Cytoskeletal Protein Palladin in Adult Gliomas Predicts Disease Incidence, Progression, and Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Mutation, Bulk Gene Expression, Clinical Data, and Survival Analysis
2.2. Cell Culture
2.3. RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.4. Animal Studies and Fluorescent Confocal Microscopy of Mouse Glioblastoma Tumors
2.5. Immunohistochemical Staining of Palladin in Human Tissue Microarrays
2.6. Single-Cell Gene Expression Data
2.7. In-Silico Flow Cytometry of Astrocytoma Tumors
2.8. Enrichment Analysis of Gene Co-Expression with Palladin
2.9. Statistical Analyses
3. Results
3.1. Palladin Is Overexpressed in Adult Glioma Tumors and Is Correlated with Shorter Overall Survival
3.2. Palladin Isoform 4 Is Specifically Overexpressed in Adult Glioma Tumors
3.3. Somatic Mutations in Palladin’s Genomic Sequence Are Extremely Rare
3.4. Palladin Is Not Overexpressed in Pediatric Glioma Tumors
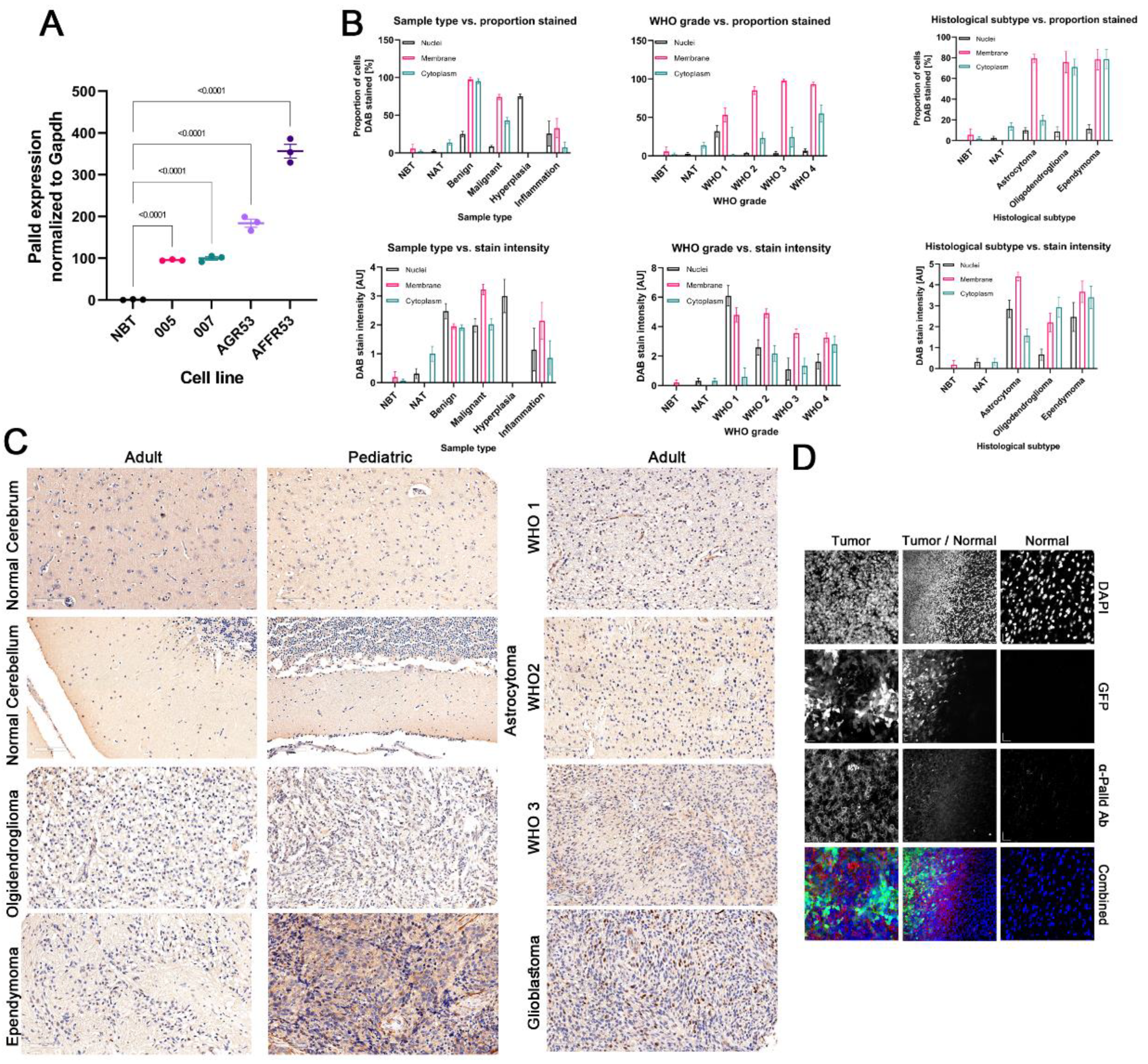
3.5. Palladin Expression Is Confined Predominantly to the Cancerous Tissue in the Brain
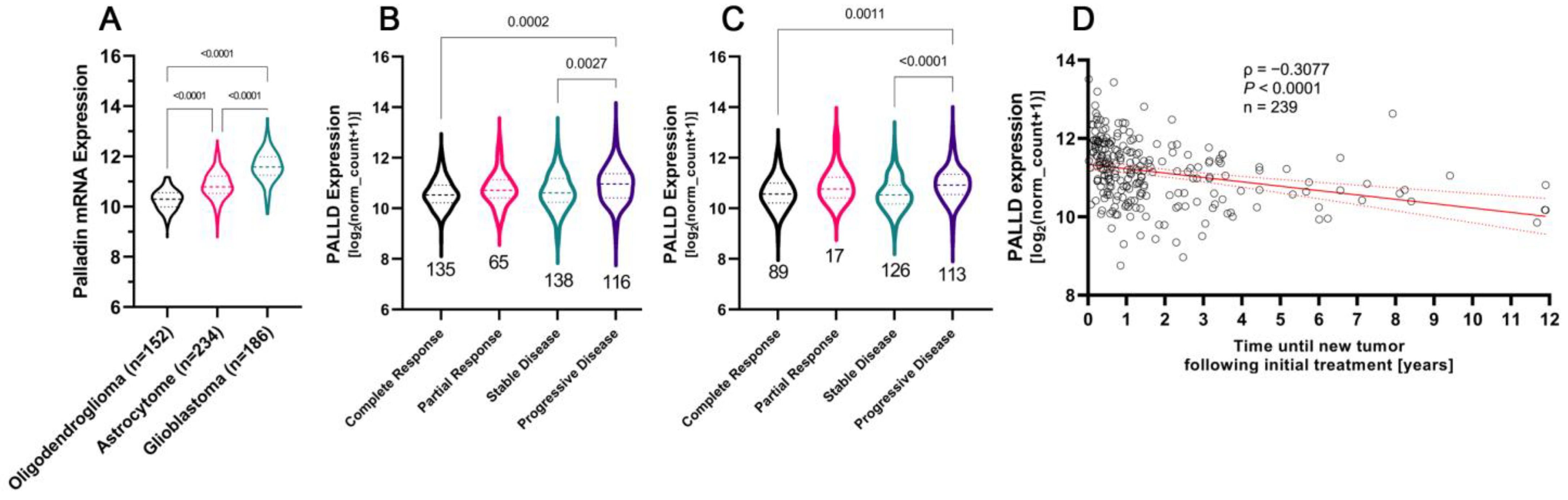
3.6. Glioblastoma Tumors Are Characterized by the Highest Levels of Palladin Expression
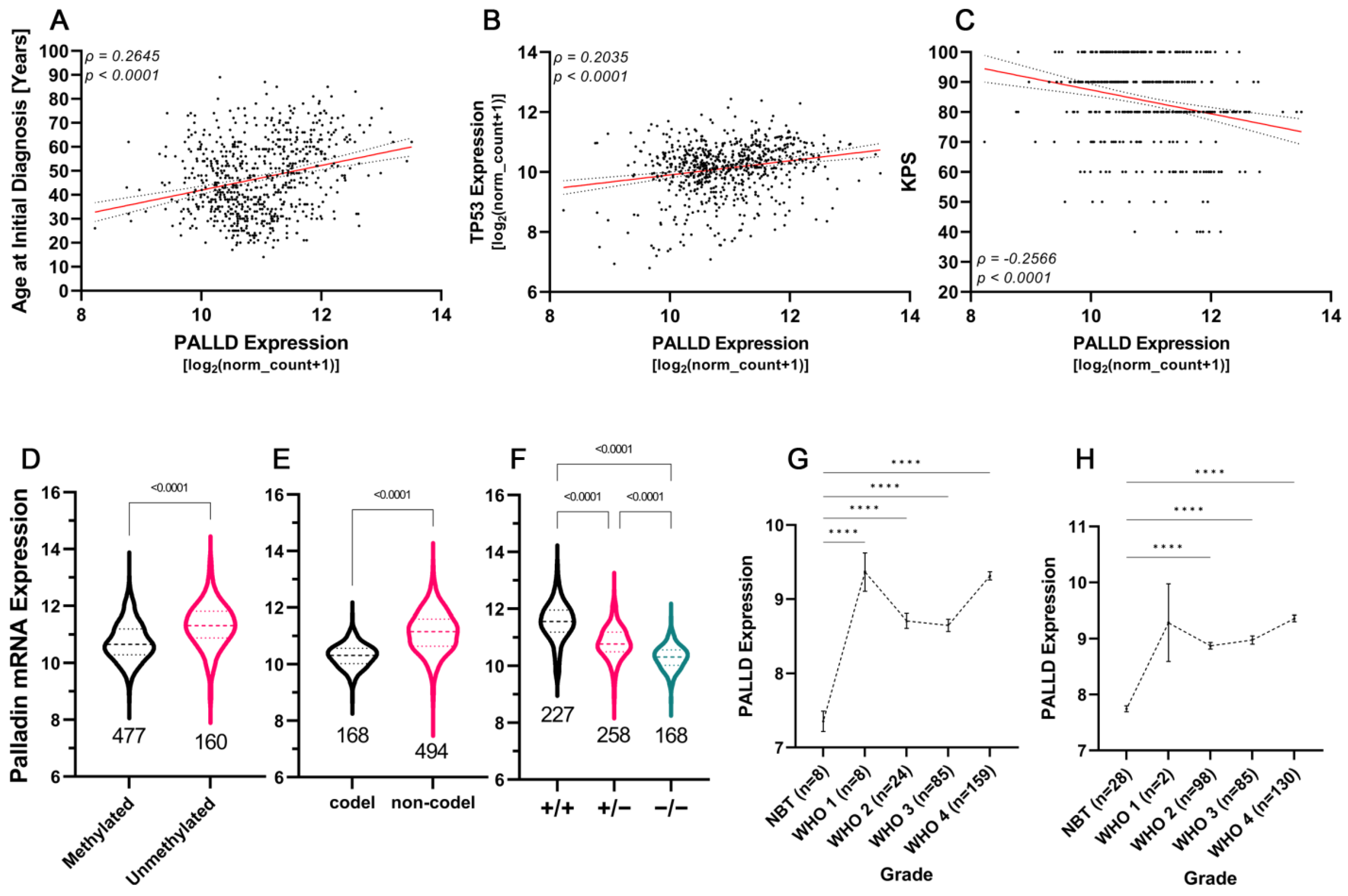
3.7. Aggressive Glioma Tumors Are Characterized by Higher Levels of Palladin
3.8. Palladin Expression Compared to Commonly Used Diagnostic and Prognostic Markers
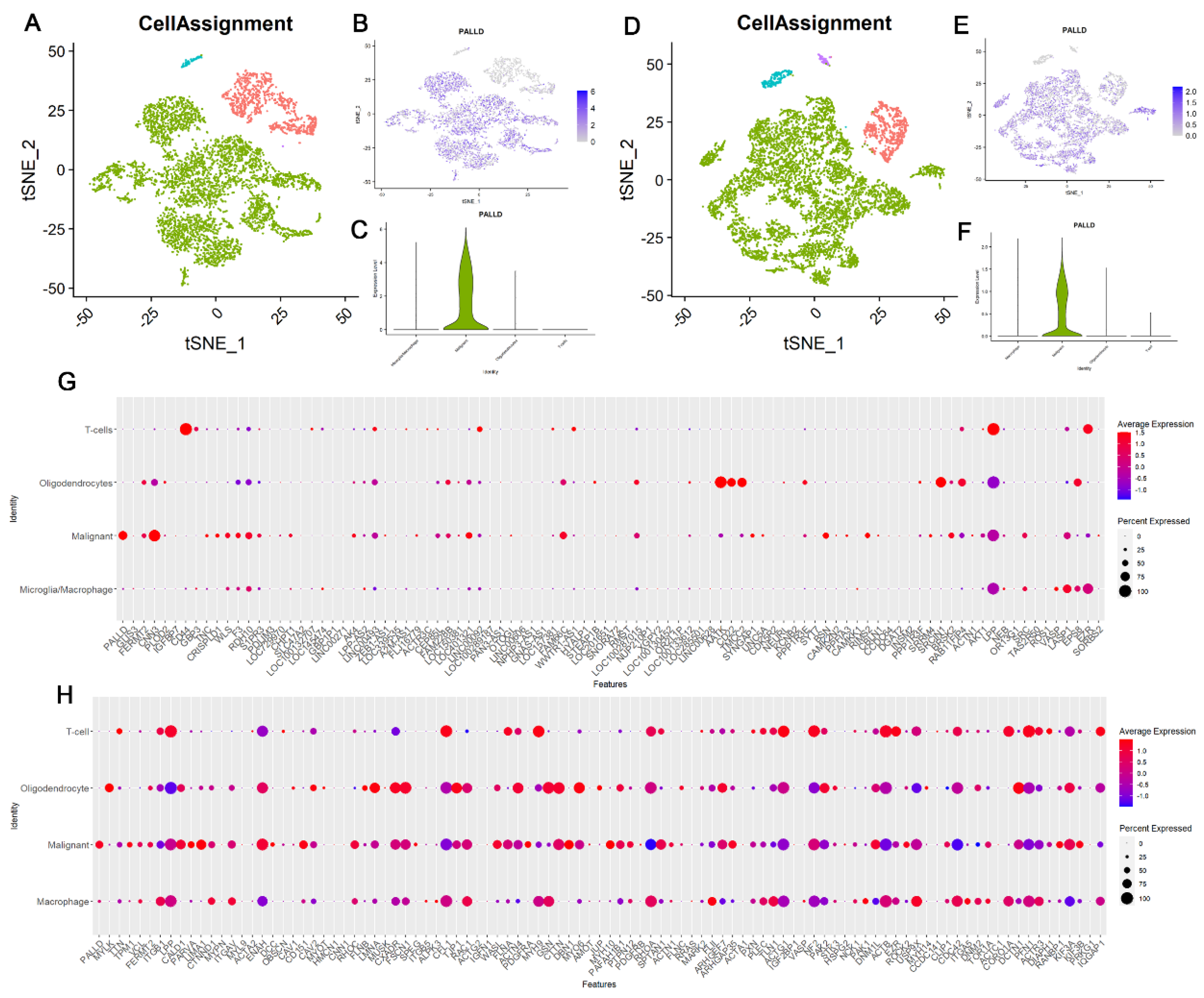
3.9. Palladin Is Overexpressed Principally in Malignant Cells and Not in Other Glioma-Related Cell Types
3.10. Palladin Is Related to a Transcriptional Program Involved in Cellular Motility and the Extracellular Matrix
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Francis, S.S.; Barnholtz-Sloan, J.S. Epidemiology of Brain and Other CNS Tumors. Curr. Neurol. Neurosci. Rep. 2021, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Lv, X.; Lu, C.; Ye, X.; Chen, X.; Fu, J.; Luo, C.; Zhao, Y. Prognostic Factors of Patients with Gliomas—An Analysis on 335 Patients with Glioblastoma and Other Forms of Gliomas. BMC Cancer 2020, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, G.; Quan, X. Analysis of the Factors Affecting the Prognosis of Glioma Patients. Open Med. 2019, 14, 331–335. [Google Scholar] [CrossRef]
- Smith, J.S.; Perry, A.; Borell, T.J.; Lee, H.K.; O’Fallon, J.; Hosek, S.M.; Kimmel, D.; Yates, A.; Burger, P.C.; Scheithauer, B.W.; et al. Alterations of Chromosome Arms 1p and 19q as Predictors of Survival in Oligodendrogliomas, Astrocytomas, and Mixed Oligoastrocytomas. J. Clin. Oncol. 2000, 18, 636–645. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Genetic Profile of Astrocytic and Oligodendroglial Gliomas. Brain Tumor. Pathol. 2011, 28, 177–183. [Google Scholar] [CrossRef]
- McNamara, M.G.; Sahebjam, S.; Mason, W.P. Emerging Biomarkers in Glioblastoma. Cancers 2013, 5, 1103–1119. [Google Scholar] [CrossRef]
- Zhang, Y.; Dube, C.; Gibert, M.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The P53 Pathway in Glioblastoma. Cancers 2018, 10, 297. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Reifenberger, G.; Hentschel, B.; Felsberg, J.; Schackert, G.; Simon, M.; Schnell, O.; Westphal, M.; Wick, W.; Pietsch, T.; Loeffler, M.; et al. Predictive Impact of MGMT Promoter Methylation in Glioblastoma of the Elderly. Int. J. Cancer 2012, 131, 1342–1350. [Google Scholar] [CrossRef]
- Kaina, B.; Christmann, M. DNA Repair in Personalized Brain Cancer Therapy with Temozolomide and Nitrosoureas. DNA Repair 2019, 78, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, P.; Mansouri, A.; Das, S. The Role of ATRX in Glioma Biology. Front. Oncol. 2017, 7, 236. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, M.; Shibahara, J.; Mukasa, A.; Takayanagi, S.; Aihara, K.; Saito, N.; Aburatani, H.; Fukayama, M. Utility of ATRX Immunohistochemistry in Diagnosis of Adult Diffuse Gliomas. Histopathology 2016, 69, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tang, L.; Li, X.; Fan, F.; Liu, Z. Immunotherapy for Glioma: Current Management and Future Application. Cancer Lett. 2020, 476, 1–12. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Hottinger, A.F.; Pacheco, P.; Stupp, R. Tumor Treating Fields: A Novel Treatment Modality and Its Use in Brain Tumors. Neuro Oncol. 2016, 18, 1338–1349. [Google Scholar] [CrossRef]
- Cohen, M.H.; Shen, Y.L.; Keegan, P.; Pazdur, R. FDA Drug Approval Summary: Bevacizumab (Avastin®) as Treatment of Recurrent Glioblastoma Multiforme. Oncologist 2009, 14, 1131–1138. [Google Scholar] [CrossRef]
- Goicoechea, S.M.; Arneman, D.; Otey, C.A. The Role of Palladin in Actin Organization and Cell Motility. Eur. J. Cell Biol. 2008, 87, 517–525. [Google Scholar] [CrossRef]
- Nguyen, N.-U.-N.; Wang, H.-V. Dual Roles of Palladin Protein in In Vitro Myogenesis: Inhibition of Early Induction but Promotion of Myotube Maturation. PLoS ONE 2015, 10, e0124762. [Google Scholar] [CrossRef]
- Luo, H.; Liu, X.; Wang, F.; Huang, Q.; Shen, S.; Wang, L.; Xu, G.; Sun, X.; Kong, H.; Gu, M.; et al. Disruption of Palladin Results in Neural Tube Closure Defects in Mice. Mol. Cell. Neurosci. 2005, 29, 507–515. [Google Scholar] [CrossRef]
- Boukhelifa, M.; Moza, M.; Johansson., T.; Rachlin, A.; Parast, M.; Huttelmaier, S.; Roy, P.; Jockusch, B.M.; Carpen, O.; Karlsson, R.; et al. The Proline-rich Protein Palladin Is a Binding Partner for Profilin. FEBS J. 2005, 273, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Boukhelifa, M.; Parast, M.M.; Bear, J.E.; Gertler, F.B.; Otey, C.A. Palladin Is a Novel Binding Partner for Ena/VASP Family Members. Cell Motil. 2004, 58, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Rönty, M.; Taivainen, A.; Moza, M.; Kruh, G.D.; Ehler, E.; Carpen, O. Involvement of Palladin and α-Actinin in Targeting of the Abl/Arg Kinase Adaptor ArgBP2 to the Actin Cytoskeleton. Exp. Cell Res. 2005, 310, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Mykkänen, O.-M.; Grönholm, M.; Rönty, M.; Lalowski, M.; Salmikangas, P.; Suila, H.; Carpén, O.; Salmon, T. Characterization of Human Palladin, a Microfilament-Associated Protein. MBoC 2001, 12, 3060–3073. [Google Scholar] [CrossRef]
- Goicoechea, S.; Arneman, D.; Disanza, A.; Garcia-Mata, R.; Scita, G.; Otey, C.A. Palladin Binds to Eps8 and Enhances the Formation of Dorsal Ruffles and Podosomes in Vascular Smooth Muscle Cells. J. Cell Sci. 2006, 119, 3316–3324. [Google Scholar] [CrossRef][Green Version]
- Rachlin, A.S.; Otey, C.A. Identification of Palladin Isoforms and Characterization of an Isoform-Specific Interaction between Lasp-1 and Palladin. J. Cell Sci. 2006, 119, 995–1004. [Google Scholar] [CrossRef][Green Version]
- Maeda, M.; Asano, E.; Ito, D.; Ito, S.; Hasegawa, Y.; Hamaguchi, M.; Senga, T. Characterization of Interaction between CLP36 and Palladin. FEBS J. 2009, 276, 2775–2785. [Google Scholar] [CrossRef]
- Gurung, R.; Yadav, R.; Brungardt, J.G.; Orlova, A.; Egelman, E.H.; Beck, M.R. Actin Polymerization Is Stimulated by Actin Cross-Linking Protein Palladin. Biochem. J. 2016, 473, 383–396. [Google Scholar] [CrossRef]
- Vattepu, R.; Yadav, R.; Beck, M.R. Actin-Induced Dimerization of Palladin Promotes Actin-Bundling. Protein Sci. 2015, 24, 70–80. [Google Scholar] [CrossRef]
- Beck, M.R.; Dixon, R.D.S.; Goicoechea, S.M.; Murphy, G.S.; Brungardt, J.G.; Beam, M.T.; Srinath, P.; Patel, J.; Mohiuddin, J.; Otey, C.A.; et al. Structure and function of palladin’s actin binding domain. J. Mol. Biol. 2013, 425, 3325–3337. [Google Scholar] [CrossRef]
- Dixon, R.D.S.; Arneman, D.K.; Rachlin, A.S.; Sundaresan, N.R.; Costello, M.J.; Campbell, S.L.; Otey, C.A. Palladin Is an Actin Cross-Linking Protein That Uses Immunoglobulin-like Domains to Bind Filamentous Actin. J. Biol. Chem. 2008, 283, 6222–6231. [Google Scholar] [CrossRef]
- Goicoechea, S.; Bednarski, B.; García-Mata, R.; Prentice-Dunn, H.; Kim, H.; Otey, C. Palladin Contributes to Invasive Motility in Human Breast Cancer Cells. Oncogene 2009, 28, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, S.; García-Mata, R.; Staub, J.; Valdivia, A.; Sharek, L.; McCulloch, C.; Hwang, R.; Urrutia, R.; Yeh, J.; Kim, H.; et al. Palladin Promotes Invasion of Pancreatic Cancer Cells by Enhancing Invadopodia Formation in Cancer-Associated Fibroblasts. Oncogene 2014, 33, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Bock, A.J.; Holth, A.; Nymoen, D.A. Expression of Palladin Is Associated with Disease Progression in Metastatic High-Grade Serous Carcinoma. Cytopathology 2020, 31, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Tay, P.N.; Tan, P.; Lan, Y.; Leung, C.H.-W.; Laban, M.; Tan, T.C.; Ni, H.; Manikandan, J.; Rashid, S.B.A.; Yan, B.; et al. Palladin, an Actin-Associated Protein, Is Required for Adherens Junction Formation and Intercellular Adhesion in HCT116 Colorectal Cancer Cells. Int. J. Oncol. 2010, 37, 909–926. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Bassi, D.E.; Simons, J.D.; Devarajan, K.; Al-Saleem, T.; Uzzo, R.G.; Cukierman, E. Elevated Expression of Stromal Palladin Predicts Poor Clinical Outcome in Renal Cell Carcinoma. PLoS ONE 2011, 6, e21494. [Google Scholar] [CrossRef] [PubMed]
- Gilam, A.; Conde, J.; Weissglas-Volkov, D.; Oliva, N.; Friedman, E.; Artzi, N.; Shomron, N. Local MicroRNA Delivery Targets Palladin and Prevents Metastatic Breast Cancer. Nat. Commun. 2016, 7, 12868. [Google Scholar] [CrossRef]
- Boukhelifa, M.; Parast, M.M.; Valtschanoff, J.G.; LaMantia, A.S.; Meeker, R.B.; Otey, C.A.; Drubin, D. A Role for the Cytoskeleton-Associated Protein Palladin in Neurite Outgrowth. MBoC 2001, 12, 2721–2729. [Google Scholar] [CrossRef][Green Version]
- Boukhelifa, M.; Hwang, S.J.; Valtschanoff, J.G.; Meeker, R.B.; Rustioni, A.; Otey, C.A. A Critical Role for Palladin in Astrocyte Morphology and Response to Injury. Mol. Cell. Neurosci. 2003, 23, 661–668. [Google Scholar] [CrossRef]
- Hwang, S.J.; Pagliardini, S.; Boukhelifa, M.; Parast, M.M.; Otey, C.A.; Rustioni, A.; Valtschanoff, J.G. Palladin Is Expressed Preferentially in Excitatory Terminals in the Rat Central Nervous System. J. Comp. Neurol. 2001, 436, 211–224. [Google Scholar] [CrossRef]
- Tan, J.; Chen, X.-J.; Shen, C.-L.; Zhang, H.-X.; Tang, L.-Y.; Lu, S.-Y.; Wu, W.-T.; Kuang, Y.; Fei, J.; Wang, Z.-G. Lacking of Palladin Leads to Multiple Cellular Events Changes Which Contribute to NTD. Neural Dev. 2017, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, X.; Liu, J.; Xu, X.; Zhang, Y.; Ruan, Z.; Xie, Y.; Huang, Q.; Yin, T.; Chen, Z.; et al. Palladin Is a Novel Microtubule-Associated Protein Responsible for Spindle Orientation. Sci. Rep. 2017, 7, 11806. [Google Scholar] [CrossRef] [PubMed]
- Umegaki, Y.; Brotons, A.M.; Nakanishi, Y.; Luo, Z.; Zhang, H.; Bonni, A.; Ikeuchi, Y. Palladin Is a Neuron-Specific Translational Target of MTOR Signaling That Regulates Axon Morphogenesis. J. Neurosci. 2018, 38, 4985–4995. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Program—NCI. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 5 June 2022).
- GTEx Portal. Available online: https://www.gtexportal.org/home/ (accessed on 5 June 2022).
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.W.; Squatrito, M. GlioVis Data Portal for Visualization and Analysis of Brain Tumor Expression Datasets. Neuro-Oncology 2017, 19, 139–141. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Tirosh, I.; Hebert, C.; Yizhak, K.; Neftel, C.; Filbin, M.G.; Hovestadt, V.; Escalante, L.E.; Shaw, M.L.; Rodman, C.; et al. Decoupling Genetics, Lineages, and Microenvironment in IDH-Mutant Gliomas by Single-Cell RNA-Seq. Science 2017, 355, eaai8478. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef]
- Jin, L. The Actin Associated Protein Palladin in Smooth Muscle and in the Development of Diseases of the Cardiovasculature and in Cancer. J. Muscle Res. Cell Motil. 2011, 32, 7–17. [Google Scholar] [CrossRef]
- Jin, L.; Gan, Q.; Zieba, B.J.; Goicoechea, S.M.; Owens, G.K.; Otey, C.A.; Somlyo, A.V. The Actin Associated Protein Palladin Is Important for the Early Smooth Muscle Cell Differentiation. PLoS ONE 2010, 5, e12823. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Cai, R.; Liu, M.; Wang, R. Identification of a Tumor Microenvironment-Associated Prognostic Gene Signature in Bladder Cancer by Integrated Bioinformatic Analysis. Int. J. Clin. Exp. Pathol. 2021, 14, 551–566. [Google Scholar]
- Niedenberger, B.A.; Chappell, V.K.; Kaye, E.P.; Renegar, R.H.; Geyer, C.B. Nuclear Localization of the Actin Regulatory Protein Palladin in Sertoli Cells. Mol. Reprod. Dev. 2013, 80, 403–413. [Google Scholar] [CrossRef]
- Niedenberger, B.A.; Chappell, V.A.; Otey, C.A.; Geyer, C.B. Actin Dynamics Regulate Subcellular Localization of the F-Actin-Binding Protein PALLD in Mouse Sertoli Cells. Reproduction 2014, 148, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Hasan, M.N.; Maniar, S.; Jia, W.; Sun, D. Reactive Astrocytes in Glioblastoma Multiforme. Mol. Neurobiol. 2018, 55, 6927–6938. [Google Scholar] [CrossRef] [PubMed]
- Stummer, W.; Pichlmeier, U.; Meinel, T.; Wiestler, O.D.; Zanella, F.; Reulen, H.-J. Fluorescence-Guided Surgery with 5-Aminolevulinic Acid for Resection of Malignant Glioma: A Randomised Controlled Multicentre Phase III Trial. Lancet Oncol. 2006, 7, 392–401. [Google Scholar] [CrossRef]
- Torrisi, F.; Alberghina, C.; D’Aprile, S.; Pavone, A.M.; Longhitano, L.; Giallongo, S.; Tibullo, D.; Di Rosa, M.; Zappalà, A.; Cammarata, F.P.; et al. The Hallmarks of Glioblastoma: Heterogeneity, Intercellular Crosstalk and Molecular Signature of Invasiveness and Progression. Biomedicines 2022, 10, 806. [Google Scholar] [CrossRef]
- Rönty, M.; Taivainen, A.; Heiska, L.; Otey, C.; Ehler, E.; Song, W.K.; Carpen, O. Palladin Interacts with SH3 Domains of SPIN90 and Src and Is Required for Src-Induced Cytoskeletal Remodeling. Exp. Cell Res. 2007, 313, 2575–2585. [Google Scholar] [CrossRef][Green Version]
- Alexander, J.I.; Vendramini-Costa, D.B.; Francescone, R.; Luong, T.; Franco-Barraza, J.; Shah, N.; Gardiner, J.C.; Nicolas, E.; Raghavan, K.S.; Cukierman, E. Palladin Isoforms 3 and 4 Regulate Cancer-Associated Fibroblast pro-Tumor Functions in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2021, 11, 3802. [Google Scholar] [CrossRef]
- Rönty, M.J.; Leivonen, S.-K.; Hinz, B.; Rachlin, A.; Otey, C.A.; Kähäri, V.-M.; Carpén, O.M. Isoform-Specific Regulation of the Actin-Organizing Protein Palladin during TGF-Β1-Induced Myofibroblast Differentiation. J. Investig. Dermatol. 2006, 126, 2387–2396. [Google Scholar] [CrossRef]
- Monteiro, A.R.; Hill, R.; Pilkington, G.J.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Invasion. Cells 2017, 6, E45. [Google Scholar] [CrossRef]
- Torrisi, F.; Vicario, N.; Spitale, F.M.; Cammarata, F.P.; Minafra, L.; Salvatorelli, L.; Russo, G.; Cuttone, G.; Valable, S.; Gulino, R.; et al. The Role of Hypoxia and SRC Tyrosine Kinase in Glioblastoma Invasiveness and Radioresistance. Cancers 2020, 12, E2860. [Google Scholar] [CrossRef]
- Chin, Y.R.; Toker, A. The actin bundling protein palladin is an akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell. 2010, 38, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.R.; Toker, A. Akt2 Regulates Expression of the Actin-Bundling Protein Palladin. FEBS Lett. 2010, 584, 4769–4774. [Google Scholar] [CrossRef] [PubMed]
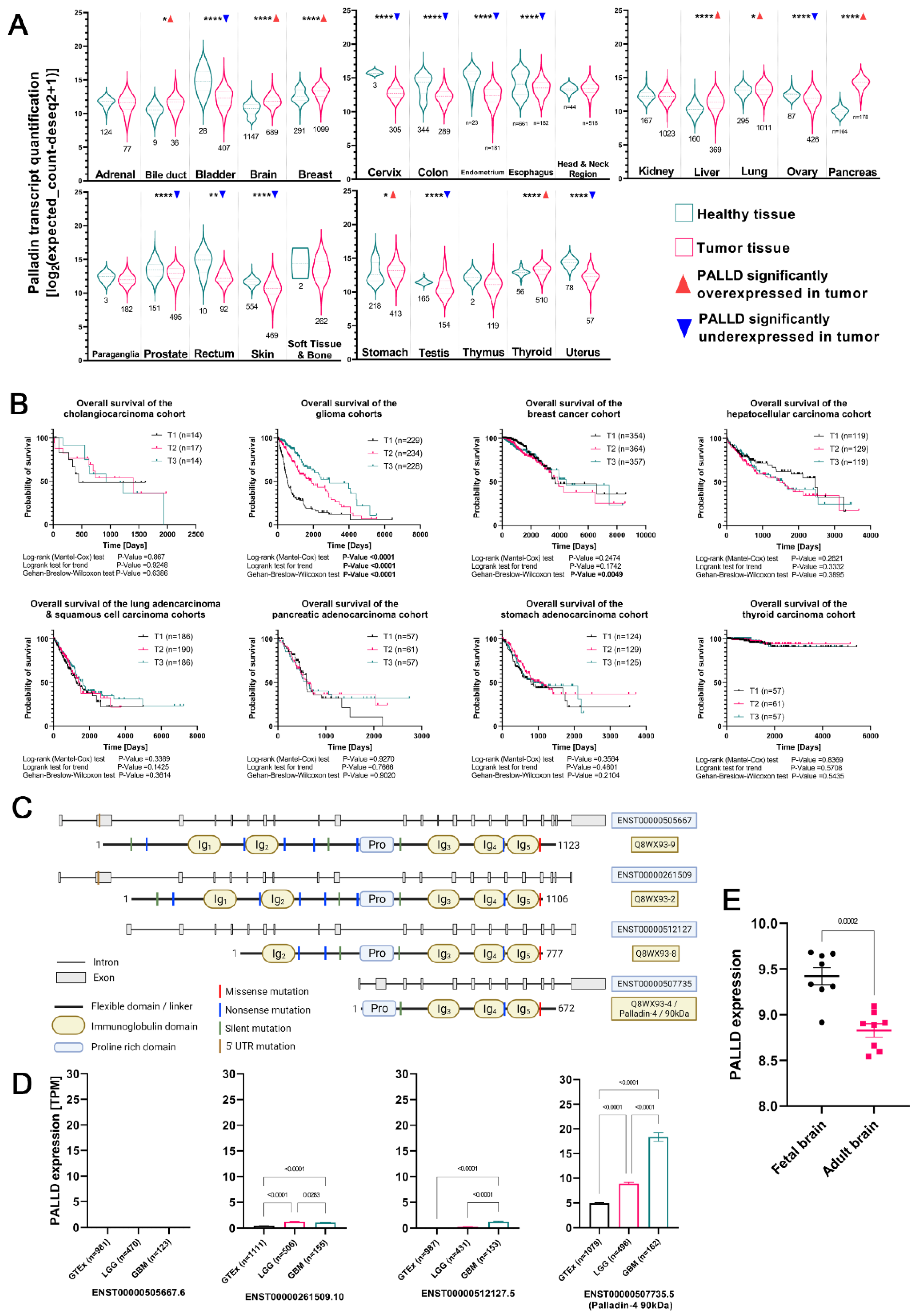

| Organ/Region | Study & Cohort | Statistic | p | PALLD in Tumor |
| Adrenal gland | TCGA-ACC + GTEx | UMann–Whitney = 4299 | 0.2364 | → |
| Bile duct | TCGA-CHOL + GTEx | tunpaired = 2.242, df = 43 | 0.0302 | ↑ |
| Bladder | TCGA-BLCA + GTEx | tunpaired = 8.511, df = 433 | <0.0001 | ↓ |
| Brain | TCGA-GBM + TCGA-LGG + GTEx | UMann–Whitney = 147,235 | <0.0001 | ↑ |
| Breast | TCGA-BRCA + GTEx | UMann–Whitney = 110,310 | <0.0001 | ↑ |
| Cervix | TCGA-CESC + GTEx | tunpaired = 5.24, df = 306 | <0.0001 | ↓ |
| Colon | TCGA-COAD + GTEx | UMann–Whitney = 21,873 | <0.0001 | ↓ |
| Endometrium | TCGA-UCEC + GTEx | UMann–Whitney = 308 | <0.0001 | ↓ |
| Esophagus | TCGA-ESCA + GTEx | UMann–Whitney = 48,610 | <0.0001 | ↓ |
| Head & neck reion | TCGA-HNSC + GTEx | UMann–Whitney = 10,902 | 0.6338 | → |
| Kidney | TCGA-KIRC + TCGA-KIRP + GTEx | UMann–Whitney = 81,033 | 0.2866 | → |
| Liver | TCGA-LIHC + GTEx | UMann–Whitney = 14,504 | <0.0001 | ↑ |
| Lung | TCGA-LUAD + TCGA-LUSC + GTEx | UMann–Whitney = 182,850 | 0.014 | ↑ |
| Ovary | TCGA-OV + GTEx | UMann–Whitney = 12,349 | <0.0001 | ↓ |
| Pancreas | TCGA-PAAD + GTEx | UMann–Whitney = 17.5 | <0.0001 | ↑ |
| Paraganglia | TCGA-PCPG + GTEx | tunpaired = 0.7294, df = 183 | 0.4667 | → |
| Prostate | TCGA-PRAD + GTEx | UMann–Whitney = 27,055 | <0.0001 | ↓ |
| Rectum | TCGA-READ + GTEx | tWelch-corrected = 4.435, df = 9.872 | 0.0013 | ↓ |
| Skin | TCGA-SKCM + GTEx | UMann–Whitney = 67,245 | <0.0001 | ↓ |
| Soft tissue & bone | TCGA-SARC + GTEx | UMann–Whitney = 215 | 0.6772 | → |
| Stomach | TCGA-STAD + GTEx | UMann–Whitney = 37,554 | 0.0105 | ↑ |
| Testis | TCGA-TGCT + GTEx | UMann–Whitney = 7660 | <0.0001 | ↓ |
| Thymus | TCGA-THYM + GTEx | UMann–Whitney = 45.5 | 0.1532 | → |
| Thyroid | TCGA-THCA + GTEx | UMann–Whitney = 10,832 | 0.0028 | ↑ |
| Uterus | TCGA-UCS + TCGA-UCEC + GTEx | tWelch-corrected = 12.95, df = 85.67 | <0.0001 | ↓ |
| Covariates | Coefficient | Standard Error | p | HR | Lower CI95% | Upper CI95% |
|---|---|---|---|---|---|---|
| TP53 | 0.42027 | 0.50473 | 0.405035 | 1.5224 | 0.5661 | 4.0939 |
| Age at diagnosis | 0.4777 | 0.0245 | 0.051199 | 1.0489 | 0.99975 | 1.1005 |
| KPS score | −0.0442 | 0.03717 | 0.234326 | 0.9568 | 0.88954 | 1.0291 |
| IDH1 mutation (0: no, 1: yes) | −2.05162 | 0.61982 | 0.000933 | 0.1285 | 0.03814 | 0.4331 |
| Covariates | Coefficient | Standard Error | p | HR | Lower CI95% | Upper CI95% |
|---|---|---|---|---|---|---|
| PALLD | 1.281586 | 0.440867 | 0.00365 | 3.6023 | 1.51817 | 8.548 |
| TP53 | 0.846903 | 0.600697 | 0.15858 | 2.3324 | 0.71861 | 7.57 |
| Age at diagnosis | 0.070992 | 0.027862 | 0.01083 | 1.0736 | 1.01652 | 1.134 |
| KPS score | −0.008419 | 0.040141 | 0.83388 | 0.9916 | 0.91659 | 1.073 |
| IDH1 mutation (0: no, 1: yes) | −1.319920 | 0.680868 | 0.05255 | 0.2672 | 0.07034 | 1.015 |
| DF | Log Likelihood | DF | χ2 | p |
|---|---|---|---|---|
| 4 | −39.743 | |||
| 5 | −34.866 | 1 | 9.7542 | 0.001789 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayer, O.; Bugis, J.; Kozlova, D.; Leemann, A.; Mansur, S.; Peerutin, I.; Mendelovich, N.; Mazin, M.; Friedmann-Morvinski, D.; Shomron, N. Cytoskeletal Protein Palladin in Adult Gliomas Predicts Disease Incidence, Progression, and Prognosis. Cancers 2022, 14, 5130. https://doi.org/10.3390/cancers14205130
Mayer O, Bugis J, Kozlova D, Leemann A, Mansur S, Peerutin I, Mendelovich N, Mazin M, Friedmann-Morvinski D, Shomron N. Cytoskeletal Protein Palladin in Adult Gliomas Predicts Disease Incidence, Progression, and Prognosis. Cancers. 2022; 14(20):5130. https://doi.org/10.3390/cancers14205130
Chicago/Turabian StyleMayer, Ori, Joshua Bugis, Daria Kozlova, Aviv Leemann, Shahar Mansur, Ilan Peerutin, Noga Mendelovich, Meital Mazin, Dinorah Friedmann-Morvinski, and Noam Shomron. 2022. "Cytoskeletal Protein Palladin in Adult Gliomas Predicts Disease Incidence, Progression, and Prognosis" Cancers 14, no. 20: 5130. https://doi.org/10.3390/cancers14205130
APA StyleMayer, O., Bugis, J., Kozlova, D., Leemann, A., Mansur, S., Peerutin, I., Mendelovich, N., Mazin, M., Friedmann-Morvinski, D., & Shomron, N. (2022). Cytoskeletal Protein Palladin in Adult Gliomas Predicts Disease Incidence, Progression, and Prognosis. Cancers, 14(20), 5130. https://doi.org/10.3390/cancers14205130





