Transcriptomic and Proteomic Profiles for Elucidating Cisplatin Resistance in Head-and-Neck Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Cell Culture
2.3. Transfection of the Small Interfering RNAs (siRNAs)
2.4. CDDP Assay
2.5. RNA Extraction
2.6. RNA-Sequencing (RNA-Seq)
2.7. Bioinformatics Workflow for the Differentially Expressed Gene Identification
2.8. Proteomic Analysis
2.9. qRT-PCR
2.10. Clinical Data Analysis (In Silico Study)
2.11. Statistical Analysis
3. Results
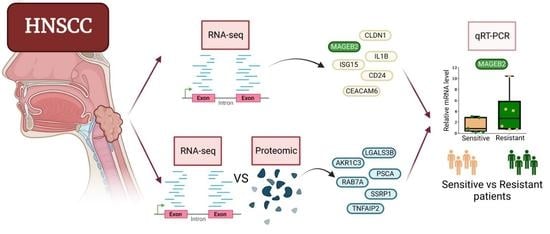
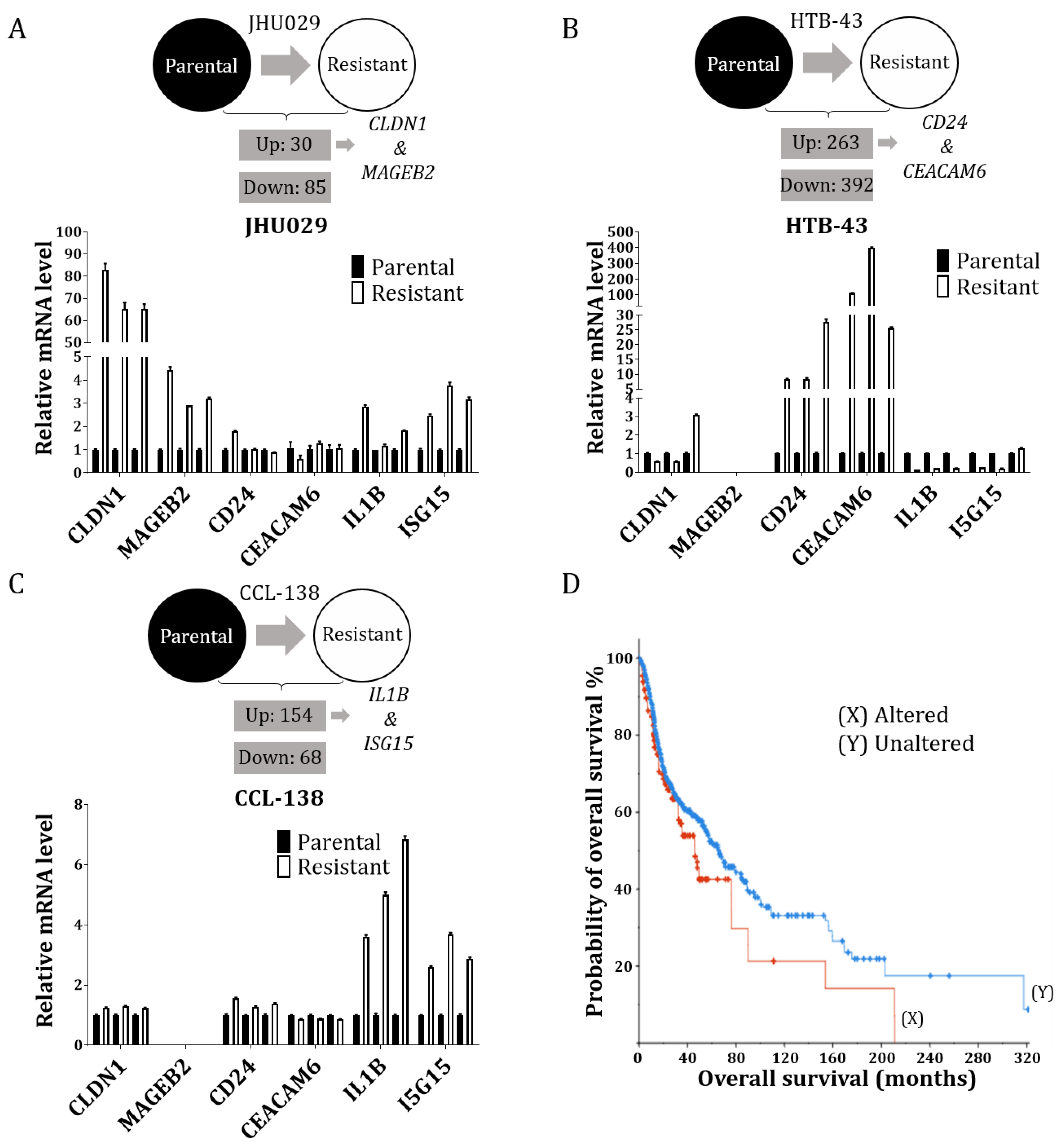
3.1. RNA-Seq Reveals Some Potential Targets of CDDP Resistance in the HNSCC Cell Lines
3.2. Proteomic Study in HTB-43 and CCL-138 Cell Lines in Relation to the RNA-Seq Results
3.3. Combinatorial Results of the Altered Genes and Proteins in the HTB-43 and CCL-138 Cell Lines
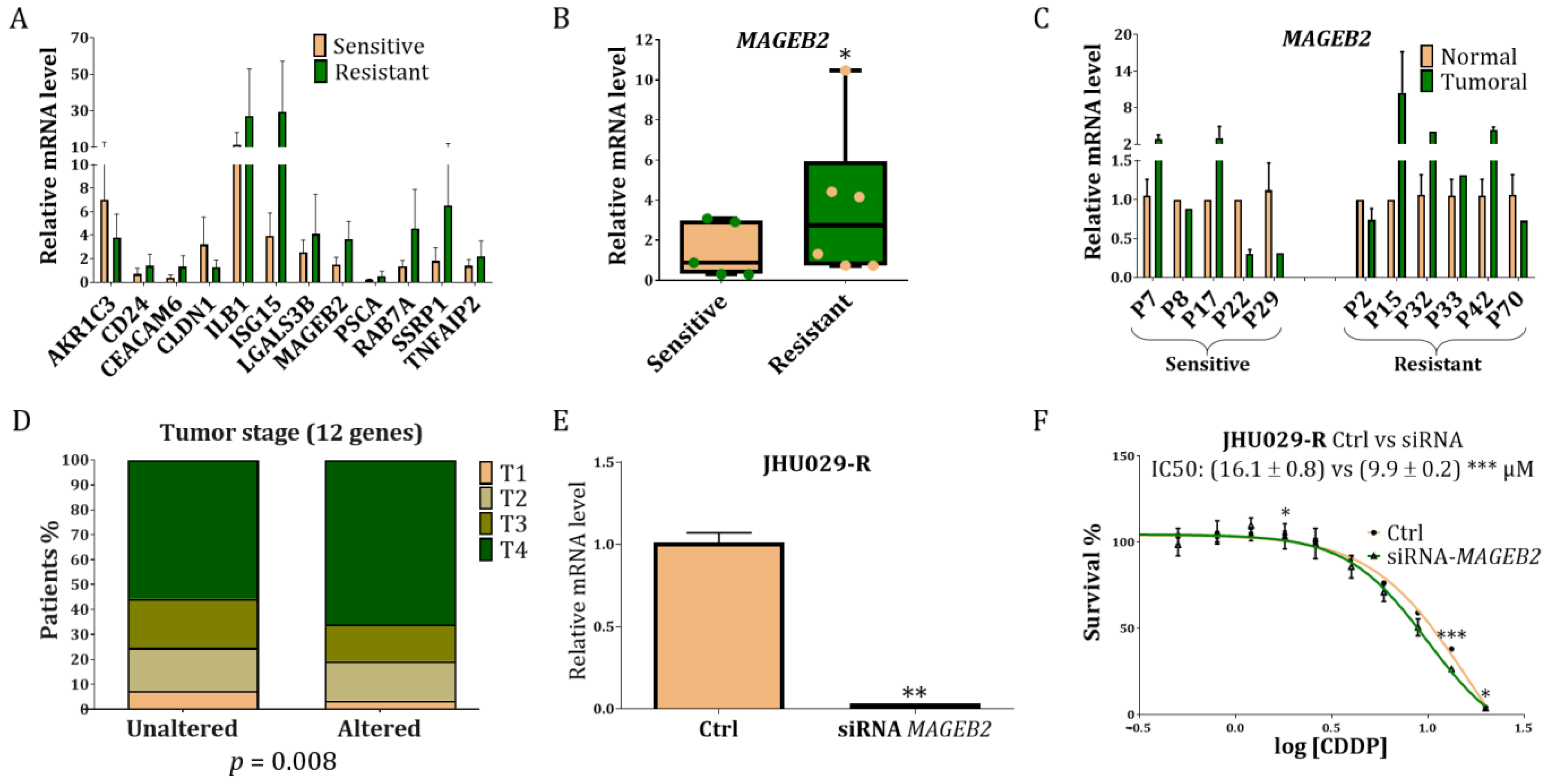
3.4. Study of the Genes Related to Resistance in Laryngeal Cancer Patients
3.5. Study of MAGEB2 in the Sensitisation of the Resistant Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Nenclares, P.; Rullan, A.; Tam, K.; Dunn, L.A.; St. John, M.; Harrington, K.J. Introducing Checkpoint Inhibitors Into the Curative Setting of Head and Neck Cancers: Lessons Learned, Future Considerations. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 511–526. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab Alone or with Chemotherapy versus Cetuximab with Chemotherapy for Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-048): A Randomised, Open-Label, Phase 3 Study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The Molecular Landscape of Head and Neck Cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Pi, C.; Jing, P.; Li, B.; Feng, Y.; Xu, L.; Xie, K.; Huang, T.; Xu, X.; Gu, H.; Fang, J. Reversing PD-1 Resistance in B16F10 Cells and Recovering Tumour Immunity Using a COX2 Inhibitor. Cancers 2022, 14, 4134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhou, Z.; Ju, W.; Liang, S.; Tang, X.; Zhu, D.; Zhang, Z.; Zhong, L. Mechanism of Sensitivity to Cisplatin, Docetaxel, and 5-fluorouracil Chemoagents and Potential ErbB2 Alternatives in Oral Cancer with Growth Differentiation Factor 15 Overexpression. Cancer Sci. 2022, 113, 478–488. [Google Scholar] [CrossRef]
- Obenauf, A.C.; Zou, Y.; Ji, A.L.; Vanharanta, S.; Shu, W.; Shi, H.; Kong, X.; Bosenberg, M.C.; Wiesner, T.; Rosen, N.; et al. Therapy-Induced Tumour Secretomes Promote Resistance and Tumour Progression. Nature 2015, 520, 368–372. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.-C.; Mansour, J.; Mollaee, M.; Wagner, K.-U.; Koduru, P.; Yopp, A.; et al. Whole-Exome Sequencing of Pancreatic Cancer Defines Genetic Diversity and Therapeutic Targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef]
- Docking, T.R.; Parker, J.D.K.; Jädersten, M.; Duns, G.; Chang, L.; Jiang, J.; Pilsworth, J.A.; Swanson, L.A.; Chan, S.K.; Chiu, R.; et al. A Clinical Transcriptome Approach to Patient Stratification and Therapy Selection in Acute Myeloid Leukemia. Nat. Commun. 2021, 12, 2474. [Google Scholar] [CrossRef]
- Kim, N.; Kim, H.K.; Lee, K.; Hong, Y.; Cho, J.H.; Choi, J.W.; Lee, J.-I.; Suh, Y.-L.; Ku, B.M.; Eum, H.H.; et al. Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma. Nat. Commun. 2020, 11, 2285. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mayea, Y.; Mir, C.; Muñoz, L.; Benavente, S.; Castellvi, J.; Temprana, J.; Maggio, V.; Lorente, J.; Paciucci, R.; LLeonart, M.E. Autophagy Inhibition as a Promising Therapeutic Target for Laryngeal Cancer. Carcinogenesis 2019, 40, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mayea, Y.; Mir, C.; Carballo, L.; Castellvi, J.; Temprana-Salvador, J.; Lorente, J.; Benavente, S.; García-Pedrero, J.M.; Allonca, E.; Rodrigo, J.P.; et al. TSPAN1: A Novel Protein Involved in Head and Neck Squamous Cell Carcinoma Chemoresistance. Cancers 2020, 12, 3269. [Google Scholar] [CrossRef] [PubMed]
- de Sena Brandine, G.; Smith, A.D. Falco: High-Speed FastQC Emulation for Quality Control of Sequencing Data. F1000Research 2021, 8, 1874. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Chang, J.W.; Seo, S.T.; Im, M.A.; Won, H.-R.; Liu, L.; Oh, C.; Jin, Y.L.; Piao, Y.; Kim, H.J.; Kim, J.T.; et al. Claudin-1 Mediates Progression by Regulating EMT through AMPK/TGF-β Signaling in Head and Neck Squamous Cell Carcinoma. Transl. Res. 2022, 247, 58–78. [Google Scholar] [CrossRef]
- Pattani, K.M.; Soudry, E.; Glazer, C.A.; Ochs, M.F.; Wang, H.; Schussel, J.; Sun, W.; Hennessey, P.; Mydlarz, W.; Loyo, M.; et al. MAGEB2 Is Activated by Promoter Demethylation in Head and Neck Squamous Cell Carcinoma. PLoS ONE 2012, 7, e45534. [Google Scholar] [CrossRef] [PubMed]
- Modur, V.; Joshi, P.; Nie, D.; Robbins, K.T.; Khan, A.U.; Rao, K. CD24 Expression May Play a Role as a Predictive Indicator and a Modulator of Cisplatin Treatment Response in Head and Neck Squamous Cellular Carcinoma. PLoS ONE 2016, 11, e0156651. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.; de Long, L.M.; Hazar-Rethinam, M.; Topkas, E.; Endo-Munoz, L.; Cumming, A.; Gannon, O.; Guminski, A.; Saunders, N. Focal Overexpression of CEACAM6 Contributes to Enhanced Tumourigenesis in Head and Neck Cancer via Suppression of Apoptosis. Mol. Cancer 2012, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Stanam, A.; Gibson-Corley, K.N.; Love-Homan, L.; Ihejirika, N.; Simons, A.L. Interleukin-1 Blockade Overcomes Erlotinib Resistance in Head and Neck Squamous Cell Carcinoma. Oncotarget 2016, 7, 76087–76100. [Google Scholar] [CrossRef]
- Govindaraj, P.K.; Kallarakkal, T.G.; Mohd Zain, R.; Tilakaratne, W.M.; Lew, H.L. Expression of Ki-67, Cornulin and ISG15 in Non-Involved Mucosal Surgical Margins as Predictive Markers for Relapse in Oral Squamous Cell Carcinoma (OSCC). PLoS ONE 2021, 16, e0261575. [Google Scholar] [CrossRef]
- Mir, C.; Garcia-Mayea, Y.; Garcia, L.; Herrero, P.; Canela, N.; Tabernero, R.; Lorente, J.; Castellvi, J.; Allonca, E.; García-Pedrero, J.; et al. SDCBP Modulates Stemness and Chemoresistance in Head and Neck Squamous Cell Carcinoma through Src Activation. Cancers 2021, 13, 4952. [Google Scholar] [CrossRef]
- Chang, W.-M.; Chang, Y.-C.; Yang, Y.-C.; Lin, S.-K.; Chang, P.M.-H.; Hsiao, M. AKR1C1 Controls Cisplatin-Resistance in Head and Neck Squamous Cell Carcinoma through Cross-Talk with the STAT1/3 Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 245. [Google Scholar] [CrossRef]
- Lu, X.; Wang, Z.; Huang, H.; Wang, H. Hedgehog Signaling Promotes Multidrug Resistance by Regulation of ABC Transporters in Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. 2020, 49, 897–906. [Google Scholar] [CrossRef]
- Sen, M.; Joyce, S.; Panahandeh, M.; Li, C.; Thomas, S.M.; Maxwell, J.; Wang, L.; Gooding, W.E.; Johnson, D.E.; Grandis, J.R. Targeting Stat3 Abrogates EGFR Inhibitor Resistance in Cancer. Clin. Cancer Res. 2012, 18, 4986–4996. [Google Scholar] [CrossRef]
- Yang, X.; Guo, Z.; Liu, Y.; Si, T.; Yu, H.; Li, B.; Tian, W. Prostate Stem Cell Antigen and Cancer Risk, Mechanisms and Therapeutic Implications. Expert Rev. Anticancer Ther. 2014, 14, 31–37. [Google Scholar] [CrossRef]
- Guerra, F.; Paiano, A.; Migoni, D.; Girolimetti, G.; Perrone, A.M.; De Iaco, P.; Fanizzi, F.P.; Gasparre, G.; Bucci, C. Modulation of RAB7A Protein Expression Determines Resistance to Cisplatin through Late Endocytic Pathway Impairment and Extracellular Vesicular Secretion. Cancers 2019, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Niwa, N.; Tanaka, N.; Hongo, H.; Miyazaki, Y.; Takamatsu, K.; Mizuno, R.; Kikuchi, E.; Mikami, S.; Kosaka, T.; Oya, M. TNFAIP2 Expression Induces Epithelial-to-Mesenchymal Transition and Confers Platinum Resistance in Urothelial Cancer Cells. Lab. Investig. 2019, 99, 1702–1713. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.A.N.; Biondini, M.; Curiel, R.; Siegel, P.M. Targeting GPNMB with Glembatumumab Vedotin: Current Developments and Future Opportunities for the Treatment of Cancer. Pharmacol. Ther. 2017, 179, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Yim, T.W.; Perling, D.; Polcz, M.; Komalavilas, P.; Brophy, C.; Cheung-Flynn, J. A Cell Permeant Phosphopeptide Mimetic of Niban Inhibits P38 MAPK and Restores Endothelial Function after Injury. FASEB J. 2020, 34, 9180–9191. [Google Scholar] [CrossRef]
- Ai, J.; Tan, G.; Li, W.; Liu, H.; Li, T.; Zhang, G.; Zhou, Z.; Gan, Y. Exosomes Loaded with CircPARD3 Promotes EBV-MiR-BART4-Induced Stemness and Cisplatin Resistance in Nasopharyngeal Carcinoma Side Population Cells through the MiR-579-3p/SIRT1/SSRP1 Axis. Cell Biol. Toxicol. 2022. [Google Scholar] [CrossRef]
- Zhao, J.; Ning, S.; Lou, W.; Yang, J.C.; Armstrong, C.M.; Lombard, A.P.; D’Abronzo, L.S.; Evans, C.P.; Gao, A.C.; Liu, C. Cross-Resistance Among Next-Generation Antiandrogen Drugs Through the AKR1C3/AR-V7 Axis in Advanced Prostate Cancer. Mol. Cancer Ther. 2020, 19, 1708–1718. [Google Scholar] [CrossRef]
- Zhang, W.; Pan, R.; Lu, M.; Zhang, Q.; Lin, Z.; Qin, Y.; Wang, Z.; Gong, S.; Lin, H.; Chong, S.; et al. Epigenetic Induction of Lipocalin 2 Expression Drives Acquired Resistance to 5-Fluorouracil in Colorectal Cancer through Integrin Β3/SRC Pathway. Oncogene 2021, 40, 6369–6380. [Google Scholar] [CrossRef]
- Pan, J.; Yu, H.; Guo, Z.; Liu, Q.; Ding, M.; Xu, K.; Mao, L. Emerging Role of Sperm-Associated Antigen 9 in Tumorigenesis. Biomed. Pharmacother. 2018, 103, 1212–1216. [Google Scholar] [CrossRef]
- Woo, J.K.; Jang, J.-E.; Kang, J.-H.; Seong, J.K.; Yoon, Y.S.; Kim, H.-C.; Lee, S.-J.; Oh, S.H. Lectin, Galactoside-Binding Soluble 3 Binding Protein Promotes 17-N-Allylamino-17-Demethoxygeldanamycin Resistance through PI3K/Akt Pathway in Lung Cancer Cell Line. Mol. Cancer Ther. 2017, 16, 1355–1365. [Google Scholar] [CrossRef]
- Jia, L.; Ge, X.; Du, C.; Chen, L.; Zhou, Y.; Xiong, W.; Xiang, J.; Li, G.; Xiao, G.; Fang, L.; et al. EEF1A2 Interacts with HSP90AB1 to Promote Lung Adenocarcinoma Metastasis via Enhancing TGF-β/SMAD Signalling. Br. J. Cancer 2021, 124, 1301–1311. [Google Scholar] [CrossRef]
- Smith, T.A.D.; AbdelKarem, O.A.; Irlam-Jones, J.J.; Lane, B.; Valentine, H.; Bibby, B.A.S.; Denley, H.; Choudhury, A.; West, C.M.L. Selection of Endogenous Control Genes for Normalising Gene Expression Data Derived from Formalin-Fixed Paraffin-Embedded Tumour Tissue. Sci. Rep. 2020, 10, 17258. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Noie Alamdari, A.; Noee Alamdari, Y.; Abak, A.; Hussen, B.M.; Taheri, M.; Jamali, E. Role of PI3K/AKT Pathway in Squamous Cell Carcinoma with an Especial Focus on Head and Neck Cancers. Cancer Cell Int. 2022, 22, 254. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.; Young, R.J.; Rischin, D. Head and Neck Squamous Cell Carcinoma: Genomics and Emerging Biomarkers for Immunomodulatory Cancer Treatments. Semin. Cancer Biol. 2018, 52, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Bonner, J.A.; Bredel, M. EGFR Mutations in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3818. [Google Scholar] [CrossRef] [PubMed]
- Boguszewicz, Ł. Predictive Biomarkers for Response and Toxicity of Induction Chemotherapy in Head and Neck Cancers. Front. Oncol. 2022, 12, 900903. [Google Scholar] [CrossRef] [PubMed]
- Schinke, H.; Shi, E.; Lin, Z.; Quadt, T.; Kranz, G.; Zhou, J.; Wang, H.; Hess, J.; Heuer, S.; Belka, C.; et al. A Transcriptomic Map of EGFR-Induced Epithelial-to-Mesenchymal Transition Identifies Prognostic and Therapeutic Targets for Head and Neck Cancer. Mol. Cancer 2022, 21, 178. [Google Scholar] [CrossRef]
- Huang, R.; Sun, H.; Lin, R.; Zhang, J.; Yin, H.; Xian, S.; Li, M.; Wang, S.; Li, Z.; Qiao, Y.; et al. The Role of Tetraspanins Pan-Cancer. iScience 2022, 25, 104777. [Google Scholar] [CrossRef]
- Weon, J.L.; Potts, P.R. The MAGE Protein Family and Cancer. Curr. Opin. Cell Biol. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Jin, M.L.; Yang, L.; Jeong, K.W. SETD1A-SOX2 Axis Is Involved in Tamoxifen Resistance in Estrogen Receptor α-Positive Breast Cancer Cells. Theranostics 2022, 12, 5761–5775. [Google Scholar] [CrossRef]
- Fletcher, J.I.; Williams, R.T.; Henderson, M.J.; Norris, M.D.; Haber, M. ABC Transporters as Mediators of Drug Resistance and Contributors to Cancer Cell Biology. Drug Resist. Updat. 2016, 26, 1–9. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer Metabolism: Looking Forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Abad, E.; Garcia-Mayea, Y.; Mir, C.; Sebastian, D.; Zorzano, A.; Potesil, D.; Zdrahal, Z.; Lyakhovich, A.; Lleonart, M.E. Common Metabolic Pathways Implicated in Resistance to Chemotherapy Point to a Key Mitochondrial Role in Breast Cancer. Mol. Cell. Proteom. 2019, 18, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Teixidó, C.; Giménez-Capitán, A.; Molina-Vila, M.Á.; Peg, V.; Karachaliou, N.; Rodríguez-Capote, A.; Castellví, J.; Rosell, R. RNA Analysis as a Tool to Determine Clinically Relevant Gene Fusions and Splice Variants. Arch. Pathol. Lab. Med. 2018, 142, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, F.; Yang, Y.; Yu, N.; Weng, X.; Yang, Y.; Gong, Z.; Huang, S.; Gan, L.; Sun, S.; et al. Integrated DNA and RNA Sequencing Reveals Early Drivers Involved in Metastasis of Gastric Cancer. Cell Death Dis. 2022, 13, 392. [Google Scholar] [CrossRef]
- Shukla, S.; Evans, J.R.; Malik, R.; Feng, F.Y.; Dhanasekaran, S.M.; Cao, X.; Chen, G.; Beer, D.G.; Jiang, H.; Chinnaiyan, A.M. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J. Natl. Cancer Inst. 2017, 109, djw200. [Google Scholar] [CrossRef]
- Nin, D.S.; Wujanto, C.; Tan, T.Z.; Lim, D.; Damen, J.M.A.; Wu, K.-Y.; Dai, Z.M.; Lee, Z.-W.; Idres, S.B.; Leong, Y.H.; et al. GAGE Mediates Radio Resistance in Cervical Cancers via the Regulation of Chromatin Accessibility. Cell Rep. 2021, 36, 109621. [Google Scholar] [CrossRef]
- Cui, J.; Chen, Y.; Ou, Y.; Liu, G.; Wen, Q.; Zhu, W.; Liang, L.; Chen, Z.; Yang, H.; Wang, L.; et al. Cancer Germline Antigen Gene MAGEB2 Promotes Cell Invasion and Correlates with Immune Microenvironment and Immunotherapeutic Efficiency in Laryngeal Cancer. Clin. Immunol. 2022, 240, 109045. [Google Scholar] [CrossRef]
- Gjerstorff, M.F.; Johansen, L.E.; Nielsen, O.; Kock, K.; Ditzel, H.J. Restriction of GAGE Protein Expression to Subpopulations of Cancer Cells Is Independent of Genotype and May Limit the Use of GAGE Proteins as Targets for Cancer Immunotherapy. Br. J. Cancer 2006, 94, 1864–1873. [Google Scholar] [CrossRef]
- Peche, L.Y.; Ladelfa, M.F.; Toledo, M.F.; Mano, M.; Laiseca, J.E.; Schneider, C.; Monte, M. Human MageB2 Protein Expression Enhances E2F Transcriptional Activity, Cell Proliferation, and Resistance to Ribotoxic Stress. J. Biol. Chem. 2015, 290, 29652–29662. [Google Scholar] [CrossRef]
- Ladelfa, M.F.; Peche, L.Y.; Amato, G.E.; Escalada, M.C.; Zampieri, S.; Pascucci, F.A.; Benevento, A.F.; Do Porto, D.F.; Dardis, A.; Schneider, C.; et al. Expression of the Tumor-Expressed Protein MageB2 Enhances RRNA Transcription. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 119015. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Mayea, Y.; Benítez-Álvarez, L.; Sánchez-García, A.; Bataller, M.; Companioni, O.; Mir, C.; Benavente, S.; Lorente, J.; Canela, N.; Fernández-Rozadilla, C.; et al. Transcriptomic and Proteomic Profiles for Elucidating Cisplatin Resistance in Head-and-Neck Squamous Cell Carcinoma. Cancers 2022, 14, 5511. https://doi.org/10.3390/cancers14225511
Garcia-Mayea Y, Benítez-Álvarez L, Sánchez-García A, Bataller M, Companioni O, Mir C, Benavente S, Lorente J, Canela N, Fernández-Rozadilla C, et al. Transcriptomic and Proteomic Profiles for Elucidating Cisplatin Resistance in Head-and-Neck Squamous Cell Carcinoma. Cancers. 2022; 14(22):5511. https://doi.org/10.3390/cancers14225511
Chicago/Turabian StyleGarcia-Mayea, Yoelsis, Lisandra Benítez-Álvarez, Almudena Sánchez-García, Marina Bataller, Osmel Companioni, Cristina Mir, Sergi Benavente, Juan Lorente, Nuria Canela, Ceres Fernández-Rozadilla, and et al. 2022. "Transcriptomic and Proteomic Profiles for Elucidating Cisplatin Resistance in Head-and-Neck Squamous Cell Carcinoma" Cancers 14, no. 22: 5511. https://doi.org/10.3390/cancers14225511
APA StyleGarcia-Mayea, Y., Benítez-Álvarez, L., Sánchez-García, A., Bataller, M., Companioni, O., Mir, C., Benavente, S., Lorente, J., Canela, N., Fernández-Rozadilla, C., Carracedo, A., & LLeonart, M. E. (2022). Transcriptomic and Proteomic Profiles for Elucidating Cisplatin Resistance in Head-and-Neck Squamous Cell Carcinoma. Cancers, 14(22), 5511. https://doi.org/10.3390/cancers14225511