SOX4 Mediates ATRA-Induced Differentiation in Neuroblastoma Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture and Treatment
2.3. Microarray Data and Data Analysis
2.4. Cell Transfection
2.5. Quantitative RT-PCR
2.6. Western Blotting
2.7. Cell Survival Analysis
2.8. Immunocytochemistry Staining
2.9. Cell Cycle Analysis
2.10. Statistical Analyses
3. Results
3.1. The Differentially Expressed Genes Obtained from the Microarray Data Analysis
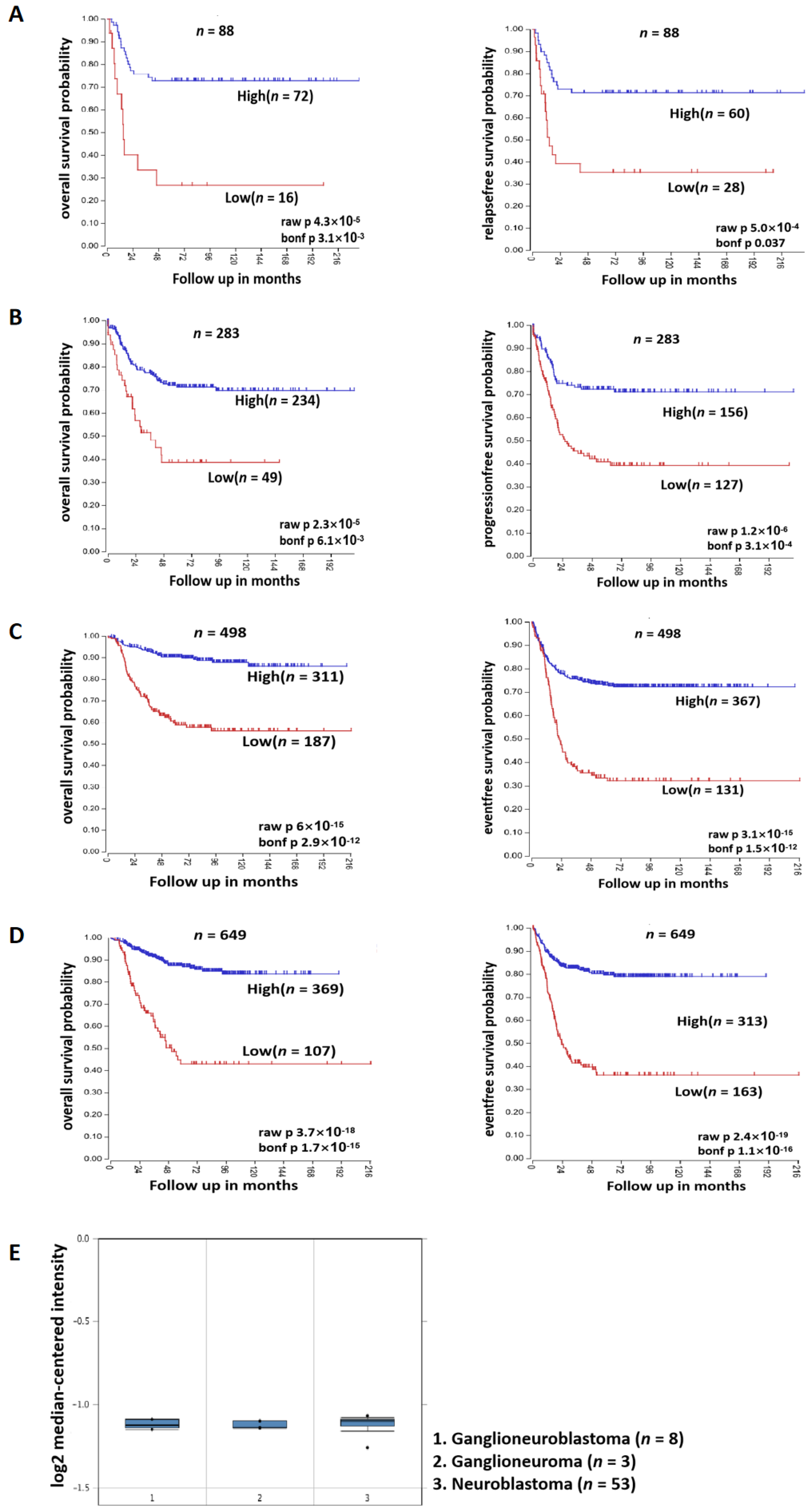
3.2. SOX4 Has a Positive Correlation with the Survival Rate of Patients with NB
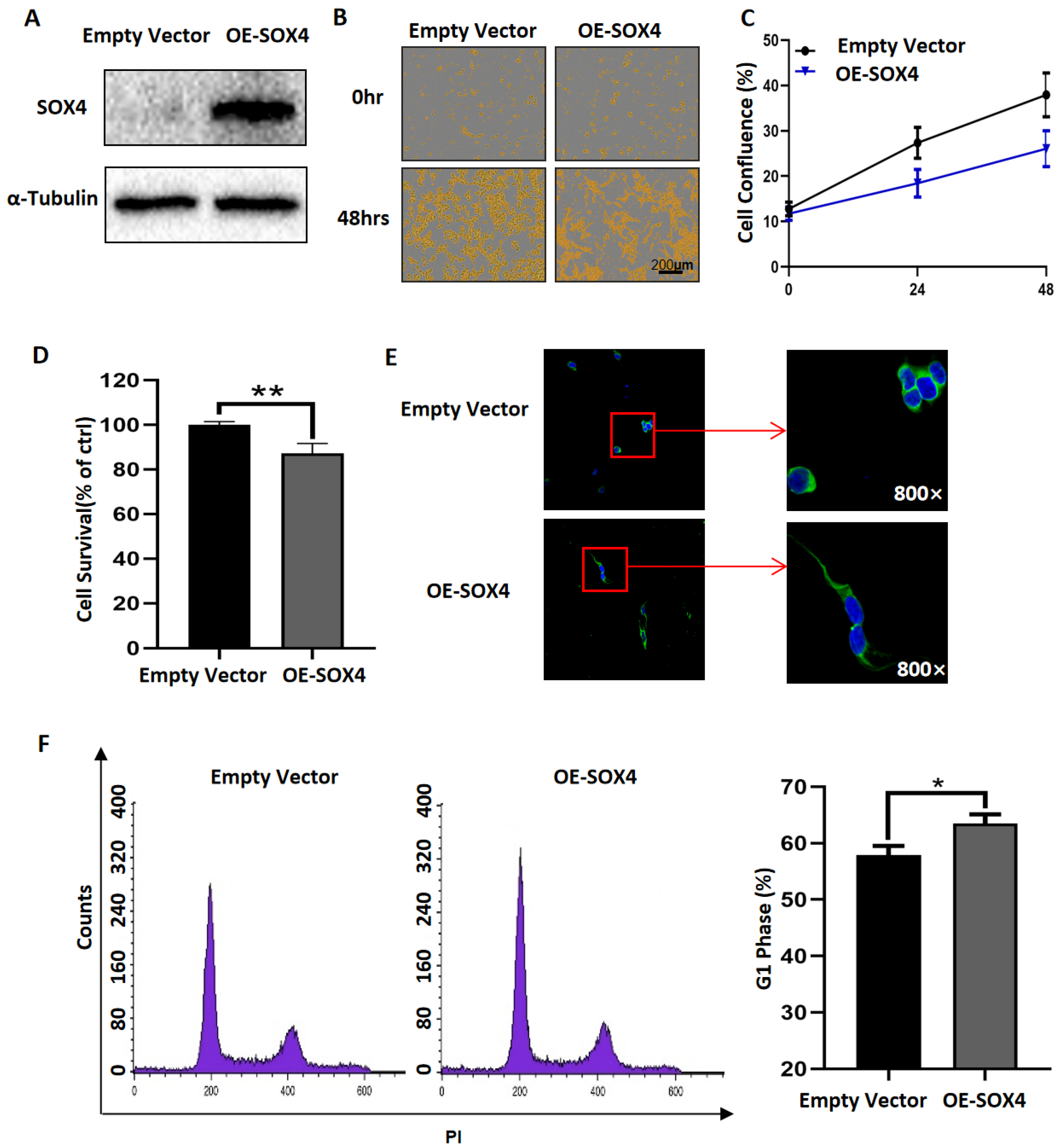
3.3. Overexpression of SOX4 Has the Potential to Induce the Differentiation of NB Cells
3.4. Downregulation of SOX4 Partially Blocks the Function of RA in NB Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Shohet, J.; Foster, J. Neuroblastoma. BMJ 2017, 357, j1863. [Google Scholar] [CrossRef] [PubMed]
- Tomolonis, J.A.; Agarwal, S.; Shohet, J.M. Neuroblastoma pathogenesis: Deregulation of embryonic neural crest development. Cell Tissue Res. 2018, 372, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Edsjo, A.; Holmquist, L.; Pahlman, S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin. Cancer Biol. 2007, 17, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Gheisari, S.; Catchpoole, D.R.; Charlton, A.; Kennedy, P.J. Convolutional Deep Belief Network with Feature Encoding for Classification of Neuroblastoma Histological Images. J. Pathol. Inform. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Bushue, N.; Wan, Y.J. Retinoid pathway and cancer therapeutics. Adv. Drug Deliv. Rev. 2010, 62, 1285–1298. [Google Scholar] [CrossRef]
- di Masi, A.; Leboffe, L.; De Marinis, E.; Pagano, F.; Cicconi, L.; Rochette-Egly, C.; Lo-Coco, F.; Ascenzi, P.; Nervi, C. Retinoic acid receptors: From molecular mechanisms to cancer therapy. Mol. Asp. Med. 2015, 41, 1–115. [Google Scholar] [CrossRef]
- Tang, X.H.; Gudas, L.J. Retinoids, retinoic acid receptors, and Cancer. Annu. Rev. Pathol. 2011, 6, 345–364. [Google Scholar] [CrossRef]
- Lammer, E.J.; Chen, D.T.; Hoar, R.M.; Agnish, N.D.; Benke, P.J.; Braun, J.T.; Curry, C.J.; Fernhoff, P.M.; Grix, A.W., Jr.; Lott, I.T.; et al. Retinoic acid embryopathy. N. Engl. J. Med. 1985, 313, 837–841. [Google Scholar] [CrossRef]
- Roenigk, H.H., Jr. Liver toxicity of retinoid therapy. Pharmacol. Ther. 1989, 40, 145–155. [Google Scholar] [CrossRef]
- Altucci, L.; Leibowitz, M.D.; Ogilvie, K.M.; de Lera, A.R.; Gronemeyer, H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007, 6, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Kagechika, H.; Shudo, K. Synthetic retinoids: Recent developments concerning structure and clinical utility. J. Med. Chem. 2005, 48, 5875–5883. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Bauer, J.; Wise, P.; Kruger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The role of SOX family members in solid tumours and metastasis. Semin. Cancer Biol. 2020, 67, 122–153. [Google Scholar] [CrossRef] [PubMed]
- Moreno, C.S. SOX4: The unappreciated oncogene. Semin. Cancer Biol. 2020, 67 Pt 1, 57–64. [Google Scholar] [CrossRef]
- Jang, S.M.; Kim, J.W.; Kim, C.H.; An, J.H.; Johnson, A.; Song, P.I.; Rhee, S.; Choi, K.H. KAT5-mediated SOX4 acetylation orchestrates chromatin remodeling during myoblast differentiation. Cell Death Dis. 2015, 6, e1857. [Google Scholar] [CrossRef]
- Inoue, H.; Takahashi, H.; Hashimura, M.; Eshima, K.; Akiya, M.; Matsumoto, T.; Saegusa, M. Cooperation of Sox4 with beta-catenin/p300 complex in transcriptional regulation of the Slug gene during divergent sarcomatous differentiation in uterine carcinosarcoma. BMC Cancer 2016, 16, 53. [Google Scholar] [CrossRef]
- Xu, E.E.; Krentz, N.A.; Tan, S.; Chow, S.Z.; Tang, M.; Nian, C.; Lynn, F.C. SOX4 cooperates with neurogenin 3 to regulate endocrine pancreas formation in mouse models. Diabetologia 2015, 58, 1013–1023. [Google Scholar] [CrossRef]
- Chen, C.; Lee, G.A.; Pourmorady, A.; Sock, E.; Donoghue, M.J. Orchestration of Neuronal Differentiation and Progenitor Pool Expansion in the Developing Cortex by SoxC Genes. J. Neurosci. 2015, 35, 10629–10642. [Google Scholar] [CrossRef]
- Liu, P.; Ramachandran, S.; Ali Seyed, M.; Scharer, C.D.; Laycock, N.; Dalton, W.B.; Williams, H.; Karanam, S.; Datta, M.W.; Jaye, D.L.; et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006, 66, 4011–4019. [Google Scholar] [CrossRef]
- Lee, H.; Goodarzi, H.; Tavazoie, S.F.; Alarcon, C.R. TMEM2 Is a SOX4-Regulated Gene That Mediates Metastatic Migration and Invasion in Breast Cancer. Cancer Res. 2016, 76, 4994–5005. [Google Scholar] [CrossRef]
- Tan, X.; Gong, W.; Chen, B.; Gong, B.; Hua, Z.; Zhang, S.; Chen, Y.; Li, Q.; Li, Z. Downregulation of fibronectin 1 attenuates ATRA-induced inhibition of cell migration and invasion in neuroblastoma cells. Mol. Cell. Biochem. 2021, 476, 3601–3612. [Google Scholar] [CrossRef] [PubMed]
- Frumm, S.M.; Fan, Z.P.; Ross, K.N.; Duvall, J.R.; Gupta, S.; VerPlank, L.; Suh, B.C.; Holson, E.; Wagner, F.F.; Smith, W.B.; et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem. Biol. 2013, 20, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Peltier, D.C.; Simms, A.; Farmer, J.R.; Miller, D.J. Human neuronal cells possess functional cytoplasmic and TLR-mediated innate immune pathways influenced by phosphatidylinositol-3 kinase signaling. J. Immunol. 2010, 184, 7010–7021. [Google Scholar] [CrossRef]
- Tsubota, S.; Kishida, S.; Shimamura, T.; Ohira, M.; Yamashita, S.; Cao, D.; Kiyonari, S.; Ushijima, T.; Kadomatsu, K. PRC2-Mediated Transcriptomic Alterations at the Embryonic Stage Govern Tumorigenesis and Clinical Outcome in MYCN-Driven Neuroblastoma. Cancer Res. 2017, 77, 5259–5271. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.G.; Barsky, L.W.; Davicioni, E.; Weinberg, K.I.; Triche, T.J.; Zhang, X.K.; Wu, L. Retinoic acid induces leukemia cell G1 arrest and transition into differentiation by inhibiting cyclin-dependent kinase-activating kinase binding and phosphorylation of PML/RARalpha. FASEB J. 2006, 20, 2142–2144. [Google Scholar] [CrossRef] [PubMed]
- Zarrilli, R.; Pignata, S.; Apicella, A.; Di Popolo, A.; Memoli, A.; Ricchi, P.; Salzano, S.; Acquaviva, A.M. Cell cycle block at G1-S or G2-M phase correlates with differentiation of Caco-2 cells: Effect of constitutive insulin-like growth factor II expression. Gastroenterology 1999, 116, 1358–1366. [Google Scholar] [CrossRef]
- Ponzoni, M.; Bachetti, T.; Corrias, M.V.; Brignole, C.; Pastorino, F.; Calarco, E.; Bensa, V.; Giusto, E.; Ceccherini, I.; Perri, P. Recent advances in the developmental origin of neuroblastoma: An overview. J. Exp. Clin. Cancer Res. 2022, 41, 92. [Google Scholar] [CrossRef]
- Hatori, Y.; Yan, Y.; Schmidt, K.; Furukawa, E.; Hasan, N.M.; Yang, N.; Liu, C.N.; Sockanathan, S.; Lutsenko, S. Neuronal differentiation is associated with a redox-regulated increase of copper flow to the secretory pathway. Nat. Commun. 2016, 7, 10640. [Google Scholar] [CrossRef]
- Chakraborty, K.; Kar, S.; Rai, B.; Bhagat, R.; Naskar, N.; Seth, P.; Gupta, A.; Bhattacharjee, A. Copper dependent ERK1/2 phosphorylation is essential for the viability of neurons and not glia. Metallomics 2022, 14, mfac005. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Mai, Y.; Yang, H.; Dong, P.Y.; Zheng, X.L.; Yang, G.S. CTSB promotes porcine preadipocytes differentiation by degrading fibronectin and attenuating the Wnt/beta-catenin signaling pathway. Mol. Cell. Biochem. 2014, 395, 53–64. [Google Scholar] [CrossRef]
- Peterson, S.; Bogenmann, E. The RET and TRKA pathways collaborate to regulate neuroblastoma differentiation. Oncogene 2004, 23, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Bohlken, A.; Kuljaca, S.; Lee, M.; Nguyen, T.; Smith, S.; Cheung, B.; Norris, M.D.; Haber, M.; Holloway, A.J.; et al. The retinoid anticancer signal: Mechanisms of target gene regulation. Br. J. Cancer 2005, 93, 310–318. [Google Scholar] [CrossRef]
- Stoney, P.N.; Fragoso, Y.D.; Saeed, R.B.; Ashton, A.; Goodman, T.; Simons, C.; Gomaa, M.S.; Sementilli, A.; Sementilli, L.; Ross, A.W.; et al. Expression of the retinoic acid catabolic enzyme CYP26B1 in the human brain to maintain signaling homeostasis. Brain Struct. Funct. 2016, 221, 3315–3326. [Google Scholar] [CrossRef]
- Backdahl, L.; Herberth, M.; Wilson, G.; Tate, P.; Campos, L.S.; Cortese, R.; Eckhardt, F.; Beck, S. Gene body methylation of the dimethylarginine dimethylamino-hydrolase 2 (Ddah2) gene is an epigenetic biomarker for neural stem cell differentiation. Epigenetics 2009, 4, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Goessler, U.R.; Bugert, P.; Bieback, K.; Deml, M.; Sadick, H.; Hormann, K.; Riedel, F. In-vitro analysis of the expression of TGFbeta -superfamily-members during chondrogenic differentiation of mesenchymal stem cells and chondrocytes during dedifferentiation in cell culture. Cell. Mol. Biol. Lett. 2005, 10, 345–362. [Google Scholar] [PubMed]
- Koli, K.; Ryynanen, M.J.; Keski-Oja, J. Latent TGF-beta binding proteins (LTBPs)-1 and -3 coordinate proliferation and osteogenic differentiation of human mesenchymal stem cells. Bone 2008, 43, 679–688. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, S.; Bai, M.; Zhang, A. The emerging role of MEIS1 in cell proliferation and differentiation. Am. J. Physiol. Cell Physiol. 2021, 320, C264–C269. [Google Scholar] [CrossRef]
- Kim, S.Y.; Volkl, S.; Ludwig, S.; Schneider, H.; Wixler, V.; Park, J. Deficiency of Fhl2 leads to delayed neuronal cell migration and premature astrocyte differentiation. J. Cell Sci. 2019, 132, jcs228940. [Google Scholar] [CrossRef]
- Lorda-Diez, C.I.; Montero, J.A.; Sanchez-Fernandez, C.; Garcia-Porrero, J.A.; Chimal-Monroy, J.; Hurle, J.M. Four and a half domain 2 (FHL2) scaffolding protein is a marker of connective tissues of developing digits and regulates fibrogenic differentiation of limb mesodermal progenitors. J. Tissue Eng. Regen. Med. 2018, 12, e2062–e2072. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Xia, H.H.; Gu, Q.; Lin, M.C.; Jiang, B.; Peng, Y.; Li, G.; An, X.; Zhang, Y.; et al. Suppression of FHL2 expression induces cell differentiation and inhibits gastric and colon carcinogenesis. Gastroenterology 2007, 132, 1066–1076. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Kimura, S.; Soga, A.; Sugiyama, M.; Ueno, A.; Kondo, H.; Zhu, Z.; Ochiai, K.; Nakayama, K.; Hakozaki, J.; et al. Plasmodium berghei Brca2 is required for normal development and differentiation in mice and mosquitoes. Parasites Vectors 2022, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Deneen, B.; Tzeng, S.F. BRCA1/BRCA2-containing complex subunit 3 controls oligodendrocyte differentiation by dynamically regulating lysine 63-linked ubiquitination. Glia 2019, 67, 1775–1792. [Google Scholar] [CrossRef] [PubMed]
- Rajan, J.V.; Wang, M.; Marquis, S.T.; Chodosh, L.A. Brca2 is coordinately regulated with Brca1 during proliferation and differentiation in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1996, 93, 13078–13083. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Su, Y.; Fassl, A.; Hinohara, K.; Qiu, X.; Harper, N.W.; Huh, S.J.; Bloushtain-Qimron, N.; Jovanovic, B.; Ekram, M.; et al. Perturbed myoepithelial cell differentiation in BRCA mutation carriers and in ductal carcinoma in situ. Nat. Commun. 2019, 10, 4182. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Shi, L.; Guo, Y.; Yu, L.; Wang, L.; Zhang, X.; Li, L.; Han, Y.; Ren, X.; Guo, Q.; et al. A novel PAD4/SOX4/PU.1 signaling pathway is involved in the committed differentiation of acute promyelocytic leukemia cells into granulocytic cells. Oncotarget 2016, 7, 3144–3157. [Google Scholar] [CrossRef]
- Yi, Y.; Zhu, H.; Klausen, C.; Leung, P.C.K. Transcription factor SOX4 facilitates BMP2-regulated gene expression during invasive trophoblast differentiation. FASEB J. 2021, 35, e22028. [Google Scholar] [CrossRef]
- Han, W.; Hu, P.; Wu, F.; Wang, S.; Hu, Y.; Li, S.; Jiang, T.; Qiang, B.; Peng, X. FHL3 links cell growth and self-renewal by modulating SOX4 in glioma. Cell Death Differ. 2019, 26, 796–811. [Google Scholar] [CrossRef]
- Chen, D.; Hu, C.; Wen, G.; Yang, Q.; Zhang, C.; Yang, H. DownRegulated SOX4 Expression Suppresses Cell Proliferation, Migration, and Induces Apoptosis in Osteosarcoma In Vitro and In Vivo. Calcif. Tissue Int. 2018, 102, 117–127. [Google Scholar] [CrossRef]

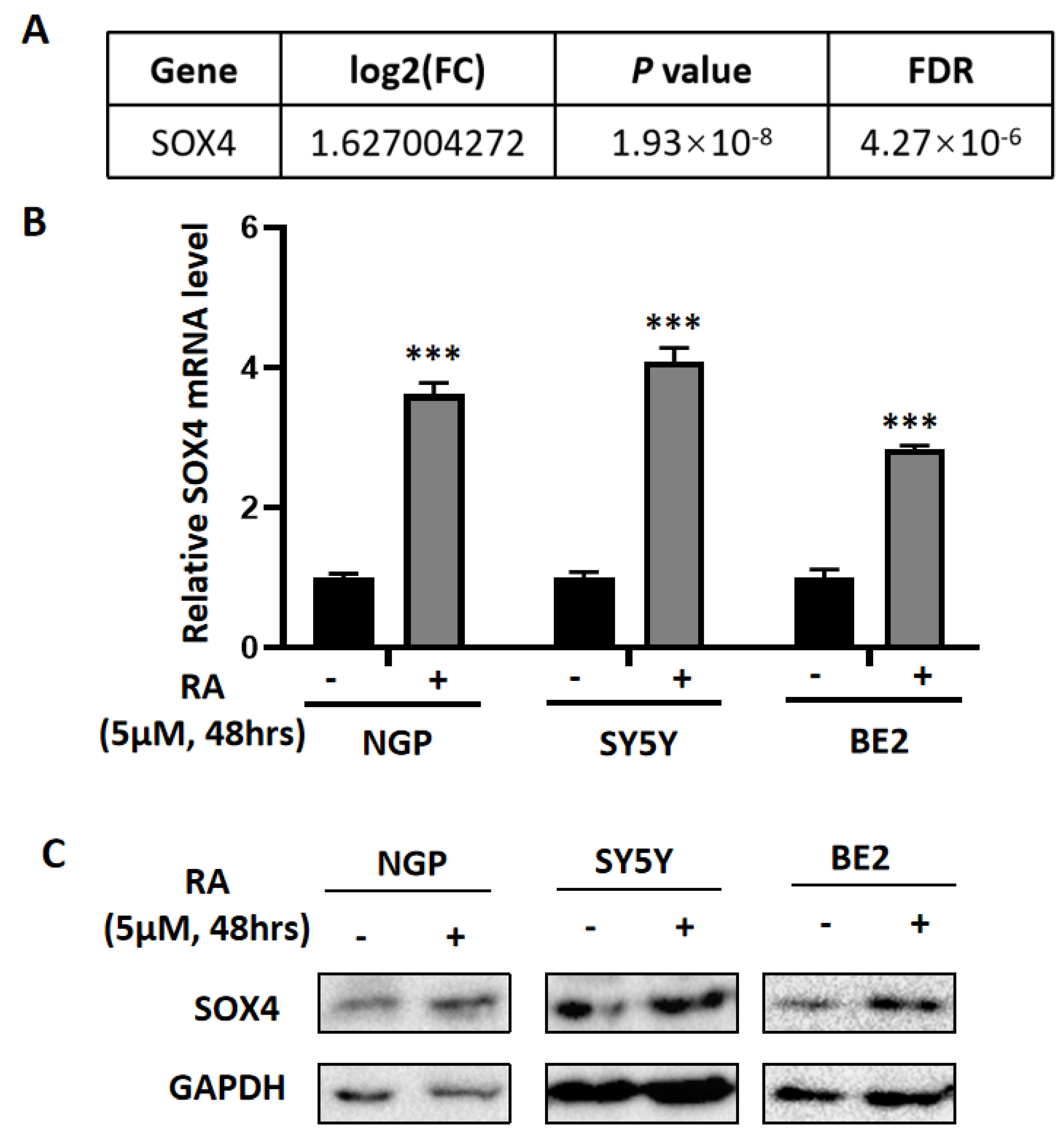

| Overlapping Upregulated-DEGs | |
|---|---|
| SOX4 | CYP26A1 |
| SOX9 | CYP26B1 |
| ADD3 | DDAH2 |
| ATP7A | LTBP3 |
| CAMK2N1 | MEIS1 |
| CTSB | NCOA3 |
| RET | |
| Overlapping Downregulated-DEGs |
|---|
| FHL2 |
| BRCA2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Gong, B.; Zhao, Q.; Li, Z.; Tan, X.; Hua, Z. SOX4 Mediates ATRA-Induced Differentiation in Neuroblastoma Cells. Cancers 2022, 14, 5642. https://doi.org/10.3390/cancers14225642
Zhang D, Gong B, Zhao Q, Li Z, Tan X, Hua Z. SOX4 Mediates ATRA-Induced Differentiation in Neuroblastoma Cells. Cancers. 2022; 14(22):5642. https://doi.org/10.3390/cancers14225642
Chicago/Turabian StyleZhang, Dongyang, Baocheng Gong, Qiang Zhao, Zhijie Li, Xiaolin Tan, and Zhongyan Hua. 2022. "SOX4 Mediates ATRA-Induced Differentiation in Neuroblastoma Cells" Cancers 14, no. 22: 5642. https://doi.org/10.3390/cancers14225642
APA StyleZhang, D., Gong, B., Zhao, Q., Li, Z., Tan, X., & Hua, Z. (2022). SOX4 Mediates ATRA-Induced Differentiation in Neuroblastoma Cells. Cancers, 14(22), 5642. https://doi.org/10.3390/cancers14225642






