survival in breast cancer patient. Oncotarget 2018, 9, 6977–6992. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, C.; Yu, Y.; Wu, W.; Cao, J.; Li, Z.; Dai, J.; Wang, C.; Tang, Y.; Zhu, Q.; et al. Systematic cancer-testis gene expression analysis identified CDCA5 as a potential therapeutic target in esophageal squamous cell carcinoma. EBioMedicine 2019, 46, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wu, J.; Chagas, C.; Du, Y.; Lyu, H.; He, Y.; Qi, S.; Peng, Y.; Hu, J. CDCA5 overexpression is an Indicator of poor prognosis in patients with hepatocellular carcinoma (HCC). BMC Cancer 2018, 18, 1187. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Dobashi, Y.; Minehara, H.; Itoman, M.; Kameya, T. Involvement of cyclins in cell proliferation and their clinical implications in soft tissue smooth muscle tumors. Am. J. Pathol. 2000, 156, 2135–2147. [Google Scholar] [CrossRef]
- Karakas, C.; Biernacka, A.; Bui, T.; Sahin, A.A.; Yi, M.; Akli, S.; Schafer, J.; Alexander, A.; Adjapong, O.; Hunt, K.K.; et al. Cytoplasmic Cyclin E and Phospho-Cyclin-Dependent Kinase 2 Are Biomarkers of Aggressive Breast Cancer. Am. J. Pathol. 2016, 186, 1900–1912. [Google Scholar] [CrossRef] [PubMed]
- Luhtala, S.; Staff, S.; Tanner, M.; Isola, J. Cyclin E amplification, over-expression, and relapse-free survival in HER-2-positive primary breast cancer. Tumour Biol. 2016, 37, 9813–9823. [Google Scholar] [CrossRef]
- Zhang, Z.; Shen, M.; Zhou, G. Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Biochem. Biophys. Res. Commun. 2018, 496, 482–489. [Google Scholar] [CrossRef]
- Alsaleem, M.; Toss, M.S.; Joseph, C.; Aleskandarany, M.; Kurozumi, S.; Alshankyty, I.; Ogden, A.; Rida, P.C.G.; Ellis, I.O.; Aneja, R.; et al. The molecular mechanisms underlying reduced E-cadherin expression in invasive ductal carcinoma of the breast: High throughput analysis of large cohorts. Mod. Pathol. 2019, 32, 967–976. [Google Scholar] [CrossRef]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef]
- Gilles, C.; Polette, M.; Mestdagt, M.; Nawrocki-Raby, B.; Ruggeri, P.; Birembaut, P.; Foidart, J.M. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 2003, 63, 2658–2664. [Google Scholar] [CrossRef]
- Schade, B.; Lesurf, R.; Sanguin-Gendreau, V.; Bui, T.; Deblois, G.; O’Toole, S.A.; Millar, E.K.; Zardawi, S.J.; Lopez-Knowles, E.; Sutherland, R.L.; et al. β-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013, 73, 4474–4487. [Google Scholar] [CrossRef]
- Falchook, G.S.; Moulder, S.L.; Wheler, J.J.; Jiang, Y.; Bastida, C.C.; Kurzrock, R. Dual HER2 inhibition in combination with anti-VEGF treatment is active in heavily pretreated HER2-positive breast cancer. Ann. Oncol. 2013, 24, 3004–3011. [Google Scholar] [CrossRef] [PubMed]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, D.; Smid, M.; Timmermans, A.M.; Sleijfer, S.; Martens, J.W.M.; Debets, R. Breast cancer genomics and immuno-oncological markers to guide immune therapies. Semin. Cancer Biol. 2018, 52, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, M.; Friedrich, K.; Wielockx, B.; Kuzmanov, A.; Kettelhake, A.; Labelle, M.; Schnittler, H.; Baretton, G.; Breier, G. Interplay between neural-cadherin and vascular endothelial-cadherin in breast cancer progression. Breast Cancer Res. 2012, 14, R154. [Google Scholar] [CrossRef]
- Ji, J.; Shen, T.; Li, Y.; Liu, Y.; Shang, Z.; Niu, Y. CDCA5 promotes the progression of prostate cancer by affecting the ERK signalling pathway. Oncol. Rep. 2021, 45, 921–932. [Google Scholar] [CrossRef]


| Parameters | METABRIC Cohort | ||
|---|---|---|---|
| Low CDCA5 | High CDCA5 | p Value | |
| N (%) | N (%) | ||
| Tumor size | |||
| ≤2.0cm | 482 (49) | 501 (51) | <0.001 |
| >2.0cm | 377 (39) | 600 (61) | |
| Nodal Status | |||
| Negative | 582 (56) | 404 (44) | <0.001 |
| Positive | 453 (43) | 534 (57) | |
| Histological Grade | |||
| Grade 1and 2 | 675 (72) | 265 (28) | <0.001 |
| Grade 3 | 265 (27) | 696 (73) | |
| Tumor Histological subtypes | |||
| Ductal NST | 691 (45) | 853 (55) | <0.001 |
| Lobular | 27 (84) | 5 (16) | |
| Medullary-like | 155 (76) | 48 (24) | |
| Special type | 105 (71) | 42 (29) | |
| Lymphovascular Invasion | |||
| Negative | 494 (64) | 284 (36) | <0.001 |
| Positive | 436 (55) | 351 (45) | |
| Estrogen receptor | |||
| Negative | 88 (9) | 902 (91) | <0.001 |
| Positive | 386 (39) | 604 (61) | |
| Progesterone receptor | |||
| Negative | 310 (31) | 680 (69) | <0.001 |
| Positive | 630 (64) | 360 (36) | |
| Human epidermal growth factor receptor 2 | |||
| Negative | 107 (95) | 140 (5) | <0.001 |
| Positive | 883 (80) | 850 (20) | |
| EGFR | |||
| Negative | 515 (52) | 475 (48%) | 0.080 |
| Positive | 475 (48) | 515 (52%) | |
| Gene Names | METABRIC Cohort | |
|---|---|---|
| Correlation Value | p Value | |
| Adhesion molecule genes | ||
| CDH1 | −0.100 | <0.001 |
| CDH2 | 0.163 | <0.001 |
| Proliferation gene | ||
| MKi-67 | 0.689 | <0.001 |
| MMPs related genes | ||
| MMP7 | 0.180 | <0.001 |
| MMP9 | 0.324 | <0.001 |
| MMP11 | 0.097 | <0.001 |
| MMP12 | 0.354 | <0.001 |
| MMP14 | 0.086 | <0.001 |
| MMP15 | 0.230 | <0.001 |
| MMP20 | 0.145 | <0.001 |
| MMP25 | 0.138 | <0.001 |
| PI3K/AKT/mTOR pathway genes | ||
| PIK3CD | 0.173 | <0.001 |
| AKT1 | 0.052 | 0.022 |
| MTOR | 0.125 | <0.001 |
| Apoptosis gens | ||
| BAX | 0.270 | <0.001 |
| MYC | 0.173 | <0.001 |
| Cyclin related genes | ||
| CDKN2A | 0.396 | <0.001 |
| CCNA1 | 0.236 | <0.001 |
| CCNA2 | 0.838 | <0.001 |
| CCNB1 | 0.614 | <0.001 |
| CCNB2 | 0.883 | <0.001 |
| CCND3 | 0.096 | <0.001 |
| CCNE1 | 0.680 | <0.001 |
| CCNE2 | 0.671 | <0.001 |
| CCNT1 | 0.128 | <0.001 |
| CDK1 | 0.726 | <0.001 |
| CDK2 | 0.572 | <0.001 |
| CDK4 | 0.442 | <0.001 |
| CDK5 | 0.191 | <0.001 |
| CDK6 | 0.221 | <0.001 |
| Factors | BCSS in METABRIC Cohort | BCSS in Nottingham BC Cohort | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p Value | Hazard Ratio | 95% CI | p Value | |
| CDCA5 | 1.70 | 1.30–2.22 | <0.001 | 1.42 | 1.01–2.01 | 0.044 |
| Tumor size | 1.52 | 1.19–1.95 | <0.001 | 1.51 | 1.03–2.20 | 0.034 |
| Tumor grade | 1.06 | 0.81–1.40 | 0.669 | 1.73 | 1.19–2.51 | 0.004 |
| Tumor Stage | 2.11 | 1.54–2.90 | <0.001 | 1.60 | 1.26–2.03 | <0.001 |
| LVI | 1.84 | 1.46–2.33 | <0.001 | 1.37 | 0.97–1.96 | 0.078 |
| ER | 0.89 | 0.67–1.20 | 0.440 | 0.83 | 0.51–1.35 | 0.453 |
| PR | 0.77 | 0.58–1.01 | 0.055 | 0.62 | 0.34–0.97 | 0.038 |
| HER2 status | 1.57 | 1.17–2.10 | 0.002 | 1.18 | 0.76–1.83 | 0.469 |
| Parameters | CDCA5 Protein Expression | ||
|---|---|---|---|
| Low N (%) | High N (%) | p Value | |
| Tumor size | |||
| ≤2.0cm | 172 (58) | 126 (42) | 0.060 |
| >2.0cm | 158 (50) | 157 (50) | |
| Nodal Status | |||
| Negative | 189 (53) | 165 (47) | 0.005 |
| Positive | 123 (48) | 133 (52) | |
| Histological Grade | |||
| Grade 1 | 49 (68) | 23 (32) | <0.001 |
| Grade 2 | 128 (64) | 73 (36) | |
| Grade 3 | 154 (45) | 187 (55) | |
| Tumor Histological Subtypes | |||
| Ductal NST | 131 (46) | 85 (33) | <0.001 |
| Lobular | 90 (30) | 40 (16) | |
| Medullary | 33 (12) | 80 (31) | |
| Special type | 33 (12) | 51 (20) | |
| Lymphovascular invasion | |||
| Negative | 196 (57) | 146 (43) | 0.004 |
| Positive | 84 (44) | 106 (56) | |
| Nottingham prognostic index | |||
| Good prognostic group | 102(65) | 55(35) | 0.005 |
| Moderate prognostic group | 167(50) | 166(50) | |
| Poor prognostic group | 59(50) | 60(50) | |
| Age | |||
| <50 | 116 (49) | 121 (51) | 0.049 |
| >50 | 212 (57) | 160 (43) | |
| Estrogen Receptor | |||
| Negative | 51 (31) | 115 (69) | 0.040 |
| Positive | 276 (62) | 168 (38) | |
| Progesterone Receptor | |||
| Negative | 105 (41) | 152 (59) | 0.001 |
| Positive | 214(63) | 128 (37) | |
| Human epidermal growth factor receptor 2 | |||
| Negative | 287 (56) | 223 (44) | 0.004 |
| Positive | 33 (39) | 51 (61) | |
| P53 | |||
| Negative | 235 (58) | 170 (42) | 0.001 |
| Positive | 80 (42) | 111 (58) | |
| Ki67 | |||
| Negative | 114(63) | 67(37) | 0.001 |
| Positive | 153(47) | 171(53) | |
| Epidermal growth factor receptor (EGFR) | |||
| Negative | 264 (56) | 208 (44) | 0.040 |
| Positive | 59 (46) | 70 (54) | |
| E-cadherin | |||
| Negative | 199 (51) | 193 (49) | 0.033 |
| Positive | 122(60) | 82 (40) | |
| N-Cadherin | |||
| Negative | 69 (55) | 56 (45) | 0.684 |
| Positive | 151 (39) | 238 (61) | |
| Basal phenotype | |||
| Negative | 361 (79) | 96 (21) | 0.010 |
| Positive | 246 (71) | 102 (29) | |
| Cyclin E | |||
| Negative | 81 (62) | 50 (38) | <0.001 |
| Positive | 9 (26) | 25 (74) | |
| Phosphoinositide 3-kinase | |||
| Negative | 56 (59) | 46 (41) | 0.215 |
| Positive | 184 (52) | 171 (48) | |
| IHC-Subtypes | |||
| Luminal A | 131 (62) | 80 (38) | 0.001 |
| Luminal B | 90 (62) | 40 (31) | |
| Her2 enriched | 33 (28) | 103 (72) | |
| TNBC | 28 (39) | 51 (61) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kariri, Y.A.; Joseph, C.; Alsaleem, M.A.; Elsharawy, K.A.; Alsaeed, S.; Toss, M.S.; Mongan, N.P.; Green, A.R.; Rakha, E.A. Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer. Cancers 2022, 14, 5643. https://doi.org/10.3390/cancers14225643
Kariri YA, Joseph C, Alsaleem MA, Elsharawy KA, Alsaeed S, Toss MS, Mongan NP, Green AR, Rakha EA. Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer. Cancers. 2022; 14(22):5643. https://doi.org/10.3390/cancers14225643
Chicago/Turabian StyleKariri, Yousif A., Chitra Joseph, Mansour A. Alsaleem, Khloud A. Elsharawy, Sami Alsaeed, Michael S. Toss, Nigel P. Mongan, Andrew R. Green, and Emad A. Rakha. 2022. "Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer" Cancers 14, no. 22: 5643. https://doi.org/10.3390/cancers14225643
APA StyleKariri, Y. A., Joseph, C., Alsaleem, M. A., Elsharawy, K. A., Alsaeed, S., Toss, M. S., Mongan, N. P., Green, A. R., & Rakha, E. A. (2022). Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer. Cancers, 14(22), 5643. https://doi.org/10.3390/cancers14225643








