Glioblastoma-Derived Three-Dimensional Ex Vivo Models to Evaluate Effects and Efficacy of Tumor Treating Fields (TTFields)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Patient-Derived Primary Cells, Cell Lines, and Cell Culture
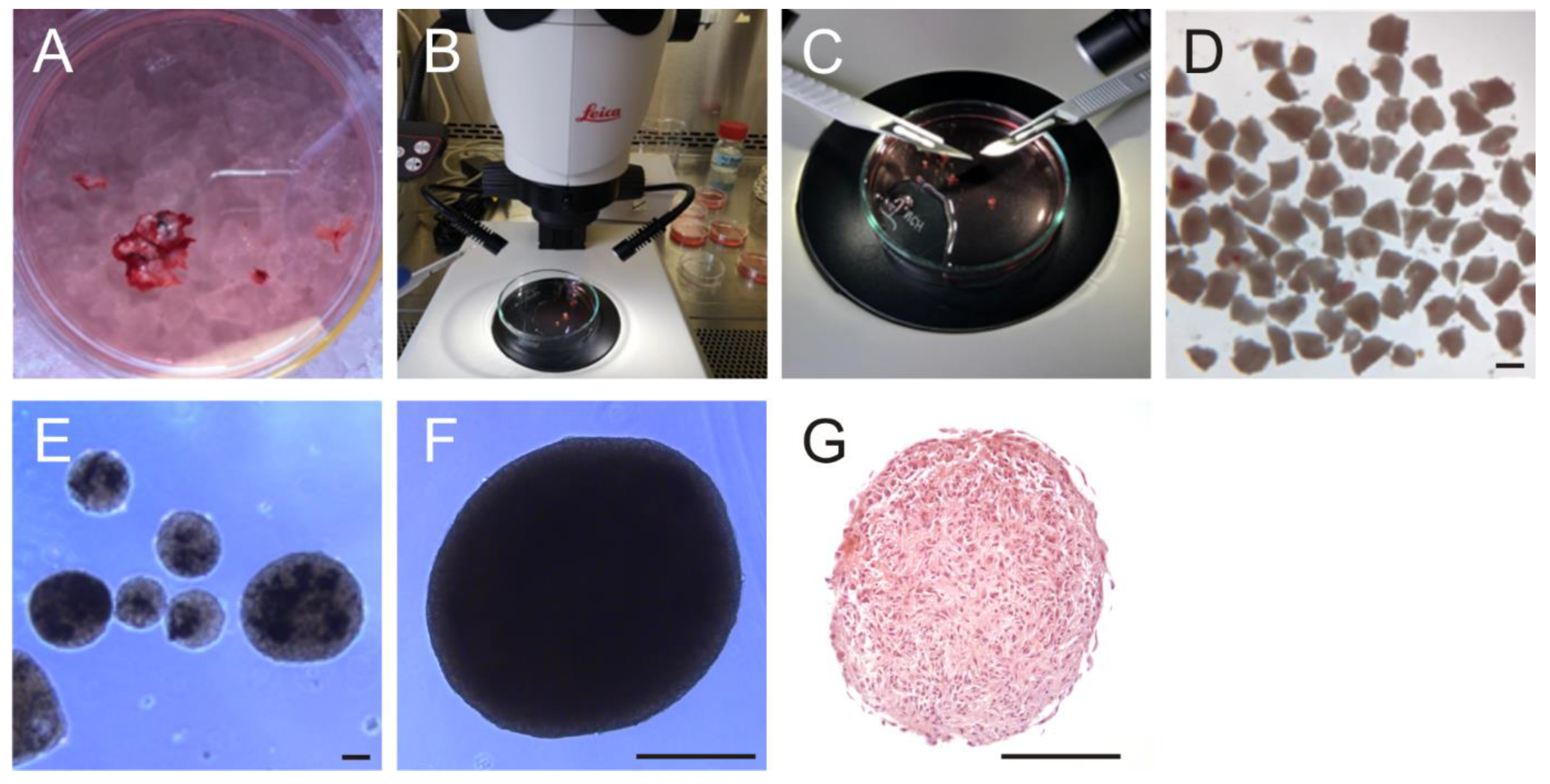
2.3. Generation of Organoids
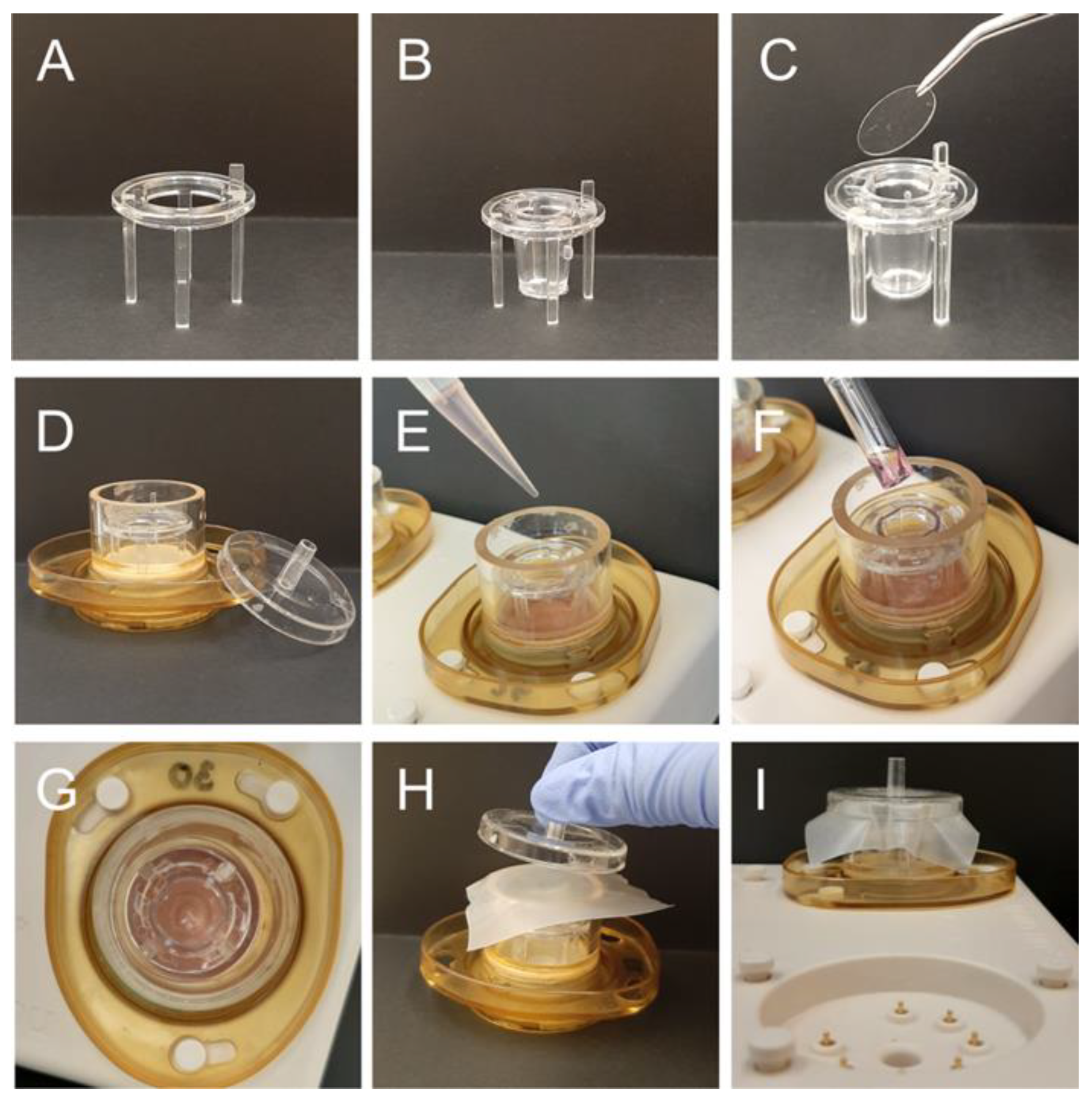
2.4. Preparation of Organotypic Hippocampal Brain Slice Cultures (OHSC)
2.5. Seeding of Fluorescence-Labeled GBM Cells and Organoids onto OHSC
2.6. Generation of Patient-Derived Organotypic Tumor Slice Cultures
2.7. TTFields Treatment
2.8. Immunohistochemical and Immunofluorescence Staining
2.9. Statistical Analysis
3. Results
3.1. Patient Cohort
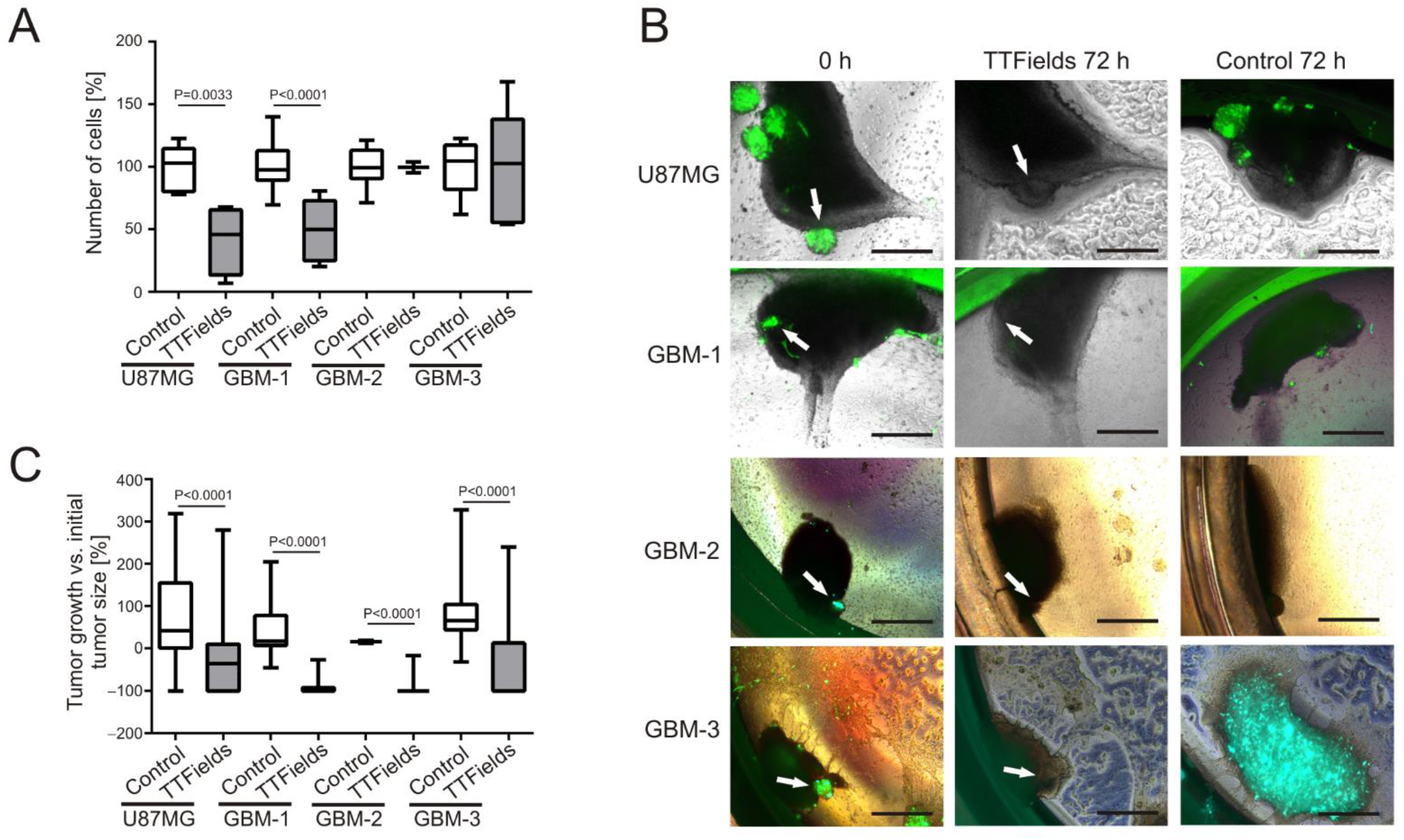
3.2. Patient-Derived GBM Primary Cells Display Variable Responses to TTFields at 200 kHz
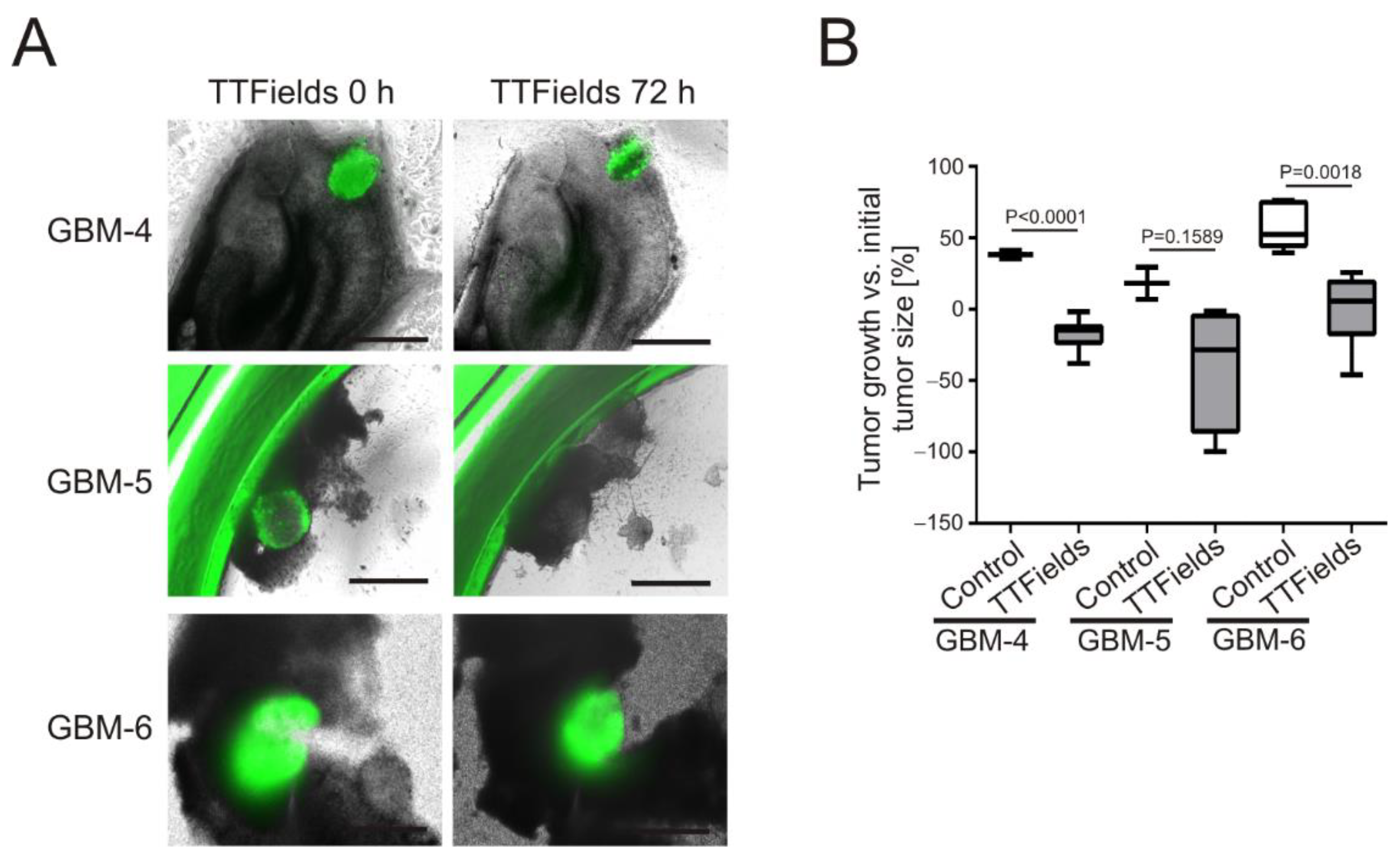
3.3. TTFields Induce Shrinkage of Patient-Derived GBM-Organoids Growing on OHSC
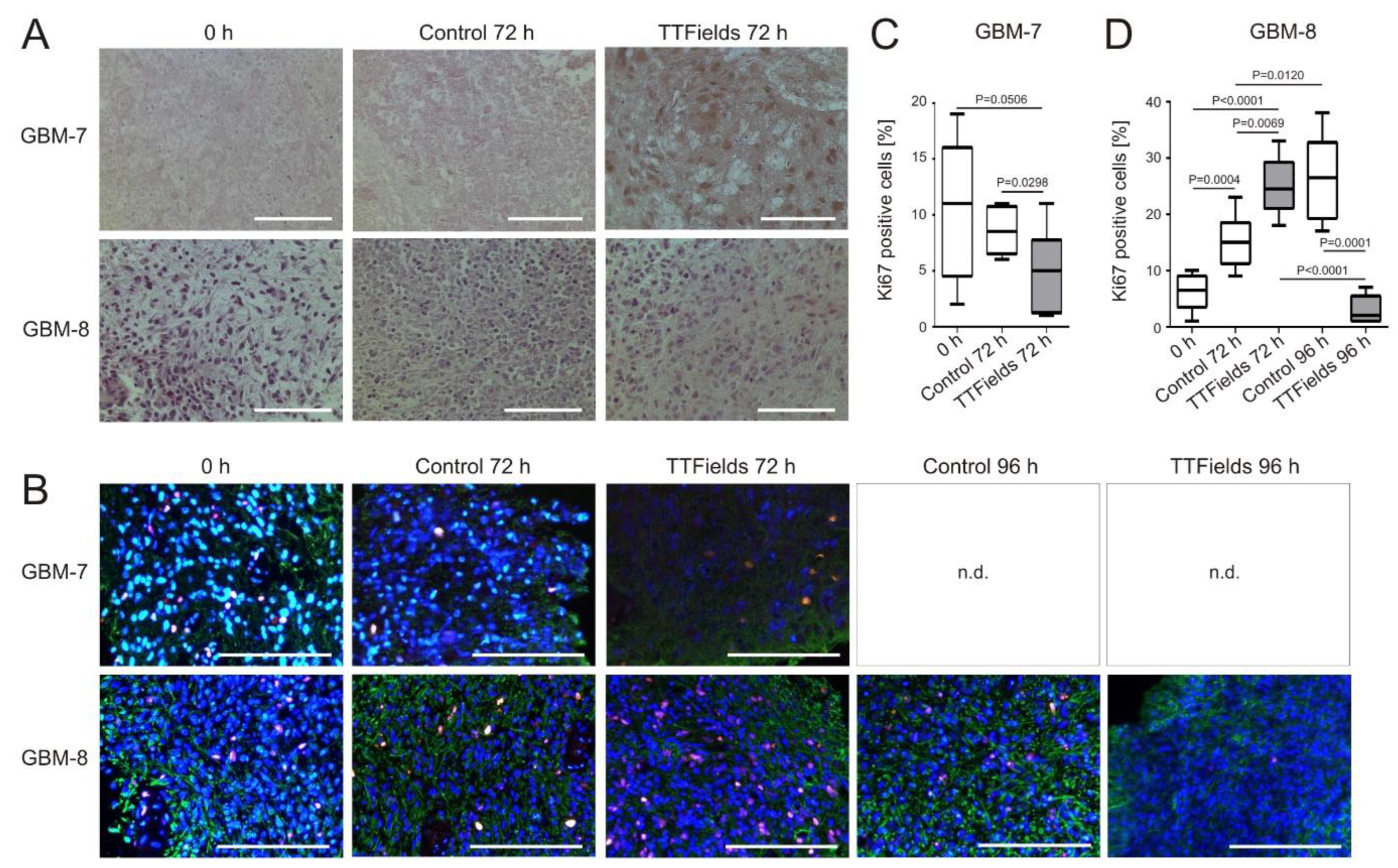
3.4. Cell Proliferation Decreases and Apoptosis Increases in Human Organotypic GBM Tumor Slice Cultures when Treated with TTFields
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wick, W.; et al. Gliome, S2k-Leitlinie. Leitlinien für Diagnostik und Therapie in der Neurologie. Deutsche Gesellschaft für Neurologie, Ed. 2021. Available online: www.dgn.org/leitlinien (accessed on 16 February 2022).
- Fritz, L.; Dirven, L.; Reijneveld, J.C.; Koekkoek, J.A.F.; Stiggelbout, A.M.; Pasman, H.R.W.; Taphoorn, M.J.B. Advance care planning in glioblastoma patients. Cancers 2016, 8, 102. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomized phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Patil, C.G.; Eboli, P.; Hu, J. Management of multifocal and multicentric gliomas. Neurosurg. Clin. N. Am. 2012, 23, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Rivera, A.L.; Pelloski, C.E.; Gilbert, M.R.; Colman, H.; De La Cruz, C.; Sulman, E.P.; Bekele, B.N.; Aldape, K.D. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010, 12, 116–121. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Monoranu, C.M.; Ernestus, R.-I.; Löhr, M.; Hagemann, C. Changes of O6-methylguanine DNA methyltransferase (MGMT) promotor methylation in glioblastoma relapse—A meta-analysis type literature review. Cancers 2019, 11, 1837. [Google Scholar] [CrossRef]
- Carieri, F.A.; Smack, C.; Siddiqui, I.; Kleinberg, L.R.; Tran, P.T. Tumor treating fields: At the crossroad between physics and biology for cancer treatment. Front. Oncol. 2020, 10, 575992. [Google Scholar] [CrossRef]
- Rominiyi, O.; Vanderlinden, A.; Clenton, S.J.; Bridgewater, C.; Al-Tamimi, Y.; Collis, S.J. Tumour treating fields therapy for glioblastoma: Current advances and future direction. Br. J. Cancer 2020, 124, 697–709. [Google Scholar] [CrossRef]
- Kirson, E.D.; Gurvich, Z.; Schneiderman, R.; Dekel, E.; Itzhaki, A.; Wasserman, Y.; Schatzberger, R.; Palti, Y. Disruption of cancer cell replication by alternating electric fields. Cancer Res. 2004, 64, 3288–3295. [Google Scholar] [CrossRef]
- Kirson, E.D.; Dbalý, V.; Tovarys, F.; Vymazal, J.; Soustiel, J.F.; Itzhaki, A.; Mordechovich, D.; Steinberg-Shapira, S.; Gurvich, Z.; Schneiderman, R.; et al. Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. Proc. Natl. Acad. Sci. USA 2007, 104, 10152–10157. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.M.; San, P.; Lok, E.; Swanson, K.D.; Wong, E.T. The clinical application of tumor treating fields therapy in glioblastoma. J. Vis. Exp. 2019, 146, e58937. [Google Scholar] [CrossRef]
- Moser, J.C.; Salvador, E.; Deniz, K.; Swanson, K.; Tuszynski, J.; Carlson, K.W.; Karanam, N.K.; Patel, C.B.; Story, M.; Lou, E.; et al. The mechanisms of action of Tumor Treating Fields. Cancer Res. 2022, CAN-22-0887, online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Kissling, C.; Di Santo, S. Tumor Treating Fields—Behind and beyond inhibiting the cancer cell. CNS Neurol Disord Drug Targets 2020, 19, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Tallibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields plus maintenance temozolomide vs. maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef]
- Gilazieva, Z.; Ponomarev, A.; Rutland, C.; Rizvnov, A.; Solovyeva, V. Promising applications of tumor spheroids and organoids for personalized medicine. Cancers 2020, 12, 2727. [Google Scholar] [CrossRef]
- Paolillo, M.; Comincini, S.; Schinelli, S. In vitro glioblastoma models: A journey into the third dimension. Cancers 2021, 13, 2449. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Nii, T.; Makina, K.; Tabata, Y. A cancer invasion model combined with cancer-associated fibroblasts aggregates incorporating gelatin hydrogel microspheres containing a p53 inhibitor. Tissue Eng. Part C Methods 2019, 25, 711–720. [Google Scholar] [CrossRef]
- Goudar, V.S.; Koduri, M.P.; Ta, Y.-N.N.; Chen, Y.; Chu, L.-A.; Lu, L.-S.; Tseng, F.-G. Impact of a desmoplastic tumor microenvironment for colon cancer drug sensitivity: A study with 3D chimeric tumor spheroids. ACS Appl. Mater Interfaces 2021, 13, 48478–48491. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Hohmann, T.; Hohmann, U.; Ernestus, R.-I.; Löhr, M.; Dehghani, F.; Hagemann, C. Preparation and culture of organotypic hippocampal slices for the analysis of brain metastasis and primary brain tumor growth. Methods Mol. Biol. 2021, 2294, 59–77. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef] [PubMed]
- Merz, F.; Gaunitz, F.; Dehghani, F.; Renner, C.; Meixensberger, J.; Gutenberg, A.; Giese, A.; Schopow, K.; Hellwig, C.; Schäfer, M.; et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol. 2013, 15, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, C.; Amend, D.; Kessler, A.F.; Linsenmann, T.; Ernestus, R.-I.; Löhr, M. High-efficiency transfection of glioblastoma cells and a simple spheroid migration assay. Methods Mol Biol 2017, 1622, 63–79. [Google Scholar] [CrossRef]
- Hagemann, C.; Meyer, C.; Stojic, J.; Eicker, S.; Gerngras, S.; Kühnel, S.; Roosen, K.; Vince, G.H. High efficiency transfection of glioma cell lines and primary cells for overexpression and RNAi experiments. J. Neurosci. Methods 2006, 156, 194–202. [Google Scholar] [CrossRef]
- Kessler, A.F.; Frömbling, G.E.; Gross, F.; Hahn, M.; Dzokou, W.; Ernestus, R.-I.; Löhr, M.; Hagemann, C. Effects of tumor treating fields (TTFields) on glioblastoma cells are augmented by mitotic checkpoint inhibition. Cell Death Discov. 2018, 4, 12. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.-I.; Hagemann, C.; Löhr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. Onco. Targets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef] [PubMed]
- Giladi, M.; Schneiderman, R.S.; Voloshin, T.; Porat, Y.; Munster, M.; Blat, R.; Sherbo, S.; Bomzon, Z.; Urman, N.; Itzhaki, A.; et al. Mitotic spindle disruption by alternating electric fields leads to inproper chromosome segregation and mitotic catastrophe in cancer cells. Sci. Rep. 2015, 5, 18046. [Google Scholar] [CrossRef]
- Thomasova, D.; Anders, H.-J. Cell cycle control in the kidney. Nephrol. Dial. Transplant. 2015, 30, 1622–1630. [Google Scholar] [CrossRef]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (Review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef]
- Voloshin, T.; Munster, M.; Blatt, R.; Shteingauz, A.; Roberts, P.C.; Schmelz, E.M.; Giladi, M.; Schneiderman, R.S.; Zeevi, E.; Porat, Y.; et al. Alternating electric fields (TTFields) in combination with paclitaxel are therapeutically effective against ovarian cancer cells in vitro and in vivo. Int. J. Cancer 2016, 139, 2850–2858. [Google Scholar] [CrossRef]
- Saeidnia, S.; Manayi, A.; Abdollahi, M. From in vitro experiments to in vivo and clinical studies; pros and cons. Curr. Drug Discov. Technol. 2015, 12, 218–224. [Google Scholar] [CrossRef]
- Wan, L.; Neumann, C.A.; LeDuc, P.R. Tumor-on-a-chip for integrating a 3D tumor microenvironment: Chemical and mechanical factors. Lab Chip 2020, 20, 873–888. [Google Scholar] [CrossRef]
- Joseph, J.V.; Blaavand, M.S.; Daubon, T.; Kruyt, F.A.E.; Thomsen, M.K. Three-dimensional culture models to study glioblastoma—Current trends and future perspectives. Curr. Opin. Pharmacol. 2021, 61, 91–97. [Google Scholar] [CrossRef]
- Orchestron-Findlay, L.; Bax, S.; Utama, R.; Engel, M.; Govender, D.; O’Neill, G. Advanced spheroid, tumouroid and 3D bioprinted in-vitro models of adult and paediatric glioblastoma. Int. J. Mol. Sci. 2021, 22, 2962. [Google Scholar] [CrossRef]
- Lacouture, M.E.; Davis, M.E.; Elzinga, G.; Butowski, N.; Tran, D.; Villano, J.L.; Di Meglio, L.; Davies, A.M.; Wong, E.T. Characterization and management of dermatologic adverse events with the novoTTF-100A system, a novel anti-mitotic electric field device for the treatment of recurrent glioblastoma. Semin. Oncol. 2014, 41 (Suppl. S4), S1–S14. [Google Scholar] [CrossRef]
- Toms, S.A.; Kim, C.Y.; Nicholas, G.; Ram, Z. Increased compliance with tumor treating fields therapy is prognostic for improved survival in the treatment of glioblastoma: A subgroup analysis of the E-14 phase III trial. J. Neurooncol. 2019, 141, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.; Schneider, J.R.; Boockvar, J.A. Quality of life in patients with glioblastoma treated with tumor- treating fields. JAMA 2018, 319, 1822–1823. [Google Scholar] [CrossRef]
- Bähr, O.; Tabatabai, G.; Fietkau, R.; Goldbrunner, R.; Glas, M. First results of the TIGER study—Treatment decision and quality of life of glioblastoma patients during TTFields therapy in routine clinical care. In Proceedings of the Jahrestagung der Deutschen Gesellschaft für Neurochirurgie (DGNC), Joint Meeting mit der Griechischen Gesellschaft für Neurochirurgie; Köln, Germany, 29 May–1 June 2022, German Medical Science GMS Publishing House: Düsseldorf, Germany, 2022; Volume DocV228, p. 73. [Google Scholar] [CrossRef]
- Xu, X.; Li, L.; Luo, L.; Shu, L.; Si, X.; Chen, Z.; Xia, W.; Huang, J.; Liu, Y.; Shao, A.; et al. Opportunities and challenges of glioma organoids. Cell Commun. Signal 2021, 19, 102. [Google Scholar] [CrossRef] [PubMed]

| ID | Sex | Age [Years] | Histology | KPI | Ki67 [%] | MGMT Promoter Methylation [%] | IDH1 Mutation | IDH2 Mutation | ATRX Expression | Experiment |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | male | 64 | GBM | 100 | 20 | yes (76) | no | no | yes | PDPC |
| 2 | female | 79 | GBM | 100 | 20 | yes (66) | no | no | yes | PDPC |
| 3 | female | 73 | GBM | 100 | 50 | yes (28) | no | no | yes | PDPC |
| 4 | female | 69 | GBM | 30 | 25 | yes (71) | no | no | yes | organoid |
| 5 | female | 65 | GBM | 90 | 20 | no (8.0) | no | no | yes | organoid |
| 6 | male | 65 | GBM | 90 | 40 | no (4.0) | no | no | yes | organoid |
| 7 | male | 64 | GBM | 40 | 30 | yes (82) | no | no | yes | tumor slice |
| 8 | male | 54 | GBM | 90 | 40 | no (3) | no | no | yes | tumor slice |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nickl, V.; Schulz, E.; Salvador, E.; Trautmann, L.; Diener, L.; Kessler, A.F.; Monoranu, C.M.; Dehghani, F.; Ernestus, R.-I.; Löhr, M.; et al. Glioblastoma-Derived Three-Dimensional Ex Vivo Models to Evaluate Effects and Efficacy of Tumor Treating Fields (TTFields). Cancers 2022, 14, 5177. https://doi.org/10.3390/cancers14215177
Nickl V, Schulz E, Salvador E, Trautmann L, Diener L, Kessler AF, Monoranu CM, Dehghani F, Ernestus R-I, Löhr M, et al. Glioblastoma-Derived Three-Dimensional Ex Vivo Models to Evaluate Effects and Efficacy of Tumor Treating Fields (TTFields). Cancers. 2022; 14(21):5177. https://doi.org/10.3390/cancers14215177
Chicago/Turabian StyleNickl, Vera, Ellina Schulz, Ellaine Salvador, Laureen Trautmann, Leopold Diener, Almuth F. Kessler, Camelia M. Monoranu, Faramarz Dehghani, Ralf-Ingo Ernestus, Mario Löhr, and et al. 2022. "Glioblastoma-Derived Three-Dimensional Ex Vivo Models to Evaluate Effects and Efficacy of Tumor Treating Fields (TTFields)" Cancers 14, no. 21: 5177. https://doi.org/10.3390/cancers14215177
APA StyleNickl, V., Schulz, E., Salvador, E., Trautmann, L., Diener, L., Kessler, A. F., Monoranu, C. M., Dehghani, F., Ernestus, R.-I., Löhr, M., & Hagemann, C. (2022). Glioblastoma-Derived Three-Dimensional Ex Vivo Models to Evaluate Effects and Efficacy of Tumor Treating Fields (TTFields). Cancers, 14(21), 5177. https://doi.org/10.3390/cancers14215177







