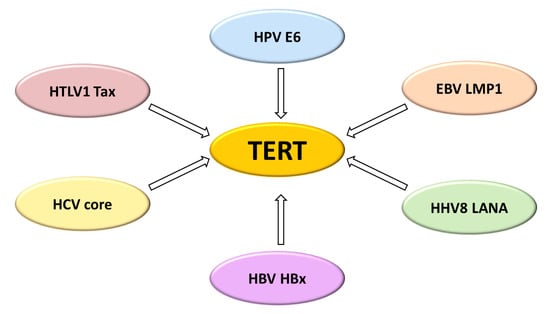
The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development
Simple Summary
Abstract
1. Introduction
2. TERT Expression and Telomerase Activities
2.1. Human Papillomavirus and Telomerase Activity in HPV-Related Tumours
2.2. Epstein-Barr Virus and Telomerase Activity in EBV-Related Tumours
2.3. Human Herpesvirus 8 and Telomerase Activity in Kaposi Sarcoma
2.4. Hepatitis B Virus and Telomerase Activity in Hepatocellular Carcinoma
2.5. Hepatitis C Virus and Telomerase Activity in Hepatocellular Carcinoma
2.6. Human T-Cell Leukemia Virus Type 1 and Telomerase Activity in T-Cell Lymphoproliferative Disorders
3. Telomerase and Immune Response in Virus-Related Cancers
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Annunziata, C.; Tornesello, A.L.; Buonaguro, L.; Buonaguro, F.M. Human Oncoviruses and p53 Tumor Suppressor Pathway Deregulation at the Origin of Human Cancers. Cancers 2018, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Crary, S.M.; Jesse Fioravanti, M.; Campbell, K.; Cech, T.R. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013, 41, 8969–8978. [Google Scholar] [CrossRef]
- Bellon, M.; Nicot, C. Regulation of telomerase and telomeres: Human tumor viruses take control. J. Natl. Cancer Inst. 2008, 100, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.S.; Cotter, M.A., 2nd; Robertson, E.S. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 2001, 276, 22971–22978. [Google Scholar] [CrossRef]
- Klingelhutz, A.J.; Foster, S.A.; McDougall, J.K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 1996, 380, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Deng, X.; Deng, L.; Gu, H.; Fan, W.; Cao, Y. Telomerase activation by Epstein-Barr virus latent membrane protein 1 is associated with c-Myc expression in human nasopharyngeal epithelial cells. J. Exp. Clin. Cancer Res. 2004, 23, 495–506. [Google Scholar] [PubMed]
- Sze, K.M.; Ho, D.W.; Chiu, Y.T.; Tsui, Y.M.; Chan, L.K.; Lee, J.M.; Chok, K.S.; Chan, A.C.; Tang, C.N.; Tang, V.W.; et al. Hepatitis B Virus-Telomerase Reverse Transcriptase Promoter Integration Harnesses Host ELF4, Resulting in Telomerase Reverse Transcriptase Gene Transcription in Hepatocellular Carcinoma. Hepatology 2021, 73, 23–40. [Google Scholar] [CrossRef]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The Telomeric Repeats of Human Herpesvirus 6A (HHV-6A) Are Required for Efficient Virus Integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk. SSSR 1971, 201, 1496–1499. [Google Scholar] [PubMed]
- Smith, E.M.; Pendlebury, D.F.; Nandakumar, J. Structural biology of telomeres and telomerase. Cell Mol. Life Sci. 2020, 77, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sfeir, A.J.; Zou, Y.; Buseman, C.M.; Chow, T.T.; Shay, J.W.; Wright, W.E. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 2009, 138, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, G.; Lingner, J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006, 25, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 2000, 1, 72–76. [Google Scholar] [CrossRef]
- Dratwa, M.; Wysoczańska, B.; Łacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, Q.L.; Sun, W.; Chandrasekharan, P.; Cheng, H.S.; Ying, Z.; Lakshmanan, M.; Raju, A.; Tenen, D.G.; Cheng, S.Y.; et al. Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 2015, 17, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, L.Y.; Allan, S.; Beach, D.; Hannon, G.J. Myc activates telomerase. Genes Dev. 1998, 12, 1769–1774. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Leão, R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolónio, J.D.; et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J. Clin. Investig. 2019, 129, 1801. [Google Scholar] [CrossRef] [PubMed]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the Human Telomerase Gene TERT by Telomere Position Effect-Over Long Distances (TPE-OLD): Implications for Aging and Cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef]
- Bell, R.J.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015, 348, 1036–1039. [Google Scholar] [CrossRef]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Stern, J.L.; Theodorescu, D.; Vogelstein, B.; Papadopoulos, N.; Cech, T.R. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015, 29, 2219–2224. [Google Scholar] [CrossRef]
- Starita, N.; Pezzuto, F.; Sarno, S.; Losito, N.S.; Perdonà, S.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Mutations in the telomerase reverse transcriptase promoter and PIK3CA gene are common events in penile squamous cell carcinoma of Italian and Ugandan patients. Int. J. Cancer 2022, 150, 1879–1888. [Google Scholar] [CrossRef]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Frequency and geographic distribution of TERT promoter mutations in primary hepatocellular carcinoma. Infect. Agent Cancer 2017, 12, 27. [Google Scholar] [CrossRef]
- Starita, N.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Telomerase promoter mutations in human immunodeficiency virus-related conjunctiva neoplasia. J. Transl. Med. 2018, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, C.; Pezzuto, F.; Greggi, S.; Ionna, F.; Losito, S.; Botti, G.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Distinct profiles of TERT promoter mutations and telomerase expression in head and neck cancer and cervical carcinoma. Int. J. Cancer 2018, 143, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Giunco, S.; Boscolo-Rizzo, P.; Rampazzo, E.; Tirelli, G.; Alessandrini, L.; Di Carlo, R.; Rossi, M.; Nicolai, P.; Menegaldo, A.; Carraro, V.; et al. TERT Promoter Mutations and rs2853669 Polymorphism: Useful Markers for Clinical Outcome Stratification of Patients with Oral Cavity Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 782658. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, A.; Paszel-Jaworska, A.; Totoń, E.; Lisiak, N.; Hołysz, H.; Królak, A.; Grodecka-Gazdecka, S.; Rubiś, B. The non-canonical functions of telomerase: To turn off or not to turn off. Mol. Biol. Rep. 2019, 46, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gaspard, J.P.; Chung, D.C. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001, 61, 6050–6054. [Google Scholar] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Park, J.I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef]
- Chen, K.; Chen, L.; Li, L.; Qu, S.; Yu, B.; Sun, Y.; Wan, F.; Chen, X.; Liang, R.; Zhu, X. A positive feedback loop between Wnt/β-catenin signaling and hTERT regulates the cancer stem cell-like traits in radioresistant nasopharyngeal carcinoma cells. J. Cell Biochem. 2020, 121, 4612–4622. [Google Scholar] [CrossRef]
- Smith, L.L.; Coller, H.A.; Roberts, J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003, 5, 474–479. [Google Scholar] [CrossRef]
- Young, J.I.; Sedivy, J.M.; Smith, J.R. Telomerase expression in normal human fibroblasts stabilizes DNA 5-methylcytosine transferase I. J. Biol. Chem. 2003, 278, 19904–19908. [Google Scholar] [CrossRef]
- Ségal-Bendirdjian, E.; Geli, V. Non-canonical Roles of Telomerase: Unraveling the Imbroglio. Front. Cell Dev. Biol. 2019, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yuan, X.; Sjöholm, L.; Liu, T.; Kong, F.; Ekström, T.J.; Björkholm, M.; Xu, D. Telomerase reverse transcriptase regulates DNMT3B expression/aberrant DNA methylation phenotype and AKT activation in hepatocellular carcinoma. Cancer Lett. 2018, 434, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.P.; Song, Z.; et al. hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef] [PubMed]
- Weng, N.; Levine, B.L.; June, C.H.; Hodes, R.J. Regulation of telomerase RNA template expression in human T lymphocyte development and activation. J. Immunol. 1997, 158, 3215–3220. [Google Scholar] [PubMed]
- Patrick, M.; Weng, N.P. Expression and regulation of telomerase in human T cell differentiation, activation, aging and diseases. Cell Immunol. 2019, 345, 103989. [Google Scholar] [CrossRef] [PubMed]
- Akbar, A.N.; Beverley, P.C.; Salmon, M. Will telomere erosion lead to a loss of T-cell memory? Nat. Rev. Immunol. 2004, 4, 737–743. [Google Scholar] [CrossRef]
- De Villiers, E.M. Cross-roads in the classification of papillomaviruses. Virology 2013, 445, 2–10. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- De Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Borghi, L.; Canali, M.; Curti, A.; Montanari, A.; Perinotto, P.; Novarini, A. [Na/K transport in red blood cells from normal subjects: Methodological problems (author’s transl)]. Ateneo Parmense Acta Biomed. 1980, 51, 23–31. [Google Scholar]
- Phelps, W.C.; Yee, C.L.; Münger, K.; Howley, P.M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 1988, 53, 539–547. [Google Scholar] [CrossRef]
- Zanier, K.; Charbonnier, S.; Sidi, A.O.; McEwen, A.G.; Ferrario, M.G.; Poussin-Courmontagne, P.; Cura, V.; Brimer, N.; Babah, K.O.; Ansari, T.; et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science 2013, 339, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Saidu, N.E.B.; Filić, V.; Thomas, M.; Sarabia-Vega, V.; Đukić, A.; Miljković, F.; Banks, L.; Tomaić, V. PDZ Domain-Containing Protein NHERF-2 Is a Novel Target of Human Papillomavirus 16 (HPV-16) and HPV-18. J. Virol. 2019, 94, e00663-19. [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Bossler, F.; Braun, J.A.; Herrmann, A.L.; Hoppe-Seyler, F. The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol. 2018, 26, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Estêvão, D.; Costa, N.R.; Gil da Costa, R.M.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef]
- Zimmermann, H.; Degenkolbe, R.; Bernard, H.U.; O’Connor, M.J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 1999, 73, 6209–6219. [Google Scholar] [CrossRef]
- Huh, K.; Zhou, X.; Hayakawa, H.; Cho, J.Y.; Libermann, T.A.; Jin, J.; Harper, J.W.; Munger, K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007, 81, 9737–9747. [Google Scholar] [CrossRef]
- White, E.A.; Sowa, M.E.; Tan, M.J.; Jeudy, S.; Hayes, S.D.; Santha, S.; Münger, K.; Harper, J.W.; Howley, P.M. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. USA 2012, 109, E260–E267. [Google Scholar] [CrossRef]
- Tomita, T.; Huibregtse, J.M.; Matouschek, A. A masked initiation region in retinoblastoma protein regulates its proteasomal degradation. Nat. Commun. 2020, 11, 2019. [Google Scholar] [CrossRef]
- Branca, M.; Giorgi, C.; Ciotti, M.; Santini, D.; Di Bonito, L.; Costa, S.; Benedetto, A.; Bonifacio, D.; Di Bonito, P.; Paba, P.; et al. Upregulation of telomerase (hTERT) is related to the grade of cervical intraepithelial neoplasia, but is not an independent predictor of high-risk human papillomavirus, virus persistence, or disease outcome in cervical cancer. Diagn. Cytopathol. 2006, 34, 739–748. [Google Scholar] [CrossRef]
- Liu, X.; Dakic, A.; Zhang, Y.; Dai, Y.; Chen, R.; Schlegel, R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc. Natl. Acad. Sci. USA 2009, 106, 18780–18785. [Google Scholar] [CrossRef] [PubMed]
- Pańczyszyn, A.; Boniewska-Bernacka, E.; Głąb, G. Telomeres and Telomerase During Human Papillomavirus-Induced Carcinogenesis. Mol. Diagn. Ther. 2018, 22, 421–430. [Google Scholar] [CrossRef]
- Katzenellenbogen, R.A. Activation of telomerase by HPVs. Virus Res. 2017, 231, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Kiyono, T.; Foster, S.A.; Koop, J.I.; McDougall, J.K.; Galloway, D.A.; Klingelhutz, A.J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 1998, 396, 84–88. [Google Scholar] [CrossRef] [PubMed]
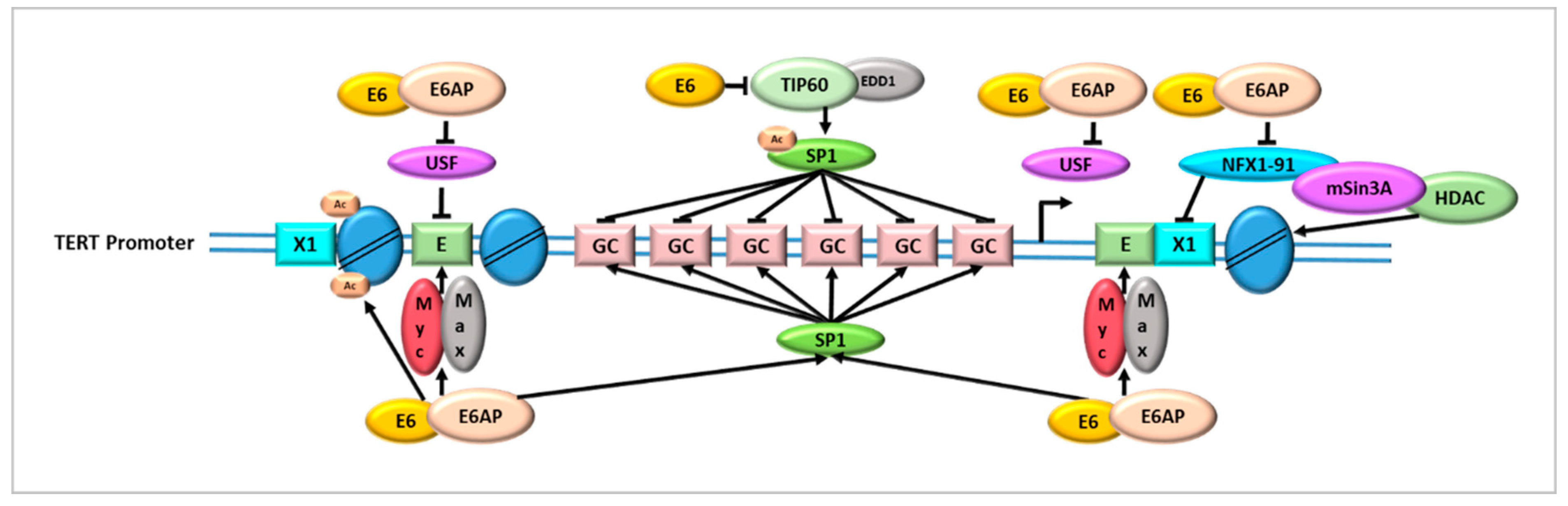
- Oh, S.T.; Kyo, S.; Laimins, L.A. Telomerase activation by human papillomavirus type 16 E6 protein: Induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 2001, 75, 5559–5566. [Google Scholar] [CrossRef]
- Takakura, M.; Kyo, S.; Kanaya, T.; Hirano, H.; Takeda, J.; Yutsudo, M.; Inoue, M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999, 59, 551–557. [Google Scholar]
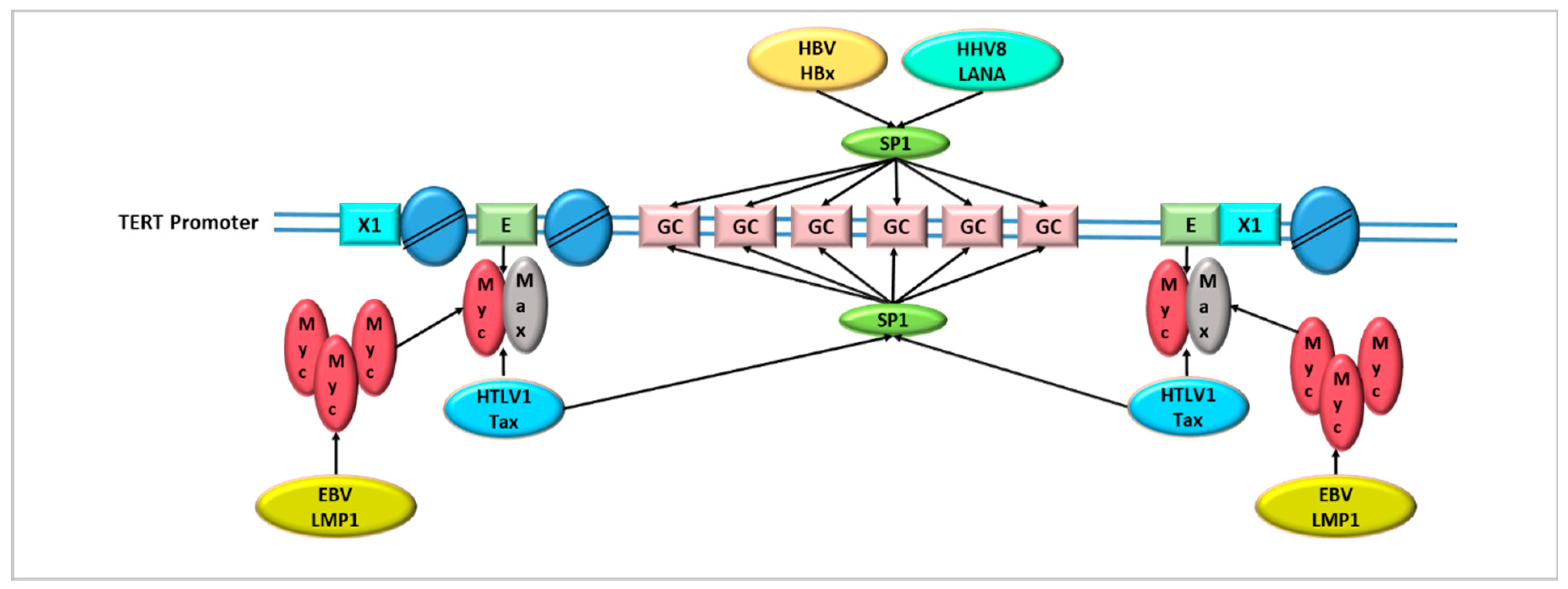
- Kyo, S.; Takakura, M.; Taira, T.; Kanaya, T.; Itoh, H.; Yutsudo, M.; Ariga, H.; Inoue, M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 2000, 28, 669–677. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Casillas, M.A.; Brotherton, S.L.; Andrews, L.G.; Ruppert, J.M.; Tollefsbol, T.O. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene 2003, 316, 57–65. [Google Scholar] [CrossRef]
- Veldman, T.; Horikawa, I.; Barrett, J.C.; Schlegel, R. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 2001, 75, 4467–4472. [Google Scholar] [CrossRef]
- McMurray, H.R.; McCance, D.J. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J. Virol. 2003, 77, 9852–9861. [Google Scholar] [CrossRef] [PubMed]
- Veldman, T.; Liu, X.; Yuan, H.; Schlegel, R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 2003, 100, 8211–8216. [Google Scholar] [CrossRef] [PubMed]
- Khattar, E.; Tergaonkar, V. Transcriptional Regulation of Telomerase Reverse Transcriptase (TERT) by MYC. Front. Cell Dev. Biol. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Gewin, L.; Myers, H.; Kiyono, T.; Galloway, D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004, 18, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yuan, H.; Fu, B.; Disbrow, G.L.; Apolinario, T.; Tomaic, V.; Kelley, M.L.; Baker, C.C.; Huibregtse, J.; Schlegel, R. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 2005, 280, 10807–10816. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.L.; Keiger, K.E.; Lee, C.J.; Huibregtse, J.M. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 2005, 79, 3737–3747. [Google Scholar] [CrossRef]
- James, M.A.; Lee, J.H.; Klingelhutz, A.J. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 2006, 119, 1878–1885. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Luo, W.; Elzi, D.J.; Grandori, C.; Galloway, D.A. NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol. Cell Biol. 2008, 28, 4819–4828. [Google Scholar] [CrossRef]
- Katzenellenbogen, R.A.; Vliet-Gregg, P.; Xu, M.; Galloway, D.A. Cytoplasmic poly(A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes. J. Virol. 2010, 84, 12934–12944. [Google Scholar] [CrossRef]
- Billingsley, C.L.; Chintala, S.; Katzenellenbogen, R.A. Post-Transcriptional Gene Regulation by HPV 16E6 and Its Host Protein Partners. Viruses 2022, 14, 1483. [Google Scholar] [CrossRef]
- Vliet-Gregg, P.A.; Robinson, K.L.; Levan, J.; Matsumoto, L.R.; Katzenellenbogen, R.A. NFX1-123 is highly expressed in cervical cancer and increases growth and telomerase activity in HPV 16E6 expressing cells. Cancer Lett. 2019, 449, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, D.; Pandey, A.K.; Xiuzhen, M.C.; Lee, K.K.; Hora, S.; Zhang, Y.; Chua, B.H.; Kwok, H.S.; Bhatia, S.S.; Deng, L.W.; et al. TIP60 represses telomerase expression by inhibiting Sp1 binding to the TERT promoter. PLoS Pathog. 2017, 13, e1006681. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Vande Pol, S.; Banerjee, N.S.; Dutta, A.B.; Chow, L.T.; Dutta, A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol. Cell 2010, 38, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, V.K.; Zhang, Y.; Rajagopalan, D.; Abdullah, L.N.; Yeo-Teh, N.S.; Tomaić, V.; Banks, L.; Myers, M.P.; Chow, E.K.; Jha, S. E3 ligase EDD1/UBR5 is utilized by the HPV E6 oncogene to destabilize tumor suppressor TIP60. Oncogene 2016, 35, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Dürst, M.; Croce, C.M.; Gissmann, L.; Schwarz, E.; Huebner, K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. USA 1987, 84, 1070–1074. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Buonaguro, L.; Giorgi-Rossi, P.; Buonaguro, F.M. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed. Res. Int. 2013, 2013, 519619. [Google Scholar] [CrossRef]
- Choo, K.B.; Pan, C.C.; Han, S.H. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: Preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 1987, 161, 259–261. [Google Scholar] [CrossRef]
- Ren, S.; Gaykalova, D.A.; Guo, T.; Favorov, A.V.; Fertig, E.J.; Tamayo, P.; Callejas-Valera, J.L.; Allevato, M.; Gilardi, M.; Santos, J.; et al. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene 2020, 39, 6327–6339. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Ferber, M.J.; Montoya, D.P.; Yu, C.; Aderca, I.; McGee, A.; Thorland, E.C.; Nagorney, D.M.; Gostout, B.S.; Burgart, L.J.; Boix, L.; et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 2003, 22, 3813–3820. [Google Scholar] [CrossRef]
- Reddy, V.G.; Khanna, N.; Jain, S.K.; Das, B.C.; Singh, N. Telomerase—A molecular marker for cervical cancer screening. Int. J. Gynecol. Cancer 2001, 11, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, L.C.; da Silva, I.D.; Corrêa, J.C.; Ribalta, J.C. Survivin and telomerase expression in the uterine cervix of women with human papillomavirus-induced lesions. Int. J. Gynecol. Cancer 2011, 21, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Rampazzo, E.; Polesel, J.; Giunco, S.; Menegaldo, A.; Mantovani, M.; Stellin, M.; Bandolin, L.; Spinato, G.; Del Mistro, A.; et al. Predictive and prognostic significance of telomerase levels/telomere length in tissues and peripheral blood in head and neck squamous cell carcinoma. Sci. Rep. 2019, 9, 17572. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Da Mosto, M.C.; Rampazzo, E.; Giunco, S.; Del Mistro, A.; Menegaldo, A.; Baboci, L.; Mantovani, M.; Tirelli, G.; De Rossi, A. Telomeres and telomerase in head and neck squamous cell carcinoma: From pathogenesis to clinical implications. Cancer Metastasis Rev. 2016, 35, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Rampazzo, E.; Perissinotto, E.; Piano, M.A.; Giunco, S.; Baboci, L.; Spinato, G.; Spinato, R.; Tirelli, G.; Da Mosto, M.C.; et al. Telomere shortening in mucosa surrounding the tumor: Biosensor of field cancerization and prognostic marker of mucosal failure in head and neck squamous cell carcinoma. Oral Oncol. 2015, 51, 500–507. [Google Scholar] [CrossRef]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef]
- Murata, T.; Tsurumi, T. Switching of EBV cycles between latent and lytic states. Rev. Med. Virol. 2014, 24, 142–153. [Google Scholar] [CrossRef]
- Young, L.S.; Murray, P.G. Epstein-Barr virus and oncogenesis: From latent genes to tumours. Oncogene 2003, 22, 5108–5121. [Google Scholar] [CrossRef]
- Chang, M.S.; Kim, W.H. Epstein-Barr virus in human malignancy: A special reference to Epstein-Barr virus associated gastric carcinoma. Cancer Res. Treat. 2005, 37, 257–267. [Google Scholar] [CrossRef]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein-Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 31–46. [Google Scholar] [CrossRef]
- Dolcetti, R.; Dal Col, J.; Martorelli, D.; Carbone, A.; Klein, E. Interplay among viral antigens, cellular pathways and tumor microenvironment in the pathogenesis of EBV-driven lymphomas. Semin. Cancer Biol. 2013, 23, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Giunco, S.; Petrara, M.R.; Zangrossi, M.; Celeghin, A.; De Rossi, A. Extra-telomeric functions of telomerase in the pathogenesis of Epstein-Barr virus-driven B-cell malignancies and potential therapeutic implications. Infect. Agent Cancer 2018, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Giunco, S.; Celeghin, A.; Gianesin, K.; Dolcetti, R.; Indraccolo, S.; De Rossi, A. Cross talk between EBV and telomerase: The role of TERT and NOTCH2 in the switch of latent/lytic cycle of the virus. Cell Death Dis. 2015, 6, e1774. [Google Scholar] [CrossRef]
- Giunco, S.; Dolcetti, R.; Keppel, S.; Celeghin, A.; Indraccolo, S.; Dal Col, J.; Mastorci, K.; De Rossi, A. hTERT inhibition triggers Epstein-Barr virus lytic cycle and apoptosis in immortalized and transformed B cells: A basis for new therapies. Clin. Cancer Res. 2013, 19, 2036–2047. [Google Scholar] [CrossRef]
- Terrin, L.; Dal Col, J.; Rampazzo, E.; Zancai, P.; Pedrotti, M.; Ammirabile, G.; Bergamin, S.; Rizzo, S.; Dolcetti, R.; De Rossi, A. Latent membrane protein 1 of Epstein-Barr virus activates the hTERT promoter and enhances telomerase activity in B lymphocytes. J. Virol. 2008, 82, 10175–10187. [Google Scholar] [CrossRef]
- Mei, Y.P.; Zhu, X.F.; Zhou, J.M.; Huang, H.; Deng, R.; Zeng, Y.X. siRNA targeting LMP1-induced apoptosis in EBV-positive lymphoma cells is associated with inhibition of telomerase activity and expression. Cancer Lett. 2006, 232, 189–198. [Google Scholar] [CrossRef]
- Ding, L.; Li, L.L.; Yang, J.; Tao, Y.G.; Ye, M.; Shi, Y.; Tang, M.; Yi, W.; Li, X.L.; Gong, J.P.; et al. Epstein-Barr virus encoded latent membrane protein 1 modulates nuclear translocation of telomerase reverse transcriptase protein by activating nuclear factor-kappaB p65 in human nasopharyngeal carcinoma cells. Int. J. Biochem. Cell Biol. 2005, 37, 1881–1889. [Google Scholar] [CrossRef]
- Chen, F.; Liu, C.; Lindvall, C.; Xu, D.; Ernberg, I. Epstein-Barr virus latent membrane 2A (LMP2A) down-regulates telomerase reverse transcriptase (hTERT) in epithelial cell lines. Int. J. Cancer 2005, 113, 284–289. [Google Scholar] [CrossRef]
- Edelman, D.C. Human herpesvirus 8—A novel human pathogen. Virol. J. 2005, 2, 78. [Google Scholar] [CrossRef]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Buonaguro, F.M.; Tornesello, M.L.; Beth-Giraldo, E.; Hatzakis, A.; Mueller, N.; Downing, R.; Biryamwaho, B.; Sempala, S.D.; Giraldo, G. Herpesvirus-like DNA sequences detected in endemic, classic, iatrogenic and epidemic Kaposi’s sarcoma (KS) biopsies. Int. J. Cancer 1996, 65, 25–28. [Google Scholar] [CrossRef]
- Ruocco, E.; Ruocco, V.; Tornesello, M.L.; Gambardella, A.; Wolf, R.; Buonaguro, F.M. Kaposi’s sarcoma: Etiology and pathogenesis, inducing factors, causal associations, and treatments: Facts and controversies. Clin. Dermatol. 2013, 31, 413–422. [Google Scholar] [CrossRef]
- Grulich, A.E.; van Leeuwen, M.T.; Falster, M.O.; Vajdic, C.M. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: A meta-analysis. Lancet 2007, 370, 59–67. [Google Scholar] [CrossRef]
- Ibrahim Khalil, A.; Franceschi, S.; de Martel, C.; Bray, F.; Clifford, G.M. Burden of Kaposi sarcoma according to HIV status: A systematic review and global analysis. Int. J. Cancer 2022, 150, 1948–1957. [Google Scholar] [CrossRef]
- Chen, Z.; Smith, K.J.; Skelton, H.G., 3rd; Barrett, T.L.; Greenway, H.T., Jr.; Lo, S.C. Telomerase activity in Kaposi’s sarcoma, squamous cell carcinoma, and basal cell carcinoma. Exp. Biol. Med. 2001, 226, 753–757. [Google Scholar] [CrossRef]
- Neipel, F.; Fleckenstein, B. The role of HHV-8 in Kaposi’s sarcoma. Semin. Cancer Biol. 1999, 9, 151–164. [Google Scholar] [CrossRef]
- Suzuki, T.; Isobe, T.; Kitagawa, M.; Ueda, K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA positively affects on ubiquitylation of p53. Biochem. Biophys. Res. Commun. 2010, 403, 194–197. [Google Scholar] [CrossRef]
- Radkov, S.A.; Kellam, P.; Boshoff, C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 2000, 6, 1121–1127. [Google Scholar] [CrossRef]
- Si, H.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 2006, 80, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Flore, O.; Rafii, S.; Ely, S.; O’Leary, J.J.; Hyjek, E.M.; Cesarman, E. Transformation of primary human endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Nature 1998, 394, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Borah, S.; Robertson, E.S. Latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 2004, 78, 10348–10359. [Google Scholar] [CrossRef] [PubMed]
- Shamay, M.; Liu, J.; Li, R.; Liao, G.; Shen, L.; Greenway, M.; Hu, S.; Zhu, J.; Xie, Z.; Ambinder, R.F.; et al. A protein array screen for Kaposi’s sarcoma-associated herpesvirus LANA interactors links LANA to TIP60, PP2A activity, and telomere shortening. J. Virol. 2012, 86, 5179–5191. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Sato, K.; Kibe, T.; Seimiya, H.; Nakamura, A.; Yukawa, M.; Tsuchiya, E.; Ueno, M. Expression of mutant RPA in human cancer cells causes telomere shortening. Biosci. Biotechnol. Biochem. 2010, 74, 382–385. [Google Scholar] [CrossRef]
- Lippert, T.P.; Marzec, P.; Idilli, A.I.; Sarek, G.; Vancevska, A.; Bower, M.; Farrell, P.J.; Ojala, P.M.; Feldhahn, N.; Boulton, S.J. Oncogenic herpesvirus KSHV triggers hallmarks of alternative lengthening of telomeres. Nat. Commun. 2021, 12, 512. [Google Scholar] [CrossRef]
- McClain, S.L.; Clippinger, A.J.; Lizzano, R.; Bouchard, M.J. Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels. J. Virol. 2007, 81, 12061–12065. [Google Scholar] [CrossRef]
- Venook, A.P.; Papandreou, C.; Furuse, J.; de Guevara, L.L. The incidence and epidemiology of hepatocellular carcinoma: A global and regional perspective. Oncologist 2010, 15 (Suppl. S4), 5–13. [Google Scholar] [CrossRef]
- Nicolini, L.A.; Orsi, A.; Tatarelli, P.; Viscoli, C.; Icardi, G.; Sticchi, L. A Global View to HBV Chronic Infection: Evolving Strategies for Diagnosis, Treatment and Prevention in Immunocompetent Individuals. Int. J. Environ. Res. Public Health 2019, 16, 3307. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Law, M.G.; Dore, G.J. Hepatitis B-related hepatocellular carcinoma: Epidemiological characteristics and disease burden. J. Viral Hepat 2009, 16, 453–463. [Google Scholar] [CrossRef]
- Nault, J.C.; Ningarhari, M.; Rebouissou, S.; Zucman-Rossi, J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 544–558. [Google Scholar] [CrossRef] [PubMed]
- In der Stroth, L.; Tharehalli, U.; Günes, C.; Lechel, A. Telomeres and Telomerase in the Development of Liver Cancer. Cancers 2020, 12, 2048. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.W.; Kim, J.S.; Kim, H.S.; Tak, K.Y.; Lee, S.K.; Nam, H.C.; Sung, P.S.; Kim, C.M.; Park, J.Y.; Bae, S.H.; et al. Significance of TERT Genetic Alterations and Telomere Length in Hepatocellular Carcinoma. Cancers 2021, 13, 2160. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, F.; Izzo, F.; De Luca, P.; Biffali, E.; Buonaguro, L.; Tatangelo, F.; Buonaguro, F.M.; Tornesello, M.L. Clinical Significance of Telomerase Reverse-Transcriptase Promoter Mutations in Hepatocellular Carcinoma. Cancers 2021, 13, 3771. [Google Scholar] [CrossRef]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef]
- Paterlini-Bréchot, P.; Saigo, K.; Murakami, Y.; Chami, M.; Gozuacik, D.; Mugnier, C.; Lagorce, D.; Bréchot, C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003, 22, 3911–3916. [Google Scholar] [CrossRef]
- Jang, J.W.; Kim, H.S.; Kim, J.S.; Lee, S.K.; Han, J.W.; Sung, P.S.; Bae, S.H.; Choi, J.Y.; Yoon, S.K.; Han, D.J.; et al. Distinct Patterns of HBV Integration and TERT Alterations between in Tumor and Non-Tumor Tissue in Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 7056. [Google Scholar] [CrossRef]
- Liu, H.; Shi, W.; Luan, F.; Xu, S.; Yang, F.; Sun, W.; Liu, J.; Ma, C. Hepatitis B virus X protein upregulates transcriptional activation of human telomerase reverse transcriptase. Virus Genes 2010, 40, 174–182. [Google Scholar] [CrossRef]
- Luan, F.; Liu, H.; Gao, L.; Liu, J.; Sun, Z.; Ju, Y.; Hou, N.; Guo, C.; Liang, X.; Zhang, L.; et al. Hepatitis B virus protein preS2 potentially promotes HCC development via its transcriptional activation of hTERT. Gut 2009, 58, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wu, W.; Wan, H.; Wu, X.; Chen, H. Knockdown of NHP2 inhibits hepatitis B virus X protein-induced hepatocarcinogenesis via repressing TERT expression and disrupting the stability of telomerase complex. Aging 2020, 12, 19365–19374. [Google Scholar] [CrossRef] [PubMed]
- Guterres, A.N.; Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Tabata, K.; Neufeldt, C.J.; Bartenschlager, R. Hepatitis C Virus Replication. Cold Spring Harb. Perspect. Med. 2020, 10, a037093. [Google Scholar] [CrossRef] [PubMed]
- Gower, E.; Estes, C.; Blach, S.; Razavi-Shearer, K.; Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 2014, 61, S45–S57. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Reimer, U.; Holenya, P.; Knaute, T.; Pezzuto, F.; Izzo, F.; Buonaguro, L.; Megna, A.S.; Buonaguro, F.M.; Tornesello, M.L. Profiling the HCV Immune Response in Patients with Chronic Liver Diseases and Hepatocellular Carcinoma by Peptide Microarray Analysis. Curr. Med. Chem. 2022, 29, 2736–2747. [Google Scholar] [CrossRef]
- Vedham, V.; Verma, M.; Mahabir, S. Early-life exposures to infectious agents and later cancer development. Cancer Med. 2015, 4, 1908–1922. [Google Scholar] [CrossRef]
- Zhu, Z.; Wilson, A.T.; Gopalakrishna, K.; Brown, K.E.; Luxon, B.A.; Schmidt, W.N. Hepatitis C virus core protein enhances Telomerase activity in Huh7 cells. J. Med. Virol. 2010, 82, 239–248. [Google Scholar] [CrossRef]
- Shiu, T.Y.; Shih, Y.L.; Feng, A.C.; Lin, H.H.; Huang, S.M.; Huang, T.Y.; Hsieh, C.B.; Chang, W.K.; Hsieh, T.Y. HCV core inhibits hepatocellular carcinoma cell replicative senescence through downregulating microRNA-138 expression. J. Mol. Med. 2017, 95, 629–639. [Google Scholar] [CrossRef]
- Zhu, Z.; Tran, H.; Mathahs, M.M.; Moninger, T.O.; Schmidt, W.N. HCV Induces Telomerase Reverse Transcriptase, Increases Its Catalytic Activity, and Promotes Caspase Degradation in Infected Human Hepatocytes. PLoS ONE 2017, 12, e0166853. [Google Scholar] [CrossRef]
- Ningarhari, M.; Caruso, S.; Hirsch, T.Z.; Bayard, Q.; Franconi, A.; Vedie, A.L.; Noblet, B.; Blanc, J.F.; Amaddeo, G.; Ganne, N.; et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J. Hepatol. 2021, 74, 1155–1166. [Google Scholar] [CrossRef]
- Verdonck, K.; González, E.; Van Dooren, S.; Vandamme, A.M.; Vanham, G.; Gotuzzo, E. Human T-lymphotropic virus 1: Recent knowledge about an ancient infection. Lancet Infect. Dis. 2007, 7, 266–281. [Google Scholar] [CrossRef]
- Coffin, J.M. The discovery of HTLV-1, the first pathogenic human retrovirus. Proc. Natl. Acad. Sci. USA 2015, 112, 15525–15529. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi Ghezeldasht, S.; Shirdel, A.; Assarehzadegan, M.A.; Hassannia, T.; Rahimi, H.; Miri, R.; Rezaee, S.A. Human T Lymphotropic Virus Type I (HTLV-I) Oncogenesis: Molecular Aspects of Virus and Host Interactions in Pathogenesis of Adult T cell Leukemia/Lymphoma (ATL). Iran. J. Basic Med. Sci. 2013, 16, 179–195. [Google Scholar]
- Bellon, M.; Datta, A.; Brown, M.; Pouliquen, J.F.; Couppie, P.; Kazanji, M.; Nicot, C. Increased expression of telomere length regulating factors TRF1, TRF2 and TIN2 in patients with adult T-cell leukemia. Int. J. Cancer 2006, 119, 2090–2097. [Google Scholar] [CrossRef]
- Kubuki, Y.; Suzuki, M.; Sasaki, H.; Toyama, T.; Yamashita, K.; Maeda, K.; Ido, A.; Matsuoka, H.; Okayama, A.; Nakanishi, T.; et al. Telomerase activity and telomere length as prognostic factors of adult T-cell leukemia. Leuk. Lymphoma 2005, 46, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Otsuka, T.; Arima, F.; Shigematsu, H.; Fukuyama, T.; Maeda, M.; Sugio, Y.; Itoh, Y.; Niho, Y. Correlation of telomerase activity with development and progression of adult T-cell leukemia. Leuk. Res. 1999, 23, 311–316. [Google Scholar] [CrossRef]
- Sinha-Datta, U.; Horikawa, I.; Michishita, E.; Datta, A.; Sigler-Nicot, J.C.; Brown, M.; Kazanji, M.; Barrett, J.C.; Nicot, C. Transcriptional activation of hTERT through the NF-kappaB pathway in HTLV-I-transformed cells. Blood 2004, 104, 2523–2531. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.K.; Lundblad, V. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 2000, 113 Pt 19, 3357–3364. [Google Scholar] [CrossRef]
- Smogorzewska, A.; van Steensel, B.; Bianchi, A.; Oelmann, S.; Schaefer, M.R.; Schnapp, G.; de Lange, T. Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 2000, 20, 1659–1668. [Google Scholar] [CrossRef]
- Terme, J.M.; Mocquet, V.; Kuhlmann, A.S.; Zane, L.; Mortreux, F.; Wattel, E.; Duc Dodon, M.; Jalinot, P. Inhibition of the hTERT promoter by the proto-oncogenic protein TAL1. Leukemia 2009, 23, 2081–2089. [Google Scholar] [CrossRef]
- Borowiak, M.; Kuhlmann, A.S.; Girard, S.; Gazzolo, L.; Mesnard, J.M.; Jalinot, P.; Dodon, M.D. HTLV-1 bZIP factor impedes the menin tumor suppressor and upregulates JunD-mediated transcription of the hTERT gene. Carcinogenesis 2013, 34, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Zane, L.; Sibon, D.; Capraro, V.; Galia, P.; Karam, M.; Delfau-Larue, M.H.; Gilson, E.; Gessain, A.; Gout, O.; Hermine, O.; et al. HTLV-1 positive and negative T cells cloned from infected individuals display telomerase and telomere genes deregulation that predominate in activated but untransformed CD4+ T cells. Int. J. Cancer 2012, 131, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Dolcetti, R.; De Rossi, A. Telomere/telomerase interplay in virus-driven and virus-independent lymphomagenesis: Pathogenic and clinical implications. Med. Res. Rev. 2012, 32, 233–253. [Google Scholar] [CrossRef]
- Valenzuela, H.F.; Effros, R.B. Divergent telomerase and CD28 expression patterns in human CD4 and CD8 T cells following repeated encounters with the same antigenic stimulus. Clin. Immunol. 2002, 105, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, Q.; Wang, Y.; Zhuang, Y.; Xu, L.; Ma, X.; Guan, D.; Zhou, J.; Liu, J.; Wu, X.; et al. TERT activates endogenous retroviruses to promote an immunosuppressive tumour microenvironment. EMBO Rep. 2022, 23, e52984. [Google Scholar] [CrossRef]
- Krump, N.A.; You, J. Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Microbiol. 2018, 16, 684–698. [Google Scholar] [CrossRef]
- Van Baarle, D.; Nanlohy, N.M.; Otto, S.; Plunkett, F.J.; Fletcher, J.M.; Akbar, A.N. Progressive telomere shortening of Epstein-Barr virus-specific memory T cells during HIV infection: Contributor to exhaustion? J. Infect. Dis. 2008, 198, 1353–1357. [Google Scholar] [CrossRef]
- Van de Berg, P.J.; Griffiths, S.J.; Yong, S.L.; Macaulay, R.; Bemelman, F.J.; Jackson, S.; Henson, S.M.; ten Berge, I.J.; Akbar, A.N.; van Lier, R.A. Cytomegalovirus infection reduces telomere length of the circulating T cell pool. J. Immunol. 2010, 184, 3417–3423. [Google Scholar] [CrossRef]
- Venuti, A.; Romero-Medina, M.C.; Melita, G.; Ceraolo, M.G.; Brancaccio, R.N.; Sirand, C.; Taverniti, V.; Steenbergen, R.; Gheit, T.; Tommasino, M. Lyon IARC Polyomavirus Displays Transforming Activities in Primary Human Cells. J. Virol. 2022, 96, e0206121. [Google Scholar] [CrossRef]
- Sharma, S.; Chowdhury, S. Emerging mechanisms of telomerase reactivation in cancer. Trends Cancer 2022, 8, 632–641. [Google Scholar] [CrossRef]
| Oncovirus | Cancer | Viral Protein | Mechanisms of Interaction with Telomerase Function | Reference |
|---|---|---|---|---|
| HPV | Cervix, anus, vagina, vulva, penis, head and neck | E6 |
| [9,64] |
| [65,70,71,72,73] | |||
| [74,75,76,77] | |||
| [79,80] | |||
| [77] | |||
| [82] | |||
| [61] | |||
| E7 |
| [65] | ||
| EBV | Burkitt’s lymphoma, nasopharyngeal carcinoma | LMP1 |
| [106,107] |
| [108] | |||
| LMP2A |
| [109] | ||
| HHV8 | Kaposi’s sarcoma, multicentric Castleman’s disease, primary effusion lymphoma | LANA |
| [8] |
| [124] | |||
| ? |
| [126] | ||
| HBV | Hepatocellular carcinoma | HBx |
| [140] |
| preS2 |
| [141] | ||
| HCV | Hepatocellular carcinoma and lymphoproliferative disorders | core |
| [148] |
| [149] | |||
| NS3-4A |
| [150] | ||
| HTLV1 | T-cell leukemia and HTLV1-associated myelopathy/tropical spastic paraparesis | Tax |
| [158] |
| [158] | |||
| HBZ |
| [162] | ||
| [162] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tornesello, M.L.; Cerasuolo, A.; Starita, N.; Tornesello, A.L.; Bonelli, P.; Tuccillo, F.M.; Buonaguro, L.; Isaguliants, M.G.; Buonaguro, F.M. The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development. Cancers 2022, 14, 5257. https://doi.org/10.3390/cancers14215257
Tornesello ML, Cerasuolo A, Starita N, Tornesello AL, Bonelli P, Tuccillo FM, Buonaguro L, Isaguliants MG, Buonaguro FM. The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development. Cancers. 2022; 14(21):5257. https://doi.org/10.3390/cancers14215257
Chicago/Turabian StyleTornesello, Maria Lina, Andrea Cerasuolo, Noemy Starita, Anna Lucia Tornesello, Patrizia Bonelli, Franca Maria Tuccillo, Luigi Buonaguro, Maria G. Isaguliants, and Franco M. Buonaguro. 2022. "The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development" Cancers 14, no. 21: 5257. https://doi.org/10.3390/cancers14215257
APA StyleTornesello, M. L., Cerasuolo, A., Starita, N., Tornesello, A. L., Bonelli, P., Tuccillo, F. M., Buonaguro, L., Isaguliants, M. G., & Buonaguro, F. M. (2022). The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development. Cancers, 14(21), 5257. https://doi.org/10.3390/cancers14215257