UM-164, a Dual Inhibitor of c-Src and p38 MAPK, Suppresses Proliferation of Glioma by Reducing YAP Activity
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Plasmids
2.2. Antibodies and Reagents
2.3. Cell Viability, Colony Formation, Cell Migration and 3D Culture Assays
2.4. Flow Cytometric Analysis
2.5. Nuclear and Cytoplasmic Extraction
2.6. Immunoblotting, Immunofluorescence and Immunohistochemistry
2.7. Quantitative Real Time PCR (qRT-PCR)
| axl: F-AACCTTCAACTCCTGCCTTCTCG |
| R-CAGCTTCTCCTTCAGCTCTTCAC |
| cyr61: F-CTTACGCTGGATGTTTGAGTGT |
| R-AGACTGGATCATCATGACGTTCT |
| ctgf: F-GCTTACCGACTGGAAGACACG |
| R-CGGATGCACTTTTTGCCCTT |
| cyclin d1: F-AGCTCCTGTGCTGCGAAGTGGAAAC |
| R-AGTGTTCAATGAAATCGTGCGGGGT |
| gapdh: F-CTTCACCACCATGGAGGAGGC |
| R-GGCATGGACTGTGGTCATGAG |
2.8. RNA-seq
2.9. Xenograft Tumor Models
2.10. Statistical Analysis
3. Results
3.1. UM-164 Inhibits Glioma Cell Growth
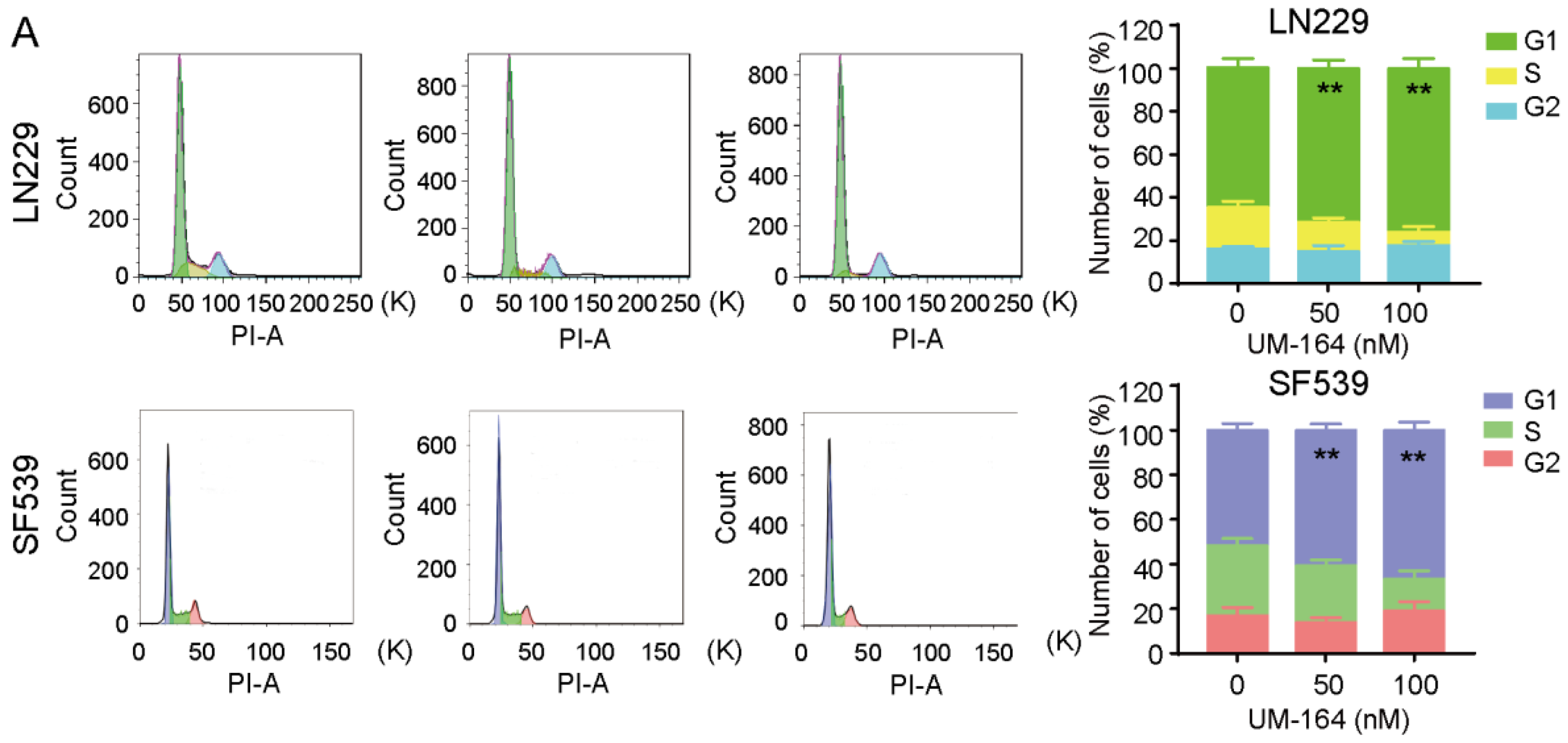
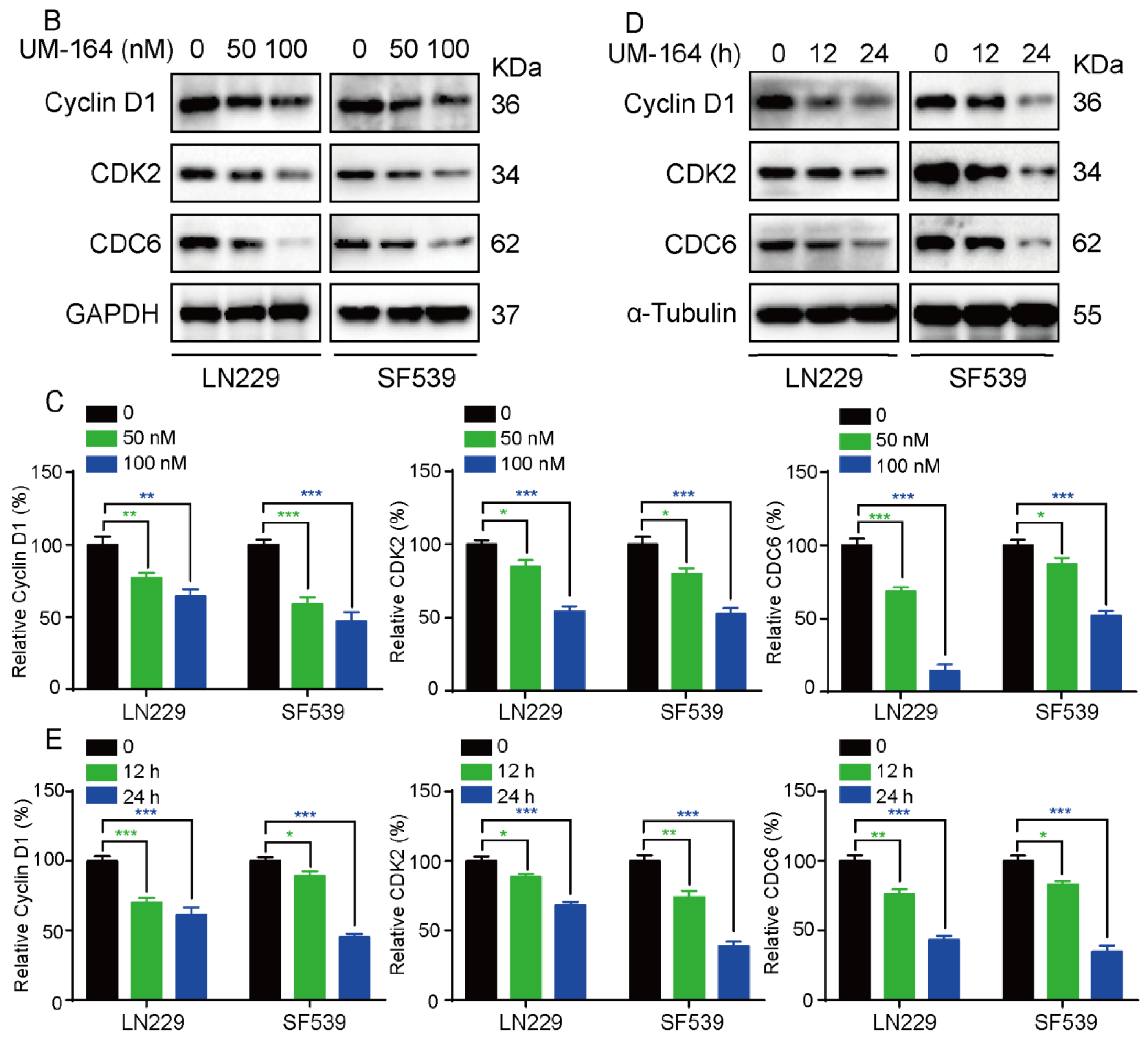
3.2. UM-164 Induces Glioma G1 Phase Cell Cycle Arrest
3.3. UM-164 Restrains YAP Nuclear Localization and Its Downstream Signaling
3.4. UM-164 Hinders Glioma Cell Proliferation via the Hippo-YAP Pathway
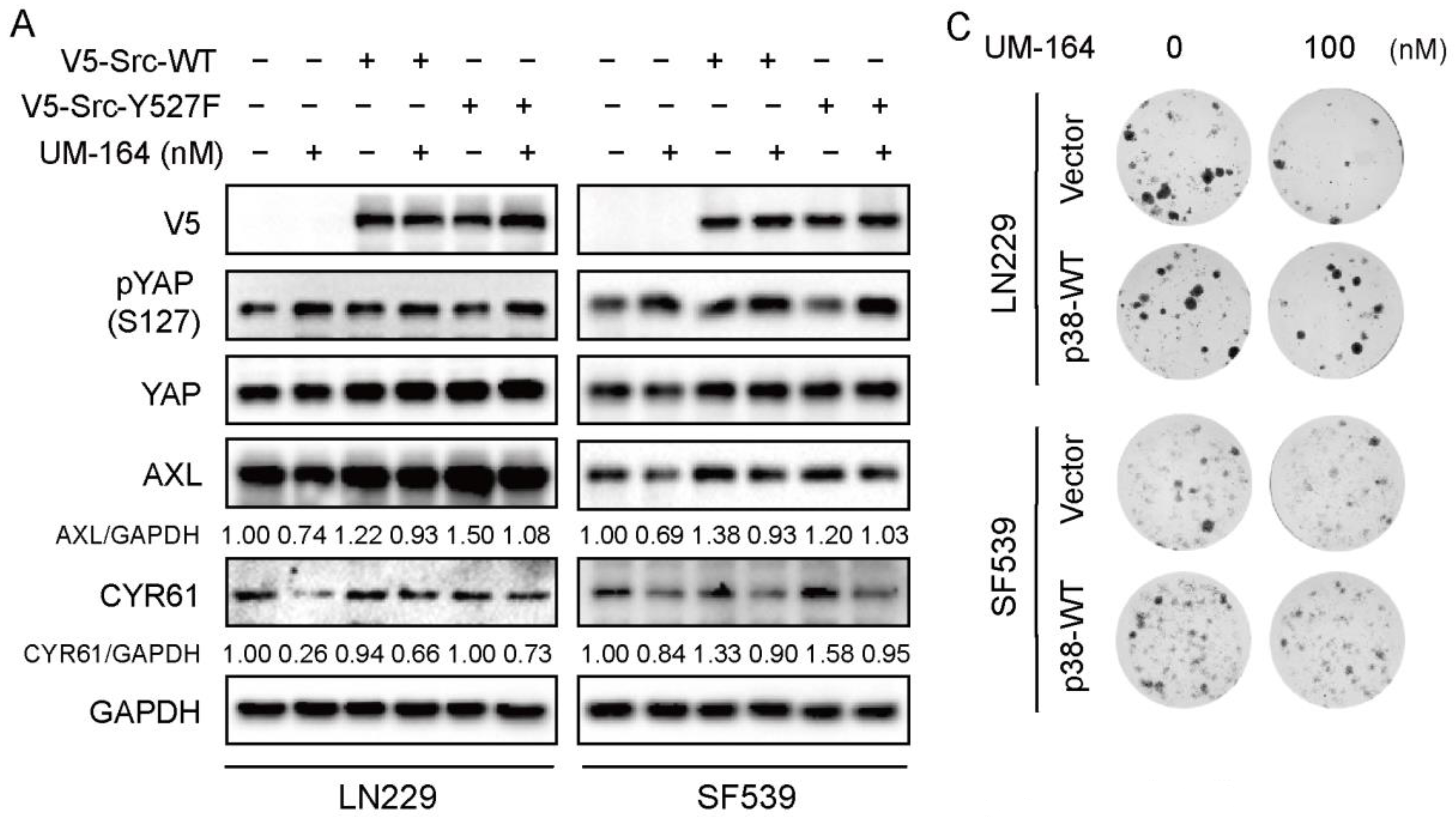
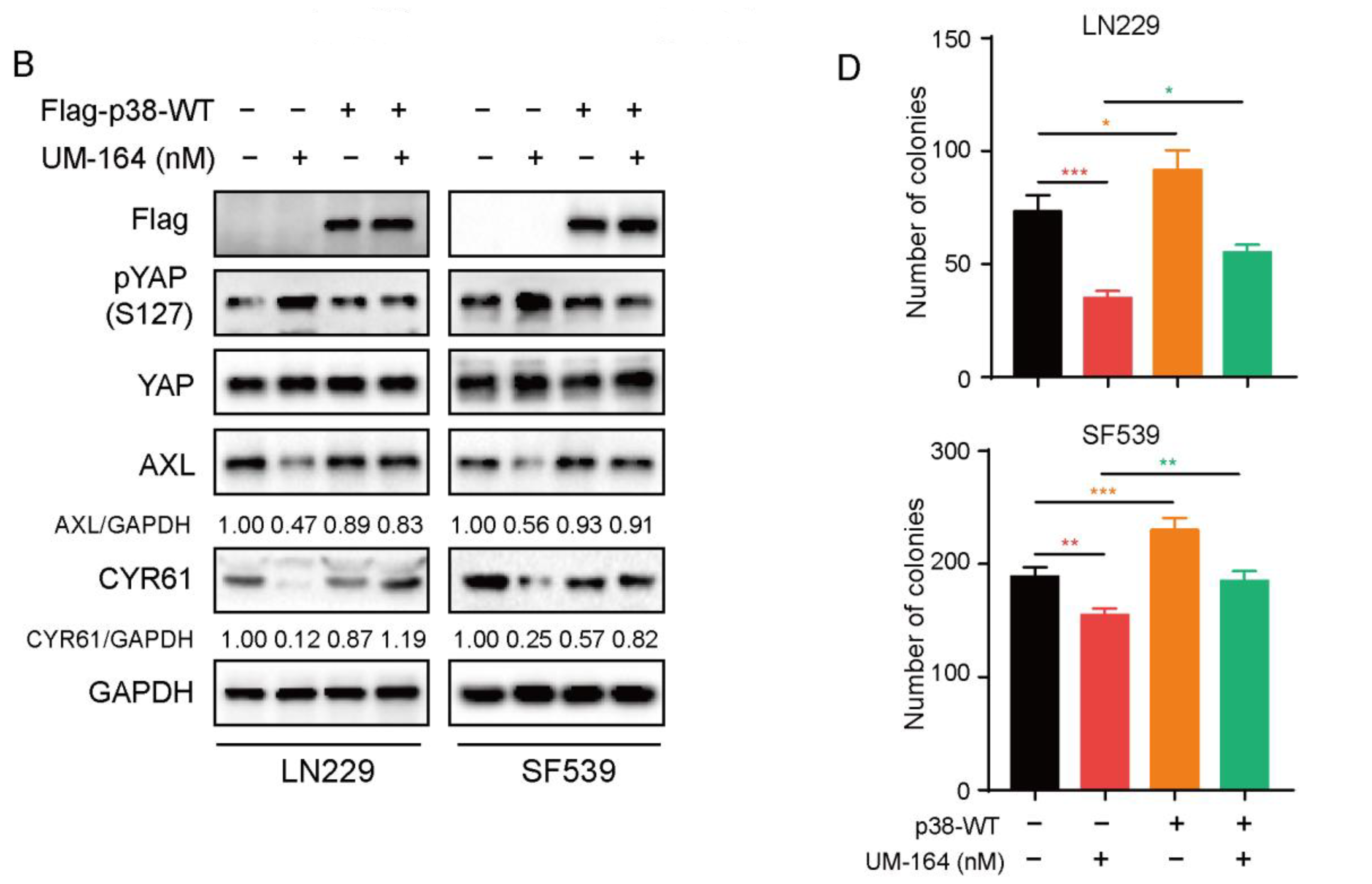
3.5. p38 MAPK Is More Prominent in Blocking UM-164-Mediated Anti-YAP Activity
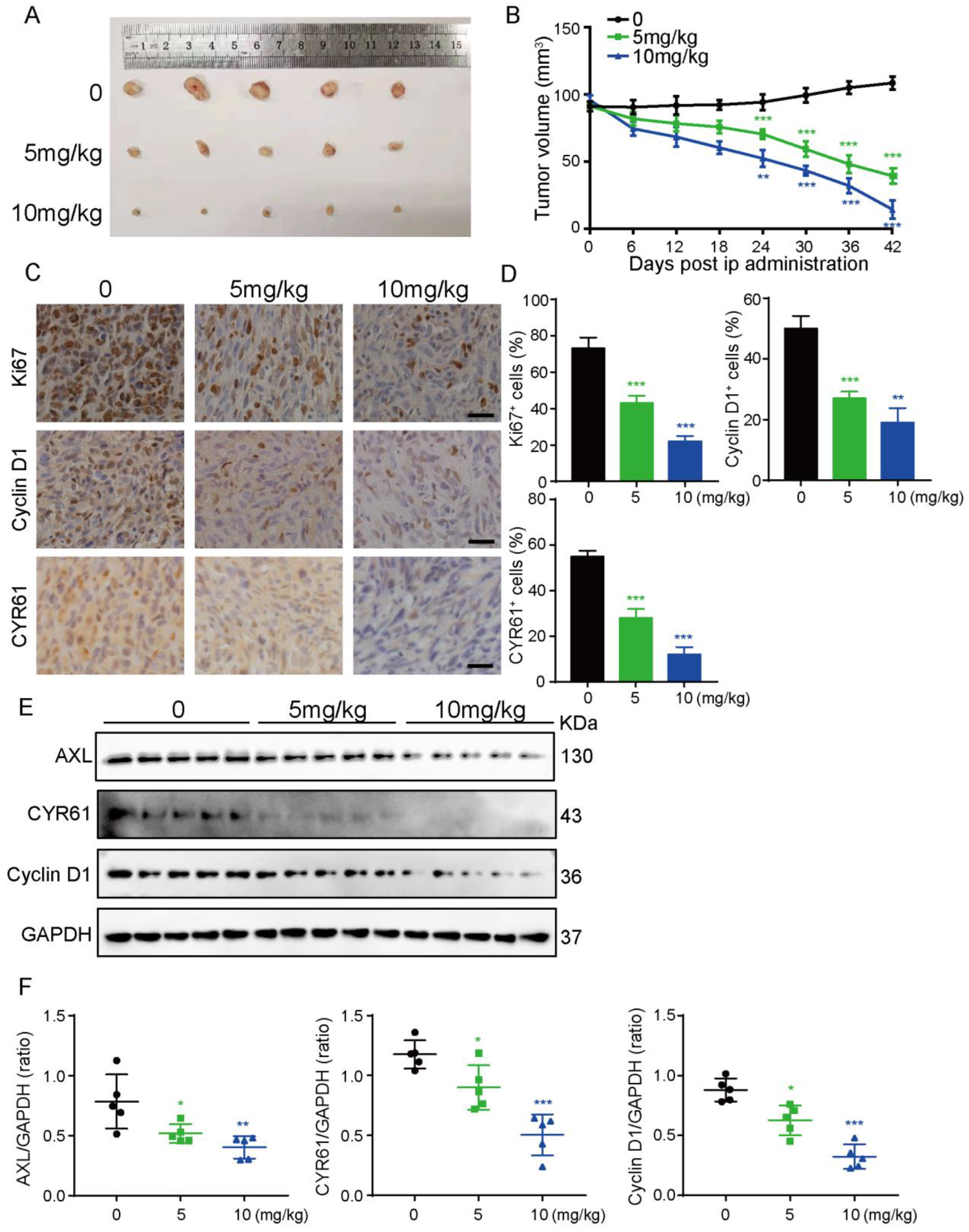
3.6. UM-164 Suppresses Glioma Xenograft Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Prim. 2015, 1, 15017. [Google Scholar] [CrossRef]
- Meng, X.; Zhao, Y.; Han, B.; Zha, C.; Zhang, Y.; Li, Z.; Wu, P.; Qi, T.; Jiang, C.; Liu, Y.; et al. Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat. Commun. 2020, 11, 594. [Google Scholar] [CrossRef]
- Lu, J.; Hu, Y.; Qian, R.; Zhang, Y.; Yang, X.; Luo, P. Enhanced proliferation inhibition and apoptosis in glioma cells elicited by combination of irinotecan and imatinib. Eur. J. Pharmacol. 2020, 874, 173022. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.J.; Ali, S.; Qadir, M.G.; De La Fuente, M.I.; Ivan, M.E.; Komotar, R.J. The role of bevacizumab in the treatment of glioblastoma. J. Neurooncol. 2017, 133, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Haberler, C.; Gelpi, E.; Marosi, C.; Rossler, K.; Birner, P.; Budka, H.; Hainfellner, J.A. Immunohistochemical analysis of platelet-derived growth factor receptor-alpha, -beta, c-kit, c-abl, and arg proteins in glioblastoma: Possible implications for patient selection for imatinib mesylate therapy. J. Neurooncol. 2006, 76, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Brandes, A.A.; Bartolotti, M.; Tosoni, A.; Poggi, R.; Franceschi, E. Practical management of bevacizumab-related toxicities in glioblastoma. Oncologist 2015, 20, 166–175. [Google Scholar] [CrossRef]
- Koekkoek, J.A.; Dirven, L.; Heimans, J.J.; Postma, T.J.; Vos, M.J.; Reijneveld, J.C.; Taphoorn, M.J. Seizure reduction in a low-grade glioma: More than a beneficial side effect of temozolomide. J. Neurol. Neurosurg. Psychiatry 2015, 86, 366–373. [Google Scholar] [CrossRef]
- He, J.; Huang, Z.; He, M.; Liao, J.; Zhang, Q.; Wang, S.; Xie, L.; Ouyang, L.; Koeffler, H.P.; Yin, D.; et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 2020, 19, 17. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, Q.; Wang, R.; Xu, G.; Li, P.; Sun, Y.; She, X.; Liu, Q.; Chen, Q.; Yu, Z.; et al. The D domain of LRRC4 anchors ERK1/2 in the cytoplasm and competitively inhibits MEK/ERK activation in glioma cells. J. Hematol. Oncol. 2016, 9, 130. [Google Scholar] [CrossRef][Green Version]
- Masliantsev, K.; Karayan-Tapon, L.; Guichet, P.O. Hippo signaling pathway in gliomas. Cells 2021, 10, 184. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440–33450. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, W.; Xu, H.; Liu, J.; Ren, L.; Yang, Y.; Li, S.; Wang, J.; Ji, T.; Du, G. Sinomenine ester derivative inhibits glioblastoma by inducing mitochondria-dependent apoptosis and autophagy by PI3K/AKT/mTOR and AMPK/mTOR pathway. Acta Pharm. Sin. B 2021, 11, 3465–3480. [Google Scholar] [CrossRef]
- Boggon, T.J.; Eck, M.J. Structure and regulation of src family kinases. Oncogene 2004, 23, 7918–7927. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, D.; Cui, Y.; Zhang, W.; Weng, J.; Yu, L.; Chen, L.; Chen, Z.; Su, H.; Yu, S.; et al. Src plays an important role in AGE-induced endothelial cell proliferation, migration, and tubulogenesis. Front. Physiol. 2018, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, C.; Huo, G.; Deng, J.; Zhao, H.; Xu, R.; Jiang, L.; Chen, S.; Wang, S. ATP1A1 Integrates AKT and ERK Signaling via Potential Interaction With Src to Promote Growth and Survival in Glioma Stem Cells. Front. Oncol. 2019, 9, 320. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.W.; Park, J.B.; Park, S.Y.; Seo, J.; Shin, S.H.; Park, J.W.; Kim, S.J.; Watanabe, M.; Chun, Y.S. The E3 ligase C-CBL inhibits cancer cell migration by neddylating the proto-oncogene c-src. Oncogene 2018, 37, 5552–5568. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Wang, P.; Liu, Y.H.; Cai, H.; Ma, J.; Liu, L.B.; Xi, Z.; Li, Z.Q.; Liu, X.B.; Xue, Y.X. Mir-383 inhibits proliferation, migration and angiogenesis of glioma-exposed endothelial cells in vitro via VEGF-mediated FAK and Src signaling pathways. Cell Signal. 2017, 30, 142–153. [Google Scholar] [CrossRef]
- Han, X.; Zhang, W.; Yang, X.; Wheeler, C.G.; Langford, C.P.; Wu, L.; Filippova, N.; Friedman, G.K.; Ding, Q.; Fathallah-Shaykh, H.M.; et al. The role of src family kinases in growth and migration of glioma stem cells. Int. J. Oncol. 2014, 45, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.G.; Jaraiz-Rodriguez, M.; Alvarez-Vazquez, A.; Talaveron, R.; Garcia-Vicente, L.; Flores-Hernandez, R.; Gomez de Cedron, M.; Tabernero, M.; Ramirez de Molina, A.; Lillo, C.; et al. Targeting metabolic plasticity in glioma stem cells in vitro and in vivo through specific inhibition of c-Src by TAT-Cx43266-283. EBioMedicine 2020, 62, 103134. [Google Scholar] [CrossRef] [PubMed]
- Alhalabi, O.T.; Fletcher, M.N.C.; Hielscher, T.; Kessler, T.; Lokumcu, T.; Baumgartner, U.; Wittmann, E.; Schlue, S.; Rahman, M.G.; Hai, L.; et al. A novel patient stratification strategy to enhance the therapeutic efficacy of dasatinib in glioblastoma. Neuro Oncol. 2022, 24, 39–51. [Google Scholar] [CrossRef]
- Taylor, J.W.; Dietrich, J.; Gerstner, E.R.; Norden, A.D.; Rinne, M.L.; Cahill, D.P.; Stemmer-Rachamimov, A.; Wen, P.Y.; Betensky, R.A.; Giorgio, D.H.; et al. Phase 2 study of bosutinib, a src inhibitor, in adults with recurrent glioblastoma. J. Neurooncol. 2015, 121, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Tuffin, L.J.; Feathers, R.; Hari, P.; Durand, N.; Li, Z.; Rodriguez, F.J.; Bakken, K.; Carlson, B.; Schroeder, M.; Sarkaria, J.N.; et al. Src family kinases differentially influence glioma growth and motility. Mol. Oncol. 2015, 9, 1783–1798. [Google Scholar] [CrossRef]
- Premkumar, D.R.; Jane, E.P.; Agostino, N.R.; Scialabba, J.L.; Pollack, I.F. Dasatinib synergizes with JSI-124 to inhibit growth and migration and induce apoptosis of malignant human glioma cells. J. Carcinog. 2010, 9, 7. [Google Scholar]
- Li, X.; Tao, Z.; Wang, H.; Deng, Z.; Zhou, Y.; Du, Z. Dual inhibition of src and plk1 regulate stemness and induce apoptosis through notch1-SOX2 signaling in egfrviii positive glioma stem cells (GSCs). Exp. Cell Res. 2020, 396, 112261. [Google Scholar] [CrossRef]
- Bello-Alvarez, C.; Moral-Morales, A.D.; Gonzalez-Arenas, A.; Camacho-Arroyo, I. Intracellular progesterone receptor and csrc protein working together to regulate the activity of proteins involved in migration and invasion of human glioblastoma cells. Front. Endocrinol. (Lausanne) 2021, 12, 640298. [Google Scholar] [CrossRef]
- Ciesielski, M.J.; Bu, Y.; Munich, S.A.; Teegarden, P.; Smolinski, M.P.; Clements, J.L.; Lau, J.Y.N.; Hangauer, D.G.; Fenstermaker, R.A. KX2-361: A novel orally bioavailable small molecule dual src/tubulin inhibitor that provides long term survival in a murine model of glioblastoma. J. Neurooncol. 2018, 140, 519–527. [Google Scholar] [CrossRef]
- Ndagi, U.; Mhlongo, N.N.; Soliman, M.E. Emergence of a promising lead compound in the treatment of triple negative breast cancer: An insight into conformational features and ligand binding landscape of c-src protein with UM-164. Appl. Biochem. Biotechnol. 2018, 185, 655–675. [Google Scholar] [CrossRef] [PubMed]
- Ndagi, U.; Abdullahi, M.; Hamza, A.N.; Soliman, M.E. An analogue of a kinase inhibitor exhibits subjective characteristics that contribute to its inhibitory activities as a potential anti-cancer candidate: Insights through computational biomolecular modelling of UM-164 binding with lyn protein. RSC Adv. 2019, 10, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Macias, M.J.; Hyvonen, M.; Baraldi, E.; Schultz, J.; Sudol, M.; Saraste, M.; Oschkinat, H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 1996, 382, 646–649. [Google Scholar] [CrossRef]
- Yagi, R.; Chen, L.F.; Shigesada, K.; Murakami, Y.; Ito, Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999, 18, 2551–2562. [Google Scholar] [CrossRef]
- Oka, T.; Sudol, M. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells 2009, 14, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Remue, E.; Meerschaert, K.; Vanloo, B.; Boucherie, C.; Gfeller, D.; Bader, G.D.; Sidhu, S.S.; Vandekerckhove, J.; Gettemans, J.; et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010, 432, 461–472. [Google Scholar] [CrossRef]
- Lai, D.; Ho, K.C.; Hao, Y.W.; Yang, X.L. Taxol resistance in breast cancer cells is mediated by the hippo pathway component taz and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011, 71, 2728–2738. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of Yap/Taz: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.C.; Yang, C.T.; Jablons, D.M.; You, L. The Crosstalk between Src and Hippo/YAP Signaling Pathways in Non-Small Cell Lung Cancer (NSCLC). Cancers 2020, 12, 1361. [Google Scholar] [CrossRef]
- Guillermin, O.; Angelis, N.; Sidor, C.M.; Ridgway, R.; Baulies, A.; Kucharska, A.; Antas, P.; Rose, M.R.; Cordero, J.; Sansom, O.; et al. Wnt and src signals converge on YAP-TEAD to drive intestinal regeneration. EMBO J. 2021, 40, e105770. [Google Scholar] [CrossRef]
- Thompson, B.J. YAP/TAZ: Drivers of tumor growth, metastasis, and resistance to therapy. Bioessays 2020, 42, e1900162. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, H.J.; Jiang, K.W.; Wang, Y.J.; Zhang, W.J.; Chu, Q.Q.; Li, J.; Huang, H.W.; Cai, T.; Ji, H.B.; et al. MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 2016, 16, 1810–1819. [Google Scholar] [CrossRef]
- Jiang, K.; Liu, P.; Xu, H.; Liang, D.; Fang, K.; Du, S.; Cheng, W.; Ye, L.; Liu, T.; Zhang, X.; et al. SASH1 suppresses triple-negative breast cancer cell invasion through YAP-ARHGAP42-actin axis. Oncogene 2020, 39, 5015–5030. [Google Scholar] [CrossRef] [PubMed]
- Limagne, E.; Nuttin, L.; Thibaudin, M.; Jacquin, E.; Aucagne, R.; Bon, M.; Revy, S.; Barnestein, R.; Ballot, E.; Truntzer, C.; et al. MEK inhibition overcomes chemoimmunotherapy resistance by inducing CXCL10 in cancer cells. Cancer Cell 2022, 40, 136–152 e112. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Babaian, A.; Nakamichi, N.; Chen, M.; Chafe, S.C.; Watanabe, A.; Forrest, D.L.; Mager, D.L.; Eaves, C.J.; Dedhar, S.; et al. Integrin-linked kinase mediates therapeutic resistance of quiescent CML stem cells to tyrosine kinase inhibitors. Cell Stem Cell 2020, 27, 110–124.e9. [Google Scholar] [CrossRef]
- Singh, K.; Pruski, M.A.; Polireddy, K.; Jones, N.C.; Chen, Q.; Yao, J.; Dar, W.A.; McAllister, F.; Ju, C.; Eltzschig, H.K.; et al. Mst1/2 kinases restrain transformation in a novel transgenic model of ras driven non-small cell lung cancer. Oncogene 2020, 39, 1152–1164. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, X.; Yu, M.; Ruan, Y.; Zhang, J.; Tian, Y.; Xiong, J.; Liu, L.; Cheng, Y.; Yang, Y.; et al. Identification, subcellular localization, and functional comparison of novel yap splicing isoforms in mouse embryonic stem cells. IUBMB Life 2021, 73, 1432–1445. [Google Scholar] [CrossRef]
- Chen, X.; Hao, A.; Li, X.; Ye, K.; Zhao, C.; Yang, H.; Ma, H.; Hu, L.; Zhao, Z.; Hu, L.; et al. Activation of JNK and p38 MAPK mediated by ZDHHC17 drives glioblastoma multiforme development and malignant progression. Theranostics 2020, 10, 998–1015. [Google Scholar] [CrossRef]
- Xu, Y.; Sun, Q.; Yuan, F.; Dong, H.; Zhang, H.; Geng, R.; Qi, Y.; Xiong, X.; Chen, Q.; Liu, B. RND2 attenuates apoptosis and autophagy in glioblastoma cells by targeting the p38 MAPK signalling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 174. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.H.; Li, X.Y.; Yang, J.; Yuan, D.T.; Zhou, Y.S.; Zhang, Y.N.; Shi, G.H.; Zhang, R.B.; Liu, J.P.; Fu, P.; et al. PPFIBP1 induces glioma cell migration and invasion through FAK/Src/JNK signaling pathway. Cell Death Dis. 2021, 12, 827. [Google Scholar] [CrossRef]
- Wang, R.; Li, B.W.; Shao, N.Y.; Deng, D.N.; Zhi, F. A novel antitumor peptide inhibits proliferation and migration and promotes apoptosis in glioma cells by regulating the MKK6/p38 signaling pathway. Neoplasma 2021, 68, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Park, J.Y.; Kang, Y.N. Tumorigenic role of YAP in hepatocellular carcinogenesis is involved in SHP2 whose function is different in vitro and in vivo. Pathol. Res. Pract. 2018, 214, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, X.; Sun, L.; Huang, P.; Ying, H.; Wang, H.; Wu, J.; Song, H. Regulation of hippo signalling by p38 signalling. J. Mol. Cell Biol. 2016, 8, 328–337. [Google Scholar] [CrossRef]
- Lin, K.C.; Moroishi, T.; Meng, Z.; Jeong, H.S.; Plouffe, S.W.; Sekido, Y.; Han, J.; Park, H.W.; Guan, K.L. Regulation of hippo pathway transcription factor TEAD by p38 MAPk-induced cytoplasmic translocation. Nat. Cell Biol. 2017, 19, 996–1002. [Google Scholar] [CrossRef]
- Salloum, S.; Jeyarajan, A.J.; Kruger, A.J.; Holmes, J.A.; Shao, T.; Sojoodi, M.; Kim, M.H.; Zhuo, Z.; Shroff, S.G.; Kassa, A.; et al. Fatty acids activate the transcriptional coactivator YAP1 to promote liver fibrosis via p38 mitogen-activated protein kinase. Cell Mol. Gastroenterol. Hepatol. 2021, 12, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.I.; Sudol, M. The WW domain of yes-associated protein binds a proline-rich ligand that differs from the consensus established for src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 1995, 92, 7819–7823. [Google Scholar] [CrossRef] [PubMed]
- Rozengurt, E.; Eibl, G. Crosstalk between KRAS, SRC and YAP signaling in pancreatic cancer: Interactions leading to aggressive disease and drug resistance. Cancers 2021, 13, 5126. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.J.; Liu, L.L.; Yang, X.; Qian, Q.F.; Du, B. c-Src promotes the growth and tumorigenesis of hepatocellular carcinoma via the hippo signaling pathway. Life Sci. 2021, 264, 118711. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Zhang, Y.; Liu, J.; Cui, J.; Gan, Y.; Wu, Z.; Chang, Y.; Sui, R.; Chen, Y.; Shi, J.; et al. UM-164, a Dual Inhibitor of c-Src and p38 MAPK, Suppresses Proliferation of Glioma by Reducing YAP Activity. Cancers 2022, 14, 5343. https://doi.org/10.3390/cancers14215343
Xu H, Zhang Y, Liu J, Cui J, Gan Y, Wu Z, Chang Y, Sui R, Chen Y, Shi J, et al. UM-164, a Dual Inhibitor of c-Src and p38 MAPK, Suppresses Proliferation of Glioma by Reducing YAP Activity. Cancers. 2022; 14(21):5343. https://doi.org/10.3390/cancers14215343
Chicago/Turabian StyleXu, Huizhe, Ye Zhang, Jia Liu, Jing Cui, Yu Gan, Zhisheng Wu, Youwei Chang, Rui Sui, Yi Chen, Ji Shi, and et al. 2022. "UM-164, a Dual Inhibitor of c-Src and p38 MAPK, Suppresses Proliferation of Glioma by Reducing YAP Activity" Cancers 14, no. 21: 5343. https://doi.org/10.3390/cancers14215343
APA StyleXu, H., Zhang, Y., Liu, J., Cui, J., Gan, Y., Wu, Z., Chang, Y., Sui, R., Chen, Y., Shi, J., Liang, H., Liu, Q., Sun, S., & Piao, H. (2022). UM-164, a Dual Inhibitor of c-Src and p38 MAPK, Suppresses Proliferation of Glioma by Reducing YAP Activity. Cancers, 14(21), 5343. https://doi.org/10.3390/cancers14215343




