Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
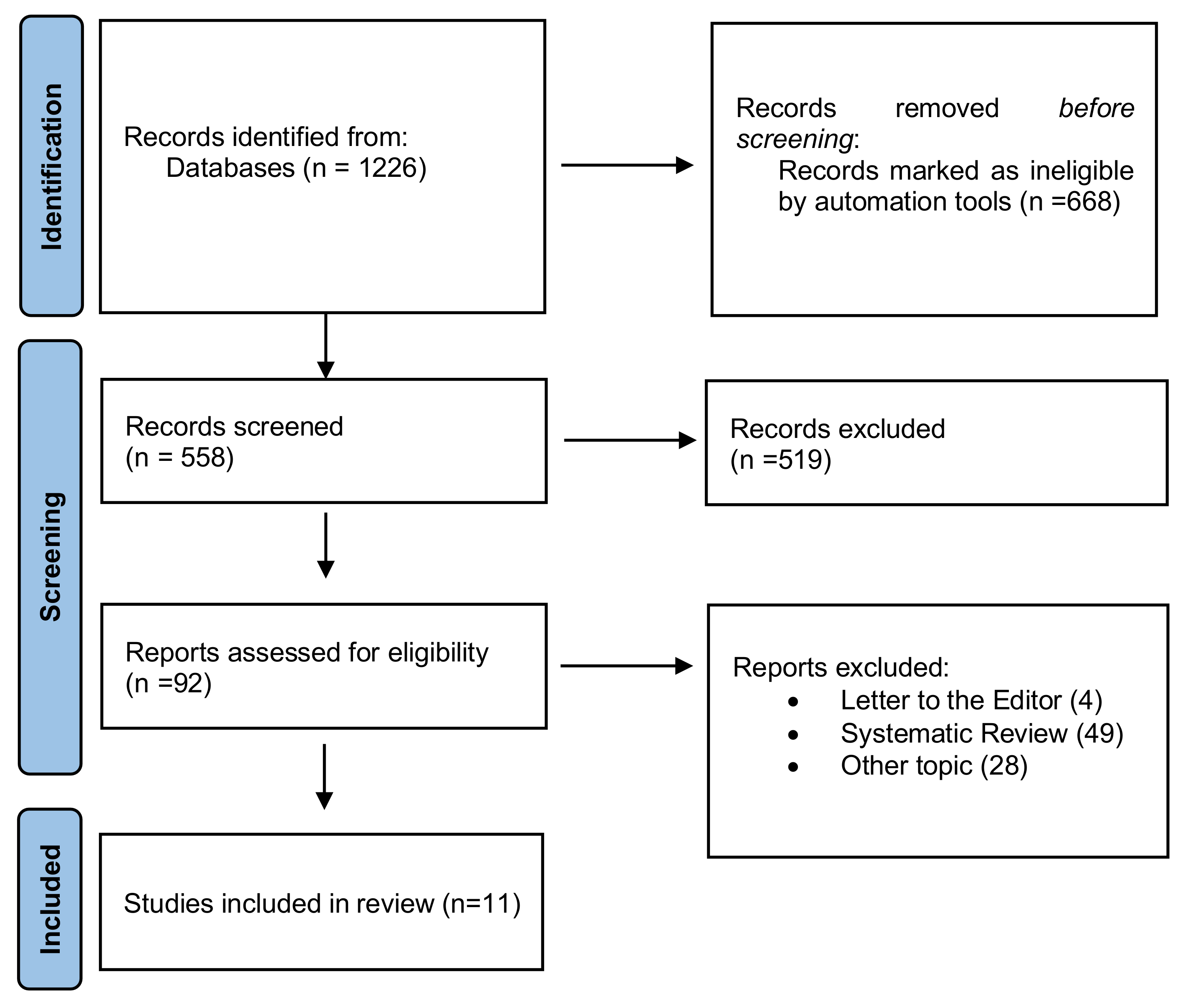
2. Materials and Methods
2.1. Protocol
2.2. PICo and Research Question
2.3. Data Sources and Search strategy
2.4. Eligibility Criteria
- -
- English language;
- -
- At least 30 patients for study;
- -
- Only diagnosis of OSCC histologically confirmed with a well-defined site classification;
- -
- Only saliva analysis;
- -
- Analysis of salivary microbiota in patients affected by OSCC, before therapy.
2.5. Study Selection and Data Collection Process
2.6. Statistical Analysis
- i.
- Study characteristics: name of the first author, year of publication, name of the country where the study was performed, study design.
- ii.
- Case-control groups characteristics: case population, age of case population, control population, age of control population, risk factors.
- iii.
- Methodology: sample collection, methods of sample collection, methods of DNA extraction, DNA amplification, sequencing platforms, reference database.
- iv.
- Outcomes: raw reds detected, microbial abundance, genera detected, species detected, phyla detected, and OTUs detected.
3. Results
3.1. Study Characteristics
3.2. Sample Collection
3.3. DNA Extraction and Amplification
3.4. DNA Sequencing and Analysis of Data
3.5. Microbial Abundance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S. Global Epidemiology of Oral and Oropharyngeal Cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Gissi, D.B.; Morandi, L.; Colella, G.; De Luca, R.; Campisi, G.; Mauceri, R.; Romeo, U.; Tenore, G.; Mignogna, M.D.; Adamo, D.; et al. Clinical Validation of 13-gene DNA Methylation Analysis in Oral Brushing Samples for Detection of Oral Carcinoma: Italian Multicenter Study. Head Neck 2021, 43, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Ong, T.K.; Murphy, C.; Smith, A.B.; Kanatas, A.N.; Mitchell, D.A. Survival after Surgery for Oral Cancer: A 30-Year Experience. Br. J. Oral Maxillofac. Surg. 2017, 55, 911–916. [Google Scholar] [CrossRef]
- Ram, H.; Sarkar, J.; Kumar, H.; Konwar, R.; Bhatt, M.L.B.; Mohammad, S. Oral Cancer: Risk Factors and Molecular Pathogenesis. J. Maxillofac. Oral Surg. 2011, 10, 132. [Google Scholar] [CrossRef]
- Cha, J.-D.; Kim, H.J.; Cha, I.-H. Genetic Alterations in Oral Squamous Cell Carcinoma Progression Detected by Combining Array-Based Comparative Genomic Hybridization and Multiplex Ligation-Dependent Probe Amplification. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011, 111, 594–607. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Pignatelli, P.; Romei, F.M.; Bondi, D.; Giuliani, M.; Piattelli, A.; Curia, M.C. Microbiota and Oral Cancer as A Complex and Dynamic Microenvironment: A Narrative Review from Etiology to Prognosis. Int. J. Mol. Sci. 2022, 23, 8323. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Ravel, J. The Vocabulary of Microbiome Research: A Proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.S.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in Abundance of Oral Microbiota Associated with Oral Cancer. PLoS ONE 2014, 9, e98741. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Ly, M.; Abeles, S.R.; Boehm, T.K.; Robles-Sikisaka, R.; Naidu, M.; Santiago-Rodriguez, T.; Pride, D.T. Altered Oral Viral Ecology in Association with Periodontal Disease. mBio 2014, 5, e01133-14. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, J. Role of Helicobacter Pylori in Gastric Cancer: Updates. World J. Gastrointest. Oncol. 2016, 8, 147. [Google Scholar] [CrossRef]
- Koshiol, J.; Wozniak, A.; Cook, P.; Adaniel, C.; Acevedo, J.; Azócar, L.; Hsing, A.W.; Roa, J.C.; Pasetti, M.F.; Miquel, J.F.; et al. Salmonella Enterica Serovar Typhi and Gallbladder Cancer: A Case–Control Study and Meta-analysis. Cancer Med. 2016, 5, 3310–3325. [Google Scholar] [CrossRef] [PubMed]
- Nosho, K. Association of Fusobacterium Nucleatum with Immunity and Molecular Alterations in Colorectal Cancer. World J. Gastroenterol. 2016, 22, 557. [Google Scholar] [CrossRef]
- Pignatelli, P.; Iezzi, L.; Pennese, M.; Raimondi, P.; Cichella, A.; Bondi, D.; Grande, R.; Cotellese, R.; Di Bartolomeo, N.; Innocenti, P.; et al. The Potential of Colonic Tumor Tissue Fusobacterium Nucleatum to Predict Staging and Its Interplay with Oral Abundance in Colon Cancer Patients. Cancers 2021, 13, 1032. [Google Scholar] [CrossRef]
- Karpiński, T. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 153303381986735. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The Oral Microbiome Diversity and Its Relation to Human Diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.B.K.; Sharma, A.; Babu, H.M. An Insight into Salivary Markers in Oral Cancer. Dent. Res. J. 2013, 10, 9. [Google Scholar]
- Cristaldi, M.; Mauceri, R.; Di Fede, O.; Giuliana, G.; Campisi, G.; Panzarella, V. Salivary Biomarkers for Oral Squamous Cell Carcinoma Diagnosis and Follow-Up: Current Status and Perspectives. Front. Physiol. 2019, 10, 1476. [Google Scholar] [CrossRef]
- Fontana, S.; Mauceri, R.; Novara, M.E.; Alessandro, R.; Campisi, G. Protein Cargo of Salivary Small Extracellular Vesicles as Potential Functional Signature of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 11160. [Google Scholar] [CrossRef] [PubMed]
- Panzarella, V.; Campisi, G.; Giardina, Y.; Maniscalco, L.; Capra, G.; Rodolico, V.; Di Fede, O.; Mauceri, R. Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review. Cancers 2021, 13, 4595. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, n71. [Google Scholar] [CrossRef] [PubMed]
- Ganly, I.; Yang, L.; Giese, R.A.; Hao, Y.; Nossa, C.W.; Morris, L.G.T.; Rosenthal, M.; Migliacci, J.; Kelly, D.; Tseng, W.; et al. Periodontal Pathogens Are a Risk Factor of Oral Cavity Squamous Cell Carcinoma, Independent of Tobacco and Alcohol and Human Papillomavirus: Periodontal Pathogens Are a Risk Factor for Oral Cancer. Int. J. Cancer 2019, 145, 775–784. [Google Scholar] [CrossRef]
- Zhou, X.; Hao, Y.; Peng, X.; Li, B.; Han, Q.; Ren, B.; Li, M.; Li, L.; Li, Y.; Cheng, G.; et al. The Clinical Potential of Oral Microbiota as a Screening Tool for Oral Squamous Cell Carcinomas. Front. Cell. Infect. Microbiol. 2021, 11, 728933. [Google Scholar] [CrossRef]
- Li, Y.; Tan, X.; Zhao, X.; Xu, Z.; Dai, W.; Duan, W.; Huang, S.; Zhang, E.; Liu, J.; Zhang, S.; et al. Composition and Function of Oral Microbiota between Gingival Squamous Cell Carcinoma and Periodontitis. Oral Oncol. 2020, 107, 104710. [Google Scholar] [CrossRef]
- Lee, W.-H.; Chen, H.-M.; Yang, S.-F.; Liang, C.; Peng, C.-Y.; Lin, F.-M.; Tsai, L.-L.; Wu, B.-C.; Hsin, C.-H.; Chuang, C.-Y.; et al. Bacterial Alterations in Salivary Microbiota and Their Association in Oral Cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef]
- Mohamed, N.; Litlekalsøy, J.; Ahmed, I.A.; Martinsen, E.M.H.; Furriol, J.; Javier-Lopez, R.; Elsheikh, M.; Gaafar, N.M.; Morgado, L.; Mundra, S.; et al. Analysis of Salivary Mycobiome in a Cohort of Oral Squamous Cell Carcinoma Patients From Sudan Identifies Higher Salivary Carriage of Malassezia as an Independent and Favorable Predictor of Overall Survival. Front. Cell. Infect. Microbiol. 2021, 11, 673465. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in Oral Microbiota Associated with Oral Cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Park, J.; Hosomi, K.; Yamada, T.; Kobayashi, A.; Yamaguchi, Y.; Iketani, S.; Kunisawa, J.; Mizuguchi, K.; Maeda, N.; et al. Analysis of Oral Microbiota in Japanese Oral Cancer Patients Using 16S RRNA Sequencing. J. Oral Biosci. 2019, 61, 120–128. [Google Scholar] [CrossRef]
- Hsiao, J.-R.; Chang, C.-C.; Lee, W.-T.; Huang, C.-C.; Ou, C.-Y.; Tsai, S.-T.; Chen, K.-C.; Huang, J.-S.; Wong, T.-Y.; Lai, Y.-H.; et al. The Interplay between Oral Microbiome, Lifestyle Factors and Genetic Polymorphisms in the Risk of Oral Squamous Cell Carcinoma. Carcinogenesis 2018, 39, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-F.; Huang, H.-D.; Fan, W.-L.; Jong, Y.-J.; Chen, M.-K.; Huang, C.-N.; Chuang, C.-Y.; Kuo, Y.-L.; Chung, W.-H.; Su, S.-C. Compositional and Functional Variations of Oral Microbiota Associated with the Mutational Changes in Oral Cancer. Oral Oncol. 2018, 77, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-W.; Wu, J.-H.; Chiang, W.-F.; Chen, Y.-L.; Wu, W.-S.; Wu, L.-W. Taxonomic and Functional Dysregulation in Salivary Microbiomes During Oral Carcinogenesis. Front. Cell. Infect. Microbiol. 2021, 11, 663068. [Google Scholar] [CrossRef] [PubMed]
- Su, S.-C.; Chang, L.-C.; Huang, H.-D.; Peng, C.-Y.; Chuang, C.-Y.; Chen, Y.-T.; Lu, M.-Y.; Chiu, Y.-W.; Chen, P.-Y.; Yang, S.-F. Oral Microbial Dysbiosis and Its Performance in Predicting Oral Cancer. Carcinogenesis 2021, 42, 127–135. [Google Scholar] [CrossRef]
- Yost, S.; Stashenko, P.; Choi, Y.; Kukuruzinska, M.; Genco, C.A.; Salama, A.; Weinberg, E.O.; Kramer, C.D.; Frias-Lopez, J. Increased Virulence of the Oral Microbiome in Oral Squamous Cell Carcinoma Revealed by Metatranscriptome Analyses. Int. J. Oral Sci. 2018, 10, 32. [Google Scholar] [CrossRef]
- Lim, Y.; Fukuma, N.; Totsika, M.; Kenny, L.; Morrison, M.; Punyadeera, C. The Performance of an Oral Microbiome Biomarker Panel in Predicting Oral Cavity and Oropharyngeal Cancers. Front. Cell. Infect. Microbiol. 2018, 8, 267. [Google Scholar] [CrossRef]
- Coletta, R.D.; Yeudall, W.A.; Salo, T. Grand Challenges in Oral Cancers. Front. Oral Health 2020, 1, 3. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Kruse, A.L.; Bredell, M.; Grätz, K.W. Oral Squamous Cell Carcinoma in Non-Smoking and Non-Drinking Patients. Head Neck Oncol. 2010, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yu, W.-H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A Web Accessible Resource for Investigating Oral Microbe Taxonomic and Genomic Information. Database 2010, 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of Oral Microbiota in Tumor and Non-Tumor Tissues of Patients with Oral Squamous Cell Carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd_Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Mascitti, M. Beyond Head and Neck Cancer: The Relationship Between Oral Microbiota and Tumour Development in Distant Organs. Front. Cell. Infect. Microbiol. 2019, 9, 8. [Google Scholar] [CrossRef]
- Perera, M.; Al-hebshi, N.N.; Speicher, D.J.; Perera, I.; Johnson, N.W. Emerging Role of Bacteria in Oral Carcinogenesis: A Review with Special Reference to Perio-Pathogenic Bacteria. J. Oral. Microbiol. 2016, 8, 10. [Google Scholar] [CrossRef]
- Voronov, E.; Shouval, D.S.; Krelin, Y.; Cagnano, E.; Benharroch, D.; Iwakura, Y.; Dinarello, C.A.; Apte, R.N. IL-1 Is Required for Tumor Invasiveness and Angiogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 2645–2650. [Google Scholar] [CrossRef]
- Carmi, Y.; Dotan, S.; Rider, P.; Kaplanov, I.; White, M.R.; Baron, R.; Abutbul, S.; Huszar, M.; Dinarello, C.A.; Apte, R.N.; et al. The Role of IL-1β in the Early Tumor Cell–Induced Angiogenic Response. J. Immunol. 2013, 190, 3500–3509. [Google Scholar] [CrossRef]
- Kossakowska, A.E.; Edwards, D.R.; Prusinkiewicz, C.; Zhang, M.C.; Guo, D.; Urbanski, S.J.; Grogan, T.; Marquez, L.A.; Janowska-Wieczorek, A. Interleukin-6 Regulation of Matrix Metalloproteinase (MMP-2 and MMP-9) and Tissue Inhibitor of Metalloproteinase (TIMP-1) Expression in Malignant Non-Hodgkin’s Lymphomas. Blood 1999, 94, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Thanan, R.; Ma, N.; Kawanishi, S. Role of Nitrative and Oxidative DNA Damage in Inflammation-Related Carcinogenesis. J. Biomed. Biotechnol. 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Sugita, H.; Kuboniwa, M.; Iwai, S.; Hamada, M.; Noda, T.; Morisaki, I.; Lamont, R.J.; Amano, A. Porphyromonas Gingivalis Promotes Invasion of Oral Squamous Cell Carcinoma through Induction of ProMMP9 and Its Activation: Promotion of Oral Cancer Invasion by P. Gingivalis. Cell. Microbiol. 2014, 16, 131–145. [Google Scholar] [CrossRef]
- Yilmaz, Ö.; Jungas, T.; Verbeke, P.; Ojcius, D.M. Activation of the Phosphatidylinositol 3-Kinase/Akt Pathway Contributes to Survival of Primary Epithelial Cells Infected with the Periodontal Pathogen Porphyromonas Gingivalis. Infect. Immun. 2004, 72, 3743–3751. [Google Scholar] [CrossRef] [PubMed]
- Uitto, V.-J.; Baillie, D.; Wu, Q.; Gendron, R.; Grenier, D.; Putnins, E.E.; Kanervo, A.; Firth, J.D. Fusobacterium Nucleatum Increases Collagenase 3 Production and Migration of Epithelial Cells. Infect. Immun. 2005, 73, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.J.; Chaudary, N.; Hill, R.P. The Tumor Microenvironment and Metastatic Disease. Clin. Exp. Metastasis 2009, 26, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Senneby, A.; Davies, J.; Svensäter, G.; Neilands, J. Acid Tolerance Properties of Dental Biofilms in Vivo. BMC Microbiol. 2017, 17, 165. [Google Scholar] [CrossRef]
- Pavlova, S.I.; Jin, L.; Gasparovich, S.R.; Tao, L. Multiple Alcohol Dehydrogenases but No Functional Acetaldehyde Dehydrogenase Causing Excessive Acetaldehyde Production from Ethanol by Oral Streptococci. Microbiology 2013, 159, 1437–1446. [Google Scholar] [CrossRef]
- Attene-Ramos, M.S.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. Evidence That Hydrogen Sulfide Is a Genotoxic Agent. Mol. Cancer Res. 2006, 4, 9–14. [Google Scholar] [CrossRef]
- Hafed, L.; Farag, H.; El-Rouby, D.; Shaker, O.; Shabaan, H.-A. Candida Albicans Alcohol Dehydrogenase 1 Gene in Oral Dysplasia and Oral Squamous Cell Carcinoma. Pol. J. Pathol. 2019, 70, 210–216. [Google Scholar] [CrossRef]
- Michela, B. Liquid Biopsy: A Family of Possible Diagnostic Tools. Diagnostics 2021, 11, 1391. [Google Scholar] [CrossRef] [PubMed]
| N. | Author, Year | Country | Study Design | N. of Case | N. of Control | Sample | Methods of DNA Extraction | DNA Amplification | Sequencing | Reference Database |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lee WH, 2017 [25] | Taiwan | Case-control | 125 | 251 | Sputum | QIAamp® DNA Blood Mini Kit | V4 | Illumina MiSeq System | SILVA |
| 2 | Zhao H, 2017 [26] | China | Case-control | 40 | 40 * | Oral swabs | QIAamp® DNA Blood Mini Kit | V4–V5 | Illumina MiSeq System | RDP |
| 3 | Hsiao JR, 2018 [24] | Taiwan | Case-control | 138 | 151 | Sputum | QIAamp® MinElute Virus Spin Kit | V3–V5 | Illumina MiSeq System | RDP |
| 4 | Yang SF, 2018 [27] | Taiwan | Cohort study | 39 | No control group | Sputum | QIAamp® DNA Blood Mini Kit | V4 | Illumina MiSeq System | Greengenes |
| 5 | Mohamed N, 2019 [28] | Sudan | Case-control | 59 | 13 | Sputum | FastDNA™ Kit | fungal ITS2 region | Illumina MiSeq System | UNITE |
| 6 | Takahashi Y, 2019 [29] | Japan | Case-control | 60 | 80 | Sputum | Gene Prep Star PI-80X device | V3–V4 | Illumina MiSeq System | SILVA 128 |
| 7 | Li Y, 2020 [30] | China | Case-control | 10 | 30 | Saliva, subgingival plaque, tumour and healthy surface | QIAampFast DNA Stool Mini Kit | V3–V4 | Illumina MiSeq System | SILVA |
| 8 | Chen JW, 2021 [31] | Taiwan | Cohort study | 27 | 48 | Sputum | QIAamp® DNA Blood Mini Kit | V3–V4 | Illumina MiSeq System | HOMD |
| 9 | Ganly I, 2021 [32] | USA | Case-control | 18 | 20 | Ora rinse | Modified QIAGEN® DNA extraction method | V3–V4 | 454 FLX platform | Greengenes |
| 10 | Su SC, 2021 [33] | Taiwan | Cross-sectional | 116 | 116 * | Oral swabs | QIAamp® DNA Blood Mini Kit | V4 | Illumina MiSeq System | SILVA |
| 11 | Zhou X, 2021 [34] | China | Case-control | 47 | 48 | Saliva, subgingival plaque, tumour and healthy surface | E.Z.N.A.® soil DNA Kit | V4–V5 | Illumina MiSeq System | SILVA |
| Author, Year | N. of Phyla Detected | Phyla | N. of Genera Detected | Genera | N. of Species Detected | Species | |||
|---|---|---|---|---|---|---|---|---|---|
| ↑ in OSCC Group | ↓ in OSCC Group | ↑ in OSCC Group | ↓ in OSCC Group | ↑ in OSCC Group | ↓ in OSCC Group | ||||
| Zhao H, 2017 [26] | 11 | Spirochaetes, Fusobacteria, Bacteroidetes | Firmicutes, Actinobacteria | 130 | Mycoplasma, Treponema, Campylobacter, Eikenella, Centipeda, Lachnospiraceae, Alloprevotella, Fusobacterium, Selenomonas, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, Parvimonas, Capnocytophaga, and Peptostreptococcaceae | Megasphaera, Stomatobaculum, Granulicatella, Lautropia, Veillonella, Streptococcus, Scardovia, Rothia, and Actinomyces | 389 | n.d. | n.d. |
| Hsiao JR, 2018 [24] | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 120 | P. tannaerae and F. nucleatum | n.d. |
| Yang SF, 2018 [27] | n.d. | n.d. | n.d. | n.d. | Capnocytophaga * | n.d. | n.d. | n.d. | n.d. |
| Takahashi Y, 2019 [29] | n.d. | n.d. | n.d. | 85 | Peptrostreptococcus, Fusobacterium, Alloprevotella, Capnocytophaga | Rothia, Haemophilus | n.d. | n.d. | n.d. |
| Ganly I, 2021 [32] | 12 | n.d. | n.d. | 116 | Fusobacterium, Prevotella, Alloprevotella | Streptococcus | 172 | n.d. | n.d. |
| Su SC, 2021 [33] | n.d. | n.d. | n.d. | n.d. | Fusobacterium, Peptostreptococcus, Campylobacter, Prevotella, Capnocytophaga | Streptococcus | n.d. | Campylobacter spp. | Streptococcus pneumoniae |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mauceri, R.; Coppini, M.; Vacca, D.; Bertolazzi, G.; Panzarella, V.; Di Fede, O.; Tripodo, C.; Campisi, G. Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review. Cancers 2022, 14, 5441. https://doi.org/10.3390/cancers14215441
Mauceri R, Coppini M, Vacca D, Bertolazzi G, Panzarella V, Di Fede O, Tripodo C, Campisi G. Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review. Cancers. 2022; 14(21):5441. https://doi.org/10.3390/cancers14215441
Chicago/Turabian StyleMauceri, Rodolfo, Martina Coppini, Davide Vacca, Giorgio Bertolazzi, Vera Panzarella, Olga Di Fede, Claudio Tripodo, and Giuseppina Campisi. 2022. "Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review" Cancers 14, no. 21: 5441. https://doi.org/10.3390/cancers14215441
APA StyleMauceri, R., Coppini, M., Vacca, D., Bertolazzi, G., Panzarella, V., Di Fede, O., Tripodo, C., & Campisi, G. (2022). Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review. Cancers, 14(21), 5441. https://doi.org/10.3390/cancers14215441










