CroReLU: Cross-Crossing Space-Based Visual Activation Function for Lung Cancer Pathology Image Recognition
Abstract
Simple Summary
Abstract
1. Introduction
- (1)
- The forced sparse processing of the ReLU AF reduces the effective capacity of the model, and the clearing of negative gradient values at x < 0 may result in neurons that are no longer activated by any data, leading to neuron ‘necrosis’.
- (2)
- The ReLU AF is not specifically used for computer vision tasks, no information about adjacent features is noted, and the AF is not spatially sensitive.
- (3)
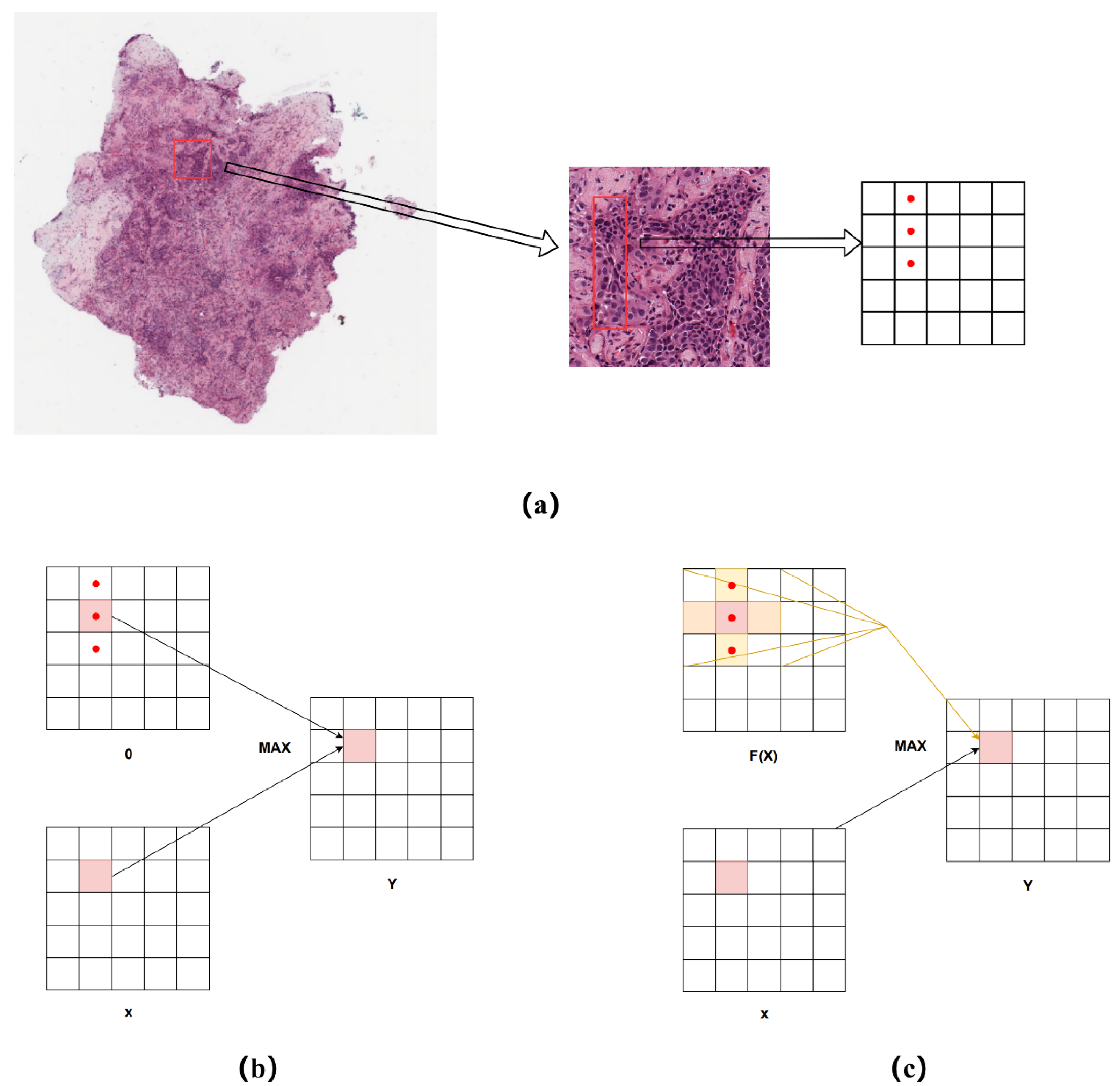
- Most of the pathological features of lung cancer show tubular morphology such as papillae, micropapillae, and apposition, which the ReLU AF may not be able to capture.
- (1)
- A novel AF called CroReLU is designed based on prior knowledge of pathology; it has the ability to model crossed spaces, and can effectively capture histological shape features such as lung cancer blisters, papillae, and micropapillae without changing the model network layer structure.
- (2)
- The proposed method uses a plug-and-play visual activation that can be applied to any state-of-the-art computer vision model for image analysis-related tasks.
- (3)
- A digital pathology image dataset for lung cancer infiltration level detection was prepared by a pathologist, and the experimental results demonstrate that CroReLU can sensitively capture infiltrative and microinfiltrative features, and possesses the potential to solve practical clinical tasks.
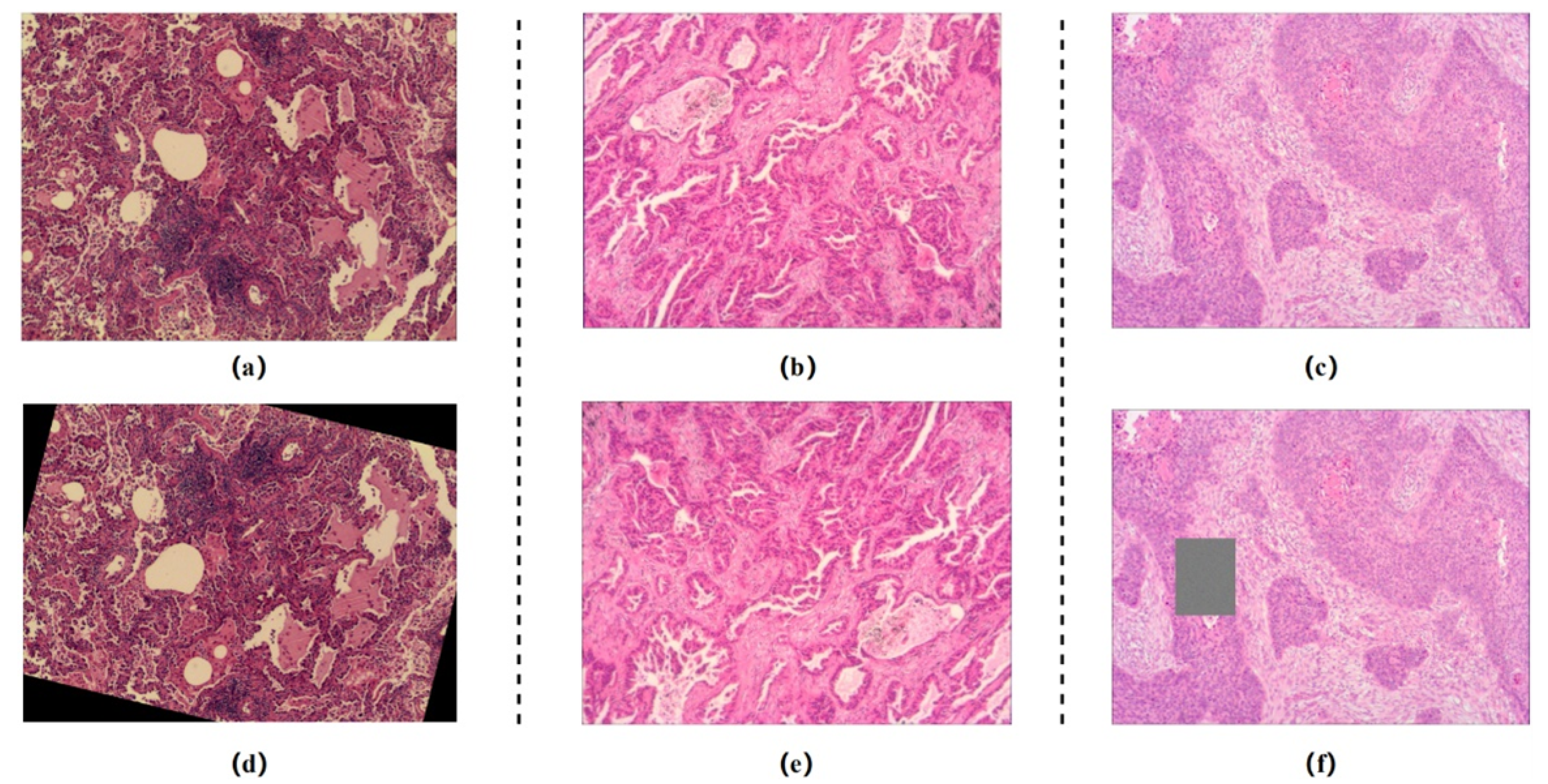
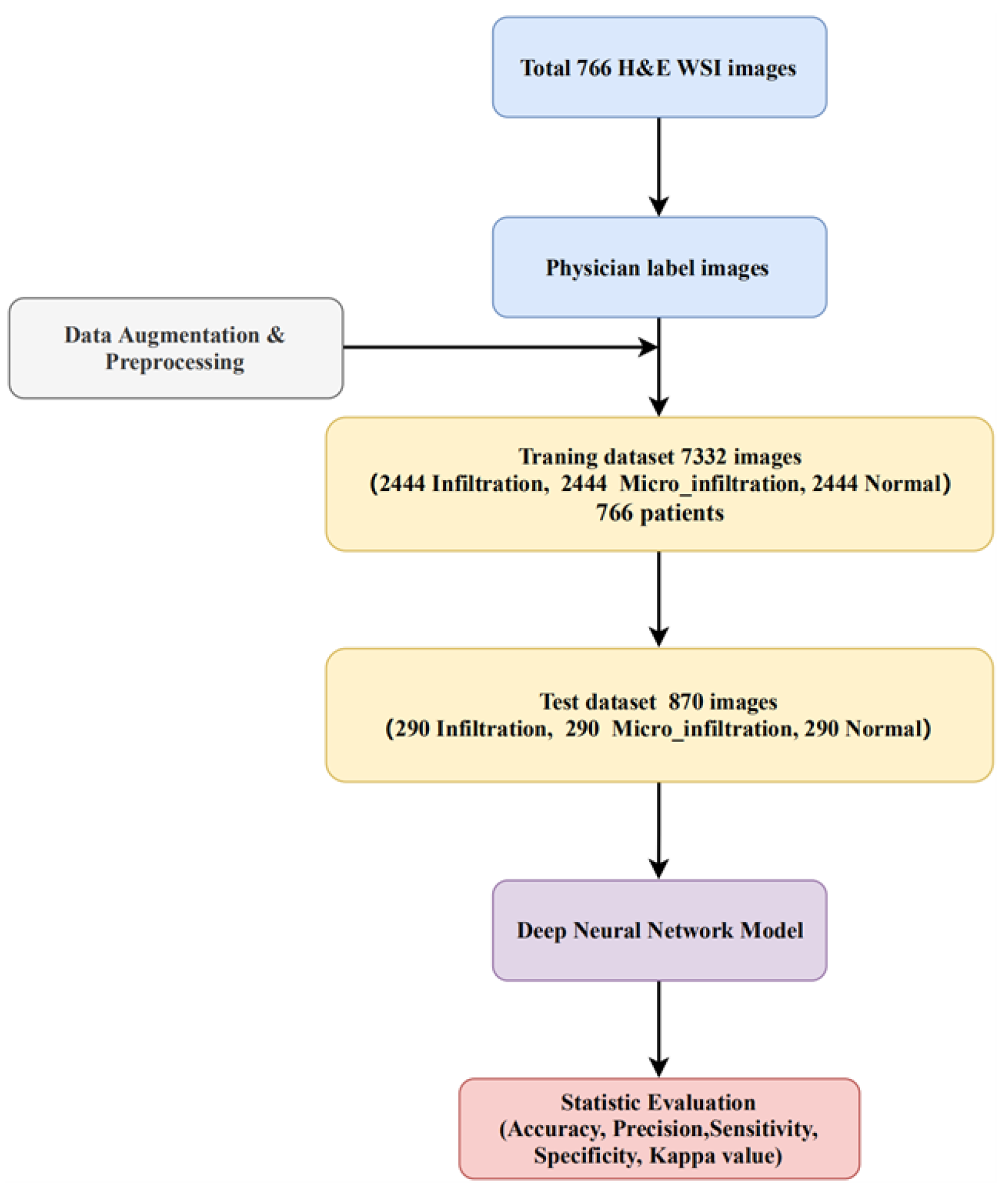
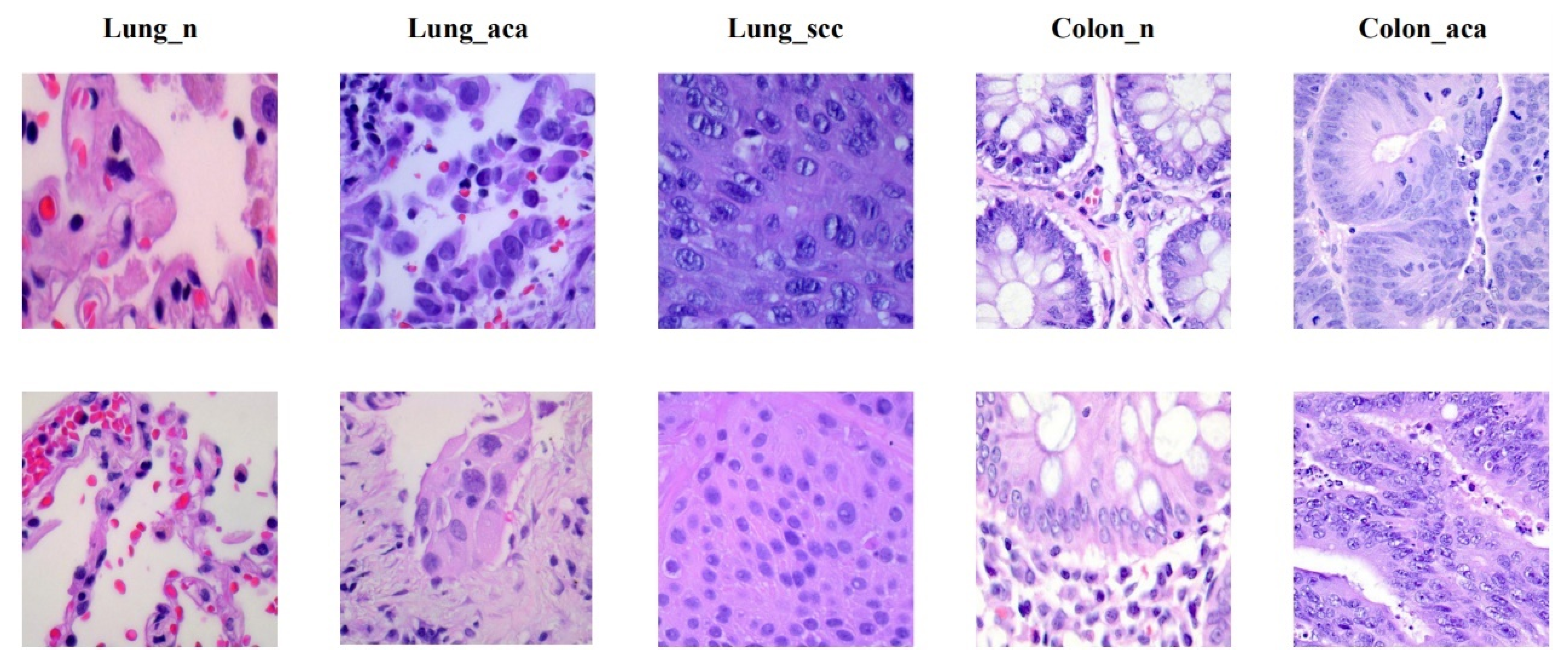
2. Dataset
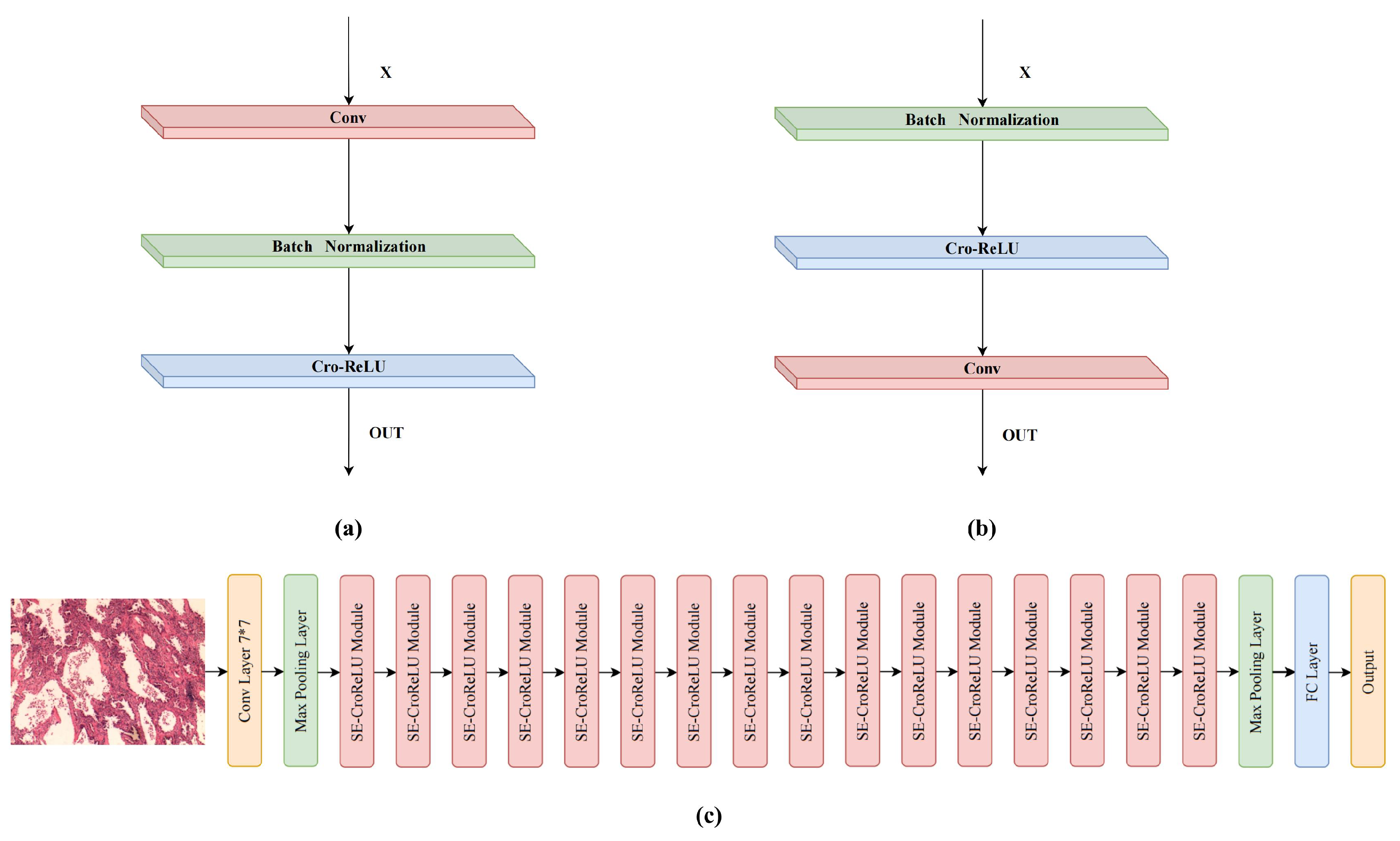
3. Neural Network Model
3.1. ReLU
3.2. CroReLU
4. Experiment
4.1. Data Augmention and Experimental Setup
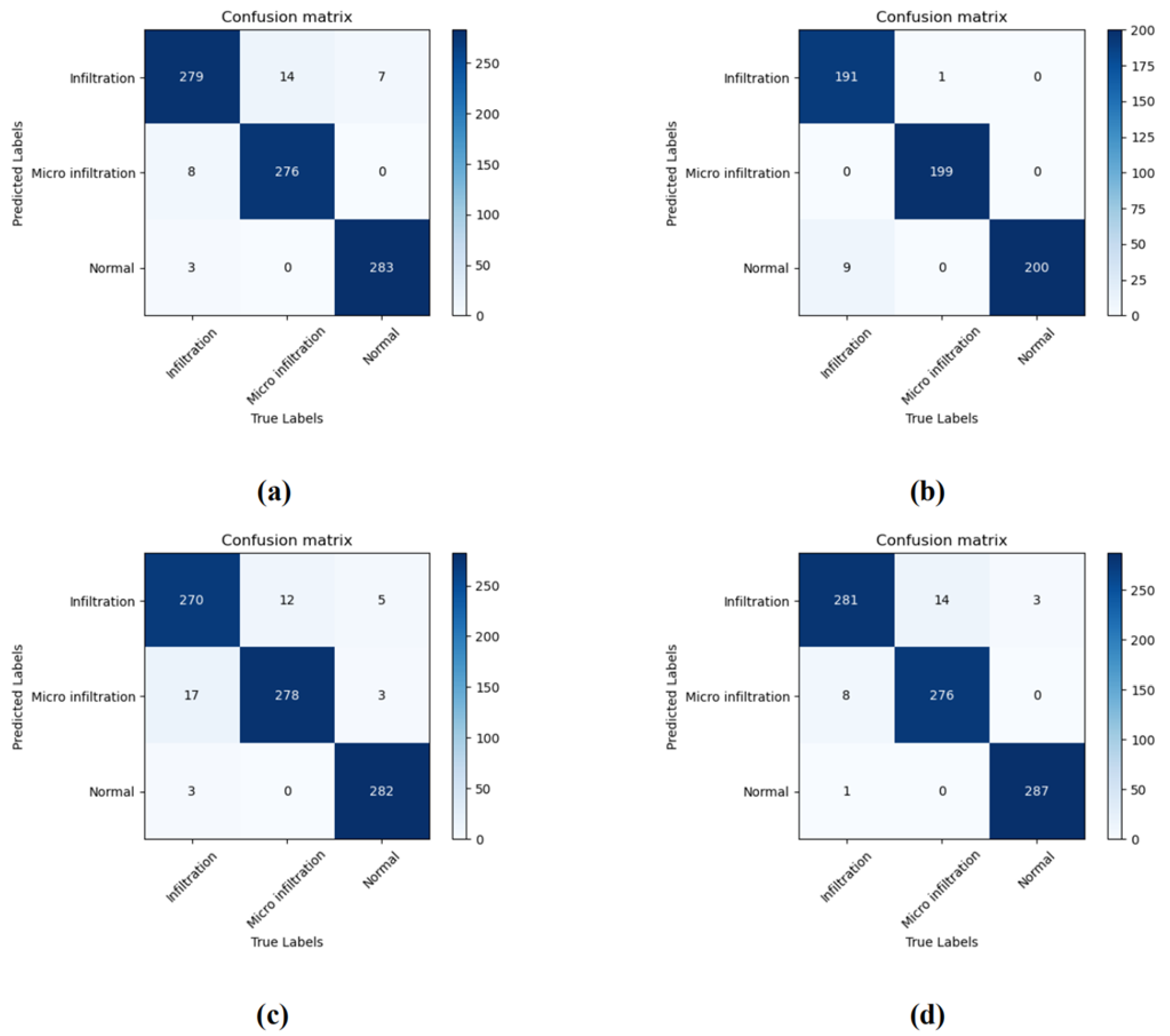
4.2. Experimental Results on a Private Dataset
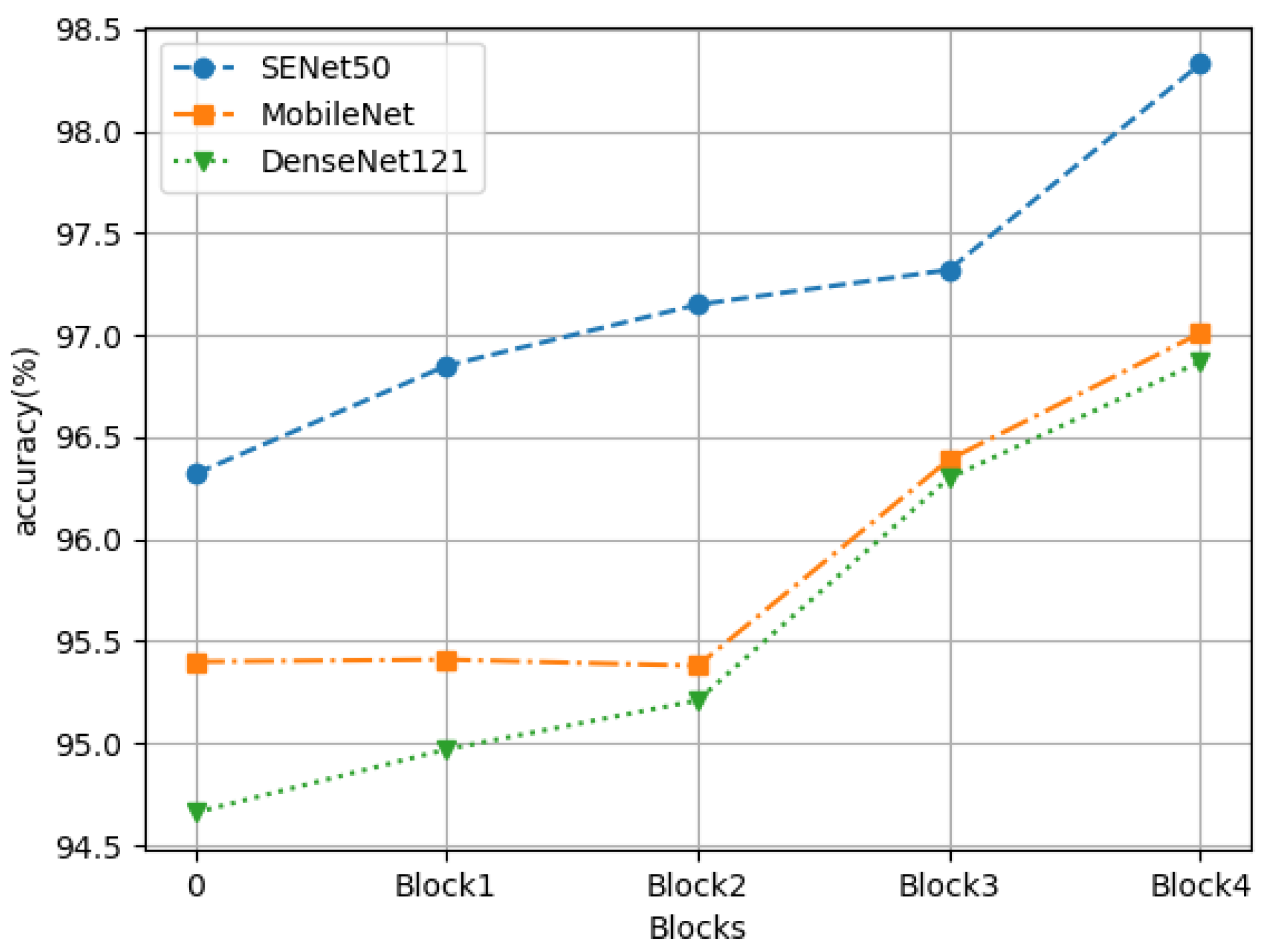
4.3. Ablation Experiment
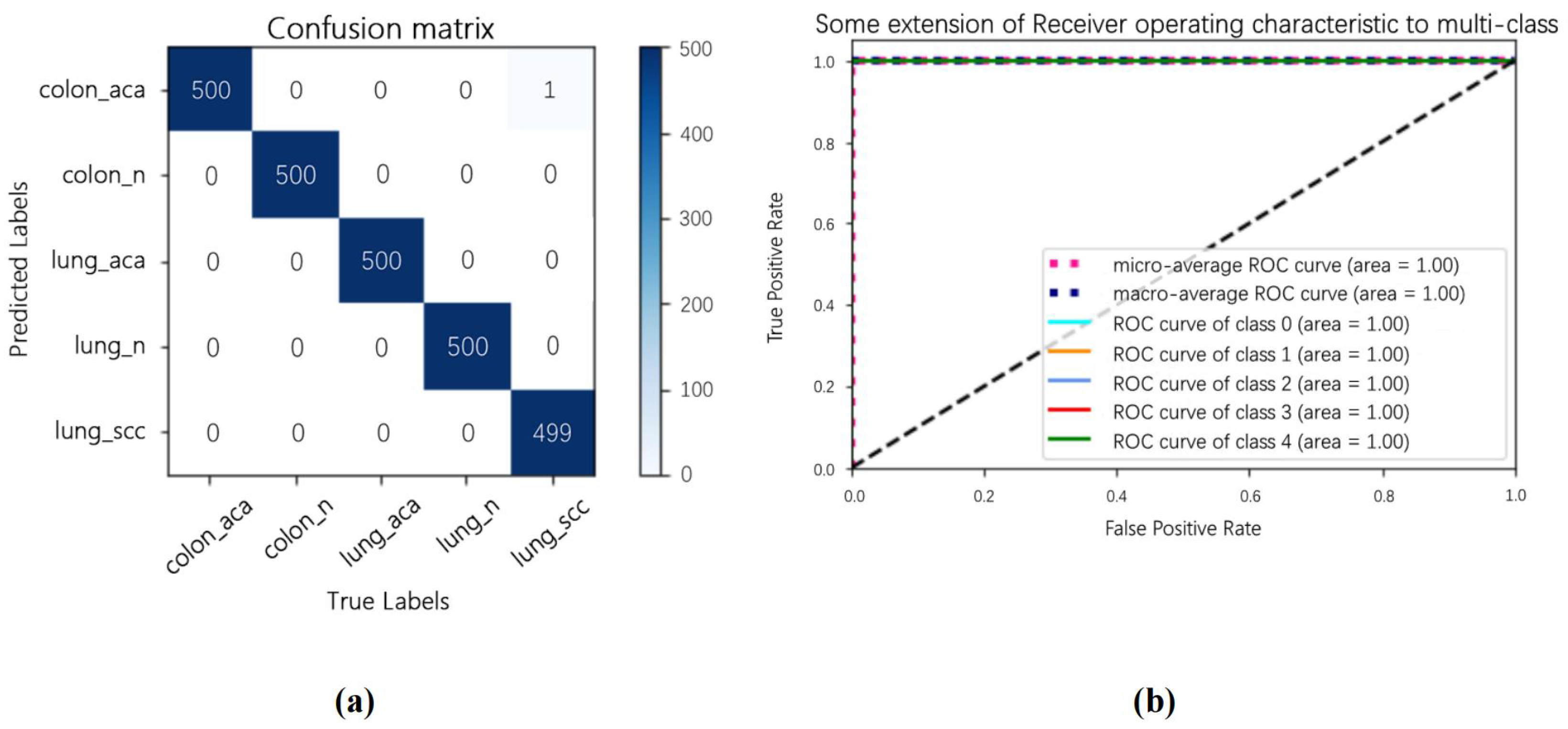
4.4. Extended Experiment
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Yang, D.M.; Rong, R.; Zhan, X.; Fujimoto, J.; Liu, H.; Minna, J.; Wistuba, I.I.; Xie, Y.; Xiao, G. Artificial intelligence in lung cancer pathology image analysis. Cancers 2019, 11, 1673. [Google Scholar] [CrossRef]
- Jara-Lazaro, A.R.; Thamboo, T.P.; Teh, M.; Tan, P.H. Digital pathology: Exploring its applications in diagnostic surgical pathology practice. Pathology 2010, 42, 512–518. [Google Scholar] [CrossRef]
- Shafiei, S.; Safarpoor, A.; Jamalizadeh, A.; Tizhoosh, H.R. Class-agnostic weighted normalization of staining in histopathology images using a spatially constrained mixture model. IEEE Trans. Med. Imaging 2020, 39, 3355–3366. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Anthimopoulos, M.; Christodoulidis, S.; Ebner, L.; Christe, A.; Mougiakakou, S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging 2016, 35, 1207–1216. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Nguyen, E.H.; Yang, H.; Deng, R.; Lu, Y.; Zhu, Z.; Roland, J.T.; Lu, L.; Landman, B.A.; Fogo, A.B.; Huo, Y. Circle Representation for Medical Object Detection. IEEE Trans. Med. Imaging 2021, 41, 746–754. [Google Scholar] [CrossRef]
- Bellver Bueno, M.; Salvador Aguilera, A.; Torres Viñals, J.; Giró Nieto, X. Budget-aware semi-supervised semantic and instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Long Beach, CA, USA, 15–20 June 2019; pp. 93–102. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Afouras, T.; Chung, J.S.; Senior, A.; Vinyals, O.; Zisserman, A. Deep audio-visual speech recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2018. [Google Scholar] [CrossRef]
- Heaton, J. Ian goodfellow, yoshua bengio, and aaron courville: Deep learning. Genet. Program. Evolvable Mach. 2018, 19, 305–307. [Google Scholar] [CrossRef]
- Harrington, P.d.B. Sigmoid transfer functions in backpropagation neural networks. Anal. Chem. 1993, 65, 2167–2168. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. Handb. Brain Theory Neural Netw. 1995, 3361, 1995. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Xu, X.; Hou, R.; Zhao, W.; Teng, H.; Sun, J.; Zhao, J. A weak supervision-based framework for automatic lung cancer classification on whole slide image. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Montreal, QC, Canada, 20–24 July 2020; pp. 1372–1375. [Google Scholar]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Adu, K.; Yu, Y.; Cai, J.; Owusu-Agyemang, K.; Twumasi, B.A.; Wang, X. DHS-CapsNet: Dual horizontal squash capsule networks for lung and colon cancer classification from whole slide histopathological images. Int. J. Imaging Syst. Technol. 2021, 31, 2075–2092. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Gao, Y.; Zhang, H.; Guan, Z.; Dong, Z.; Zheng, Y.; Jiang, J.; Yang, H.; Wang, L.; et al. Predicting gastric cancer outcome from resected lymph node histopathology images using deep learning. Nat. Commun. 2021, 12, 1637. [Google Scholar] [CrossRef]
- Khosravi, P.; Kazemi, E.; Imielinski, M.; Elemento, O.; Hajirasouliha, I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. eBioMedicine 2018, 27, 317–328. [Google Scholar] [CrossRef]
- Riasatian, A.; Babaie, M.; Maleki, D.; Kalra, S.; Valipour, M.; Hemati, S.; Zaveri, M.; Safarpoor, A.; Shafiei, S.; Afshari, M.; et al. Fine-tuning and training of densenet for histopathology image representation using tcga diagnostic slides. Med. Image Anal. 2021, 70, 102032. [Google Scholar] [CrossRef]
- Ding, X.; Guo, Y.; Ding, G.; Han, J. Acnet: Strengthening the kernel skeletons for powerful cnn via asymmetric convolution blocks. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 1911–1920. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and colon cancer histopathological image dataset (lc25000). arXiv 2019, arXiv:1912.12142. [Google Scholar]
- Masud, M.; Sikder, N.; Nahid, A.A.; Bairagi, A.K.; AlZain, M.A. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors 2021, 21, 748. [Google Scholar] [CrossRef]
- Mangal, S.; Chaurasia, A.; Khajanchi, A. Convolution Neural Networks for diagnosing colon and lung cancer histopathological images. arXiv 2020, arXiv:2009.03878. [Google Scholar]
- Hatuwal, B.K.; Thapa, H.C. Lung cancer detection using convolutional neural network on histopathological images. Int. J. Comput. Trends Technol. 2020, 68, 21–24. [Google Scholar] [CrossRef]
| Methods | Accuracy | Precision | Sensitivity |
|---|---|---|---|
| SENet50 | 96.32 | 96.51 | 96.33 |
| SENet50_CroReLU | 98.33 | 98.38 | 98.35 |
| MobileNet | 95.40 | 95.46 | 95.42 |
| MobileNet_CroReLU | 97.01 | 97.07 | 97.04 |
| Methods | Accuracy | Parameters | Test Time |
|---|---|---|---|
| SENet | 96.32 | 25.5 M | 0.372 |
| SENet_3 × 3 | 98.33 | 26.1 M | 0.393 |
| SENet_5 × 5 | 97.26 | 26.5 M | 0.478 |
| SENet_7 × 7 | 97.09 | 27.2 M | 0.505 |
| Image Type | Train | Test | Sum |
|---|---|---|---|
| Lung_n | 4500 | 500 | 5000 |
| Lung_aca | 4500 | 500 | 5000 |
| Lung_scc | 4500 | 500 | 5000 |
| Colon_n | 4500 | 500 | 5000 |
| Colon_aca | 4500 | 500 | 5000 |
| Authors | Accuracy(%) | Precision(%) | Sensitivity(%) | Remark |
|---|---|---|---|---|
| Masud M. et al. [29] | 96.33 | 96.39 | 96.37 | inter-class recognition |
| Mangal S. et al. [30] | Lung: 97.89 Colon: 96.61 | - | - | intra-class recognition |
| B.K.Hatuwal et al. [31] | 97.20 | 97.33 | 97.33 | intra-class recognition |
| Proposed | 99.96 | 99.87 | 99.86 | inter-class recognition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, H.; Song, K.; Sun, M.; Shao, Y.; Xue, S.; Li, L.; Li, Y.; Cai, H.; Jiao, Y.; et al. CroReLU: Cross-Crossing Space-Based Visual Activation Function for Lung Cancer Pathology Image Recognition. Cancers 2022, 14, 5181. https://doi.org/10.3390/cancers14215181
Liu Y, Wang H, Song K, Sun M, Shao Y, Xue S, Li L, Li Y, Cai H, Jiao Y, et al. CroReLU: Cross-Crossing Space-Based Visual Activation Function for Lung Cancer Pathology Image Recognition. Cancers. 2022; 14(21):5181. https://doi.org/10.3390/cancers14215181
Chicago/Turabian StyleLiu, Yunpeng, Haoran Wang, Kaiwen Song, Mingyang Sun, Yanbin Shao, Songfeng Xue, Liyuan Li, Yuguang Li, Hongqiao Cai, Yan Jiao, and et al. 2022. "CroReLU: Cross-Crossing Space-Based Visual Activation Function for Lung Cancer Pathology Image Recognition" Cancers 14, no. 21: 5181. https://doi.org/10.3390/cancers14215181
APA StyleLiu, Y., Wang, H., Song, K., Sun, M., Shao, Y., Xue, S., Li, L., Li, Y., Cai, H., Jiao, Y., Sun, N., Liu, M., & Zhang, T. (2022). CroReLU: Cross-Crossing Space-Based Visual Activation Function for Lung Cancer Pathology Image Recognition. Cancers, 14(21), 5181. https://doi.org/10.3390/cancers14215181





