Simple Summary
Immunotherapy is currently the backbone of new drug treatments for many cancer patients. CXC chemokine ligand 13 (CXCL13) is an important factor involved in recruiting immune cells that express CXC chemokine receptor type 5 (CXCR5) in the tumor microenvironment and serves as a key molecular determinant of tertiary lymphoid structure (TLS) formation. An increasing number of studies have identified the influence of CXCL13 on prognosis in patients with cancer, regardless of the use of immunotherapy treatment. However, no comprehensive reviews of the role of CXCL13 in cancer immunotherapy have been published to date. This review aims to provide an overview of the CXCL13/CXCR5 signaling axis to summarize its mechanisms of action in cancer cells and lymphocytes, in addition to effects on immunity and cancer pathobiology, and its potential as a biomarker for the response to cancer immunotherapy.
Abstract
Immune checkpoint inhibitors (ICIs), including antibodies that target programmed cell death protein 1 (PD-1), programmed death-ligand 1 (PD-L1), or cytotoxic T lymphocyte antigen 4 (CTLA4), represent some of the most important breakthroughs in new drug development for oncology therapy from the past decade. CXC chemokine ligand 13 (CXCL13) exclusively binds CXC chemokine receptor type 5 (CXCR5), which plays a critical role in immune cell recruitment and activation and the regulation of the adaptive immune response. CXCL13 is a key molecular determinant of the formation of tertiary lymphoid structures (TLSs), which are organized aggregates of T, B, and dendritic cells that participate in the adaptive antitumor immune response. CXCL13 may also serve as a prognostic and predictive factor, and the role played by CXCL13 in some ICI-responsive tumor types has gained intense interest. This review discusses how CXCL13/CXCR5 signaling modulates cancer and immune cells to promote lymphocyte infiltration, activation by tumor antigens, and differentiation to increase the antitumor immune response. We also summarize recent preclinical and clinical evidence regarding the ICI-therapeutic implications of targeting the CXCL13/CXCR5 axis and discuss the potential role of this signaling pathway in cancer immunotherapy.
1. Introduction
Immunotherapy is one of the most successful forms of treatment for cancer patients [1]. Understanding and identifying tumor-specific cytotoxic T cells and tumor-associated antigens have provided additional insight into the effective design of immunotherapies capable of killing cancer cells [2,3,4,5,6]. In addition to the generation of tumor-specific effector cells that can attack cancer cells, the attraction of immune cells, especially effector cells, to the tumor microenvironment represents a critical step in cancer therapy. To effectively attack established cancer cells, effector immune cells must be able to traffic to the tumor site. Chemokines were originally identified by their chemoattractant abilities to attract white blood cells [7,8,9,10]. During inflammation or an immune response, various leukocytes recognize specific chemokines, which attract them to specific sites where they mediate effects. Consequently, chemokines maintain immune system homeostasis, regulate the innate or adaptive immunity response, and recruit leukocytes to the specific site [11,12]. Additionally, chemokines can activate and prime lymphocytes, triggering their transformation into effector cells that attack tumor cells and suppress tumor proliferation [13,14]. Chemokines are also capable of suppressing immune cell function, limiting their abilities to attack tumor cells [15,16,17].
Chemokines are small peptides with molecular weights of 8–10 kDa that can specifically circulate, retain, and activate some immunocompetent cells [18,19]. Chemokines are chemotactic cytokines that mediate activity through interactions with specific G protein-coupled receptors (GPCRs) with seven transmembrane domains, known as chemokine receptors [20]. The activation of the chemokine/chemoreceptor signaling pathway induces cell proliferation, migration, homing, survival, and gene expression [21,22,23,24]. The interaction between chemokines and their receptors activates a complex and plastic signaling pathway, which is crucial for the regulation of specific leukocyte subpopulation responses. Variations in chemokine expression levels can result in a wide range of functions with consequential effects on the treatment response to immune checkpoint inhibitors (ICIs) among patients with cancer. Elevated or reduced expression levels of different chemokines or chemokine receptors have been associated with diverse responses to immunotherapy across multiple cancer types [25,26,27]. Chemokine activation and the downstream function of signaling effectors influence almost all of the processes involved in the immunotherapy treatment response in cancer.
C-X-C motif chemokine ligand 13 (CXCL13), also known as B lymphocyte chemoattractant, was initially detected in stromal cells of B cell follicles, associated with the recruitment of B cells and T cells subsets [28,29]. CXCL13 has also been identified as a critical homeostatic chemokine that is expressed in associated lymphoid tissues, and B cells and follicular T helper (TFH) cells form B and T cell connective zones according to CXCL13 gradients.
C-X-C motif chemokine receptor type 5 (CXCR5) is the main and only receptor for CXCL13 and mediates the functions of chemokines through specific downstream interactions [30,31,32]. The interaction between CXCR5 and CXCL13 can induce the entry of T or B cells into lymphoid organs and integrin expression, and CXCR5-deficient mice present with serious immune system defects [32,33]. An increasing number of studies have identified that various influences of CXCL13 expression levels prognosis in patients with cancer regardless of immunotherapy treatments [34,35,36,37,38].
The CXCL13/CXCR5 axis is linked with tumor development, progression, proliferation, and invasion. Abnormally active CXCL13/CXCR5 signaling enhances cancer cell growth through complex and different mechanisms in breast cancer [39,40], intestinal cancer [32,41], lung cancer [42], prostate cancer [43,44,45], oral squamous cell carcinoma [46,47], renal cell carcinoma [48], neuroblastoma [49,50], and lymphoma [51,52]. In this review, therefore, we discuss the CXCL13/CXCR5 axis and summarize its mechanism of action in cancer cells and lymphocytes, its contributions to immunity and cancer pathobiology, and its role in response to immunotherapy and potential to serve as a biomarker.
2. The CXCL13/CXCR5 Signaling Axis
The specific downstream signaling pathway activated by the binding of CXCR5 with CXCL13 remains unclear and is likely dependent on the GPCR activity of CXCR5 [53]. El-Haibi et al. reported that Gα13 co-immunoprecipitated with CXCR5 under CXCL13-treated conditions in prostate cancer cells [54]. However, Gαq and Gαi2 co-immunoprecipitate with CXCR5 in the absence of CXCL13 but dissociated in the presence of CXCL13. When a GPCR binds with a ligand, the GDP bound to the small G protein is replaced with a GTP, activating the small G protein and resulting in dissociation from the GPCR. The dissociated small G protein mediates further cellular signal transduction [55]. Gαq and Gαi2 but not Gα13 dissociate from the GPCR CXCR5 upon binding with CXCL13, suggesting that Gαq and Gαi2 were the major active small G proteins involved in CXCL13/CXCR5 signaling activation. Additional evidence was provided about small interfering RNA (siRNA) targeting Gαq, Gαi2, and Gα13 [54]. CXCL13-dependent cell invasion in prostate cancer (PCa) cells was significantly inhibited by the application of siRNA targeting Gαq and Gαi2, indicating that the CXCL13/CXCR5/Gαq signaling pathway regulates cellular chemotaxis. Small G protein downstream kinases Rac and RhoA were determined upon cancer cells receiving CXCL13. They found CXCL13 activated Rac but not RhoA, and this was in Gαq and Gαi2 dependent manner. Small G protein downstream kinases Rac and RhoA were determined upon cancer cells receiving CXCL13. They found CXCL13 activated Rac but not RhoA, and this was in Gαq- and Gαi2-dependent manner. The precise mechanism of CXCL13/CXCR5 signaling is still unclear in B cells and T cells. There is only rare evidence suggesting that small G proteins would play an important role in CXCL13/CXCR5 signaling. In B cells, Han et al. demonstrated that Gαi2 is the dominant factor determining B cell mobility [56]. The Gαi2−/− B cells respond poorly to CXCL13 treatment and fail to migrate toward chemokine sites in a filter plate assay. When Gαi2−/− B cells were transplanted into recipient mice, they failed to migrate to the lymph node. In T cells, Hwang et al. demonstrated that Gαi2−/− cells were characterized by poor mobility and impaired trafficking capabilities [57]. Cell migration declined, on average, approximately 4- to 7-fold in Gαi2−/− CD4+ and CD8+ T cells compared with wild-type cells when exposed to chemotaxis conditions. They assessed the morphology of secondary lymphoid structures in Gαi2−/− mice by immunohistochemical staining and identified the disrupted expression of T cells in the marginal zones, the failure of germinal center formation, and a smaller lymphoid structure size in Gαi2−/− mice compared with wild-type mice. The detailed mechanism of how CXCL13/CXCR5 signaling works is represented in Figure 1 below. In summary, the CXCL13/CXCR5 signaling in T cells, B cells, and cancer cells are similar. In T cells and B cells, CXCL13/CXCR5 stimulates their chemotaxis, and this is in Gαi2 dependent manner. The downstream signaling of Gαi2 in CXCL13-treated T cells and B cells is still not fully clear, probably are Rac1, Rac2, or other Rho GTPase [58]. In cancer cells, CXCL13/CXCR5 regulates cell invasion and migration, and those are in Gαq and Gαi2 /Rac dependent manner.
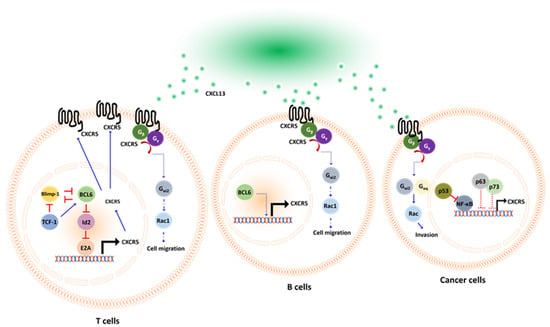
Figure 1.
Schematic representation of CXCL13/CXCR5 axis in T cells, B cells, and cancer cells, respectively. CXCL13 acts as a ligand that specifically binds to its receptor CXCR5. Upon activation, the GDP bound Gα protein subunit is replaced with a GTP molecule. This leads to the dissociation of Gαq or Gαi2 from the Gβ–Gγ dimer, mediating downstream signal activation. CXCL13 promotes chemotaxis in T cells and B cells. However, in cancer cells, CXCL13 promotes migration and invasion. The transcriptional factors involved in CXCR5 expression are diverse in T or B cells and cancer cells. In T cells, BCL6 regulates the CXCR5 expression via a repressor of repressor circuit. While in cancer cells, p53 regulates CXCR5 indirectly by suppressing NF-κB.
Transcriptional factor p52/RelB in the NF-κB pathway had been reported as a transcriptional factor of CXCL13 in macrophage [59] and B cells [60,61] in mice lacking p52 or with a RelB defect in B-cell follicles and germinal centers formation [62]. NF-κB proteins are also key transcriptional factors of immune checkpoint PD-L1 [63,64,65], suggesting that NF-κB would simultaneously control immune checkpoint blockade response and T/B cell recruitment. However, different transcriptional machinery controls the expression of CXCR5. In T cells, BCL6 activates CXCR5 expression by repressing repressor pathways [66]. Transcriptional factor E2A is directly bound to enhancer regions of CXCR5 to activate its expression [67,68]. However, Id2/Id3 dampened CXCR5 expression by antagonizing E2A [68,69]. BCL6 acts as a transcriptional repressor that directly inhibits Id2 expression in T cells, which, in turn, activates E2A activity and then stimulates CXCR5 expression [66,70]. Interestingly, BCL6 and Blimp-1 are mutually exclusive and antagonize each other [71]. If Blimp-1 dominates BCL6 in this mutual competition, naïve CD4+ T cells will differentiate into Th1 cells that do not express CXCR5 [71], in contrast, if upstream transcriptional factor T cell factor 1 (TCF-1) is triggered, naïve CD4+ T cells would differentiate into TFH and express CXCR5 [72]. This differentiation occurs because TCF-1 is a transcriptional factor that simultaneously suppresses Blimp-1 [73,74] and activates BCL6 [75]. In CD8+ T cells, transcriptional regulators TCF-1, BCL6, E2A, Blimp-1, and Id2/Id3 also work collaboratively to shape CXCR5 expression with the same mechanism, as described in TFH cells [68]. In cancer cells, Mitkin et al. identified that p53 indirectly suppressed CXCR5 expression in the MCF breast cancer cell line. Silencing of p53 with shRNA significantly restored CXCR5 expression in breast cancer cells. This was because p53 suppressed transcriptional factor NF-κB, which directly transactivates CXCR5 by interacting with the promoter region [76]. The expression of CXCR5 is regulated by the transcription factors BCL6 and Blimp-1. BCL6 drives the expression of CXCR5, whereas Blimp1 plays an inhibitory role. BCL6 and Blimp1 antagonize each other to regulate CXCR5 expression in T cells. [71] (Figure 1).
In addition to CXCL13/CXCR5, Gai2 is also involved in CXCR3 downstream signaling upon receiving its chemotaxis ligands CXCL9, CXCL10, and CXCL11. The CXCR3 has two variants, CXCR3A and CXCR3B. CXCL4, CXCL9, CXCL10, and CXCL 11 activate Gai in CXCR3A but not CXCR3B [77]. Of those Gai subunits, Gai2 is the most dispensable for T cells’ CXCR5 response to CXCL9, CXCL10, and CXCL11 [78].
The regulator of G protein signaling (RGS) acted as a GTPase-activating protein to terminate GPCR signaling by dephosphorylating the Gα•GTP subunit [78,79]. Silencing RGS1 restored chemotaxis of cytotoxic T cells and Th1 cells to enhance their infiltrating toward tumors. This process revealed that a G protein component, Gα, is essential for the chemotaxis of cytotoxic T cells and Th1 cells [80]. The regulatory proteins of GPCR would be potential drug targets to revive anti-tumor activity.
3. The Expression and Implications of CXCL13/CXCR5
CXCL13 is a chemoattractant that selectively interacts with its receptor, CXCR5, to promote the migration of CXCR5+ cells toward high CXCL13 concentration areas. CXCL13 was originally identified as B cell attracting chemokine 1(BCA-1) and was shown to act on CXCR5+ B cells to induce chemotaxis and Ca2+ mobilization [81]. In lymph nodes, the CXCL13/CXCR5 axis is essential for homing B lymphocytes to lymphoid tissue, and CXCL13/CXCR5 deficient mice show the disrupted localization of B cells [81].
CXCL13 recruits B cells to tumors, where they form tertiary lymphoid structures (TLSs) [82]. TLSs are similar to secondary lymphoid organs on the cellular, morphological, and molecular levels, and TLSs are considered with good prognostic markers, commonly associated with better survival rates. However, the mechanisms underlying TLS development remain unclear. Rodriguez et al. found that the organization of cancer-associated fibroblasts into reticular networks depends on tumor necrosis factor receptor signaling, which acts as a lymphoid tissue organizer [83]. Cancer-associated fibroblasts secrete CXCL13 to recruit CXCR5+ B cells expressing lymphotoxin-α1β2, which expands TLSs in the tumor microenvironment. Moreover, CD8+ T cells mediate cancer-associated fibroblast organization, serving as an inducer of lymphoid structures to drive the formation of TLSs.
CXCR5+ T cells were originally defined as CD4+ memory-like T cells, which function to assist B cells with antibody production. The colocalization of CXCL13, CXCR5+ T cells, and B cells was detected in the follicular mantle zone. A unique subset of T cells that assist in B cell function was identified as TFH cells [84]. High levels of BCL6 drive TFH differentiation, and Blimp-1 is able to block T cells from differentiating into TFH cells [71].
TFH cells are the major CXCL13-producing cells in the lymphoid structure. In tumor microenvironments, tumor-infiltrating CXCL13-producing TFH cells may be involved in promoting local B cell localization, differentiation, and maturation [85]. An activated germinal center of B cells then contributes to the formation of TLSs in tumors. Gu-Trantien et al. first observed CXCL13 producing TFH cells in breast cancer and found that the TFH signature strongly predicted a positive clinical outcome in breast cancer [86]. Naïve CD4+ T cells differentiate into T helper 1 (Th1)-oriented TFH cells that produce CXCL13 in tumor microenvironments after being primed by antigen-presenting cells in the lymph node [87,88]. The balance of TFH/Th1 differentiation from precursor cells is determined by the competitive expression of Bcl6 and T-bet [71,89,90,91]. Further investigation revealed that tumor-infiltrating TFH cells produce CXCL13 to attract CXCR5+ CD8+ T cells and CXCR5+ B cells toward the germinal centers within the TLS, where both T cells and B cells are primed, reinforcing their cytotoxic capacity against cancer cells [92]. These studies indicate that BCL6-dominant signaling promotes CXCL13 expression in TFH cells [93]. Another subset of TFH cells, which were PD-1hiCXCR5−CD4+ T cells that expressed CXCL13, did not show elevated BCL6 expression.
In addition to TFH cells, there are several CXCL13 producing cells in lymphoid tissue and would secrete CXCL13 to chemoattract cells that express CXCR5. Denton et al. identified pulmonary fibroblasts as CXCL13-producing cells induced by Type I interferon (Type I IFN) during Influenza A virus transfection, which facilitate the recruitment of CXCR5+ B cells and T cells to the lungs to generate pulmonary germinal centers [94].
Antigen-specific CD4+ T cells mediate and coordinate immune cell functions in antitumor activities. Although the effects of ICI have been thoroughly explored in CD8+ T cells, the roles played by CD4+ T cells in response to ICI remain poorly understood. Balança et al. studied exhausted CD4+ T cells in head and neck, cervical, and ovarian cancers [95]. With exhausted CD4+ T cells defined as those cells presenting high PD-1 and CD39 levels, which were found to secrete CXCL13 and express the transcription factor thymocyte selection-associated high mobility group box (TOX). The transcription factor SRY-box transcription factor 4 (SOX-4) mediates CXCL13 production on CD4+ T cells, enriching transforming growth factor-β (TGF-β) and reducing IL-2 expression [96]. Similar to CXCL13+ exhausted CD8+ T cells, exhausted CD4+ T cells recruit B cells and promote the formation of TLS, suggesting a good response to immunotherapy and indicating a crucial role for CD4+ T cells in mediating the functions of various immune cells.
Cytotoxic CD8+ T cells are the strongest effectors, playing important roles in the anticancer immune response. When examining the CXCL13/CXCR5 axis, Workel et al. found that TGF-β dependent CD103+CD8+ tumor-infiltrating T cells represent an important CXCL13 resource [97]. When TGF-β receptors are inhibited, CXCL13 signaling is abrogated. CXCL13+CD103+CD8+ tumor-infiltrating T cells are associated with B cell recruitment, TLS formation, and neoantigen burden. Additionally, Thommen et al. also found that PD-1highCD8+ tumor-infiltrating T cells produce more CXCL13 than PD-1-CD8+ T cells in non-small-cell lung cancer (NSCLC) [34]. They also observed that CXCL13 recruited other CXCR5-expressing immune cells and induced the formation of TLSs in NSCLC. PD-1highCD8+ tumor-infiltrating T cells are enriched inside the TLS, and their presence successfully predicts the response to anti-PD-1 and is positively correlated with overall survival.
In chronic hepatitis B infection, Li et al. found that higher levels of CXCL13 facilitate the recruitment of CXCR5+CD8+ T cells to liver tissue in patients, which is associated with a favorable response to antiviral drug treatment [98]. CXCR5+CD8+ T cells secrete hepatitis B virus (HBV)-specific cytokines, such as IL-2, IFN-γ, IL-17, and IL-21. The subset of CD8+ T cells is partially exhausted but not dysfunctional and is involved in controlling the viral infection load. Strikingly, the adoptive transfer of CXCR5+CD8+ T cells to an HBV mouse model resulted in a significant decrease in HBV antigen expression. Finally, they found that B cell-deficient mice have a lower frequency of CXCR5+CD8+ T cells and decreased the HBV-specific IFN-γ+ CXCR5+CD8+ T cells in the blood and liver. These data demonstrated that B cells are required for CXCR5+ CD8+ T cells, contributing to the functions and activities of CXCR5+CD8+ T cells.
In classical Hodgkin lymphoma, Le et al. identified a special CD8+ T cell subset expressing CXCR5 and an inducible T cell costimulator (ICOS) [99]. This subset was functionally similar to TFH cells, with low CCR7 expression and high levels of BCL6, PD-1, CD200, and OX40 expression. In addition, these cells displayed poor cytotoxic function and low interferon-secretion and produced IL-4, IL-21, and CXCL13, similar to CD4+ TFH cells. Gene profiling analyses demonstrated that CXCR5+ICOS+CD8+ T cells are significantly similar to CD4+ TFH cells and are involved in the generation of lymphoma tumors with residual germinal centers. Overall, CD8+ T cell differentiation pathways are functionally similar to those involved in CD4+ TFH cell differentiation in Hodgkin lymphoma.
Follicular dendritic cells represent the primary cellular source of cytokines in lymphoid organs [100]. Previous research has identified that various dendritic cell subsets are able to secrete CXCL13, playing an important role in the establishment of interactions between lymphocytes and dendritic cells [101]. Additionally, CXCL13 serves as a plasma biomarker that reflects germinal center activity [102]. Follicular dendritic cells represent the primary source of CXCL13 expression in reactive tonsils and lymph nodes located in the B cell zone. Moreover, dysplastic and neoplastic follicular dendritic cells are able to secrete CXCL13 to recruit lymphocytes to areas of dense inflammatory infiltration [100]. Overall, dendritic cells secrete CXCL13 to recruit CXCR5+ lymphocytes, orchestrating the immune response in lymphoid tissues.
We summarize prior findings regarding CXCL13 and CXCR5 expression. CD8+ and CD4+ T cells, cancer-associated fibroblasts, cancer cells, and dendritic cells are able to secrete CXCL13, and CD8+ and CD4+ T cells, B cells, and cancer cells express CXCR5 (Figure 2). Increasing research has reported the detection of CXCL13 in the tumor microenvironment of many different types of cancer [103,104,105,106,107,108,109]. When the cancer microenvironment is enriched in CXCL13, the recruitment of CXCR5-expressing leukocytes to the tumor microenvironment increases. B cells are recruited by CXCL13 to promote TLS formation in the tumor microenvironment (Figure 3). Previous studies have correlated TLS formation with better prognosis among patients with cancer [110,111,112,113]; therefore, treatments that promote the development of TLSs represent potential therapeutic strategies.
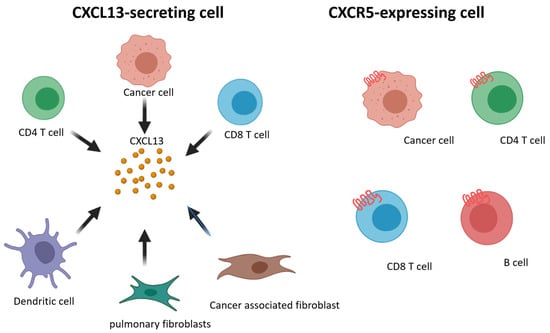
Figure 2.
Schematic representation of CXCL13 and CXCR5 expressing on different cell subsets.
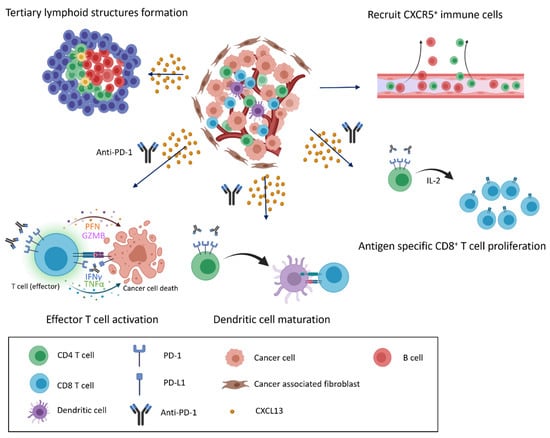
Figure 3.
CXCL13/CXCR5 signaling and response to immune checkpoint blockade in the tumor microenvironment. CXCL13 secretion and enrichment in the tumor microenvironment alter the immune cell composition. CXCL13 recruits cells that express CXCR5 to infiltrate the cancer microenvironment, inducing the formation of tertiary lymphoid structures and the further infiltration of various immune cells (top). When anti-PD-1 antibodies are present, recruited immune cells actively attack cancer cells, and exhausted cells transition into effector cells, leading to the proliferation of CD8+ T cells and the maturation of dendritic cells in response to anti-PD-1 treatment (bottom).
4. The CXCL13/CXCR5 Signaling Axis in the ICI Response of Preclinical Models
Immune checkpoint blockade therapy has been hugely successful with effective results in many cancer types [114,115,116], acting through reinforcement of T cell function and improving the cytotoxic capacity against cancer cells [1]. In this section, we summarize existing research examining the contributions of CXCL13/CXCR5 to immunotherapy in preclinical models (Table 1).

Table 1.
Cancer immunotherapy approaches related to the CXCL13/CXCR5 axis, based on preclinical models.
Li et al. used an HBV mouse model and HBV-related hepatocellular carcinoma (HCC) samples from patients to investigate the role placed by CXCL13/CXCR5 signaling in HBV and HBV-related HCC [98]. They detect CXCR5 and CXCL13 expression levels using RNA-sequencing, quantitative reverse transcriptase PCR (qPCR), immunohistochemistry (IHC), enzyme-linked immunosorbent assay (ELISA), and Western blotting methods. They identified the recruitment of CXCR5+CD8+ T cells by CXCL13. Additionally, anti-PD-1 blockade and recombinant IL-21 therapy can promote the secretion of IFN-γ from CXCR5+CD8+ T cells in patients with HBV. In an HBV mouse model, CXCR5+CD8+ T cells would inhibit hepatitis B surface antigen (HBsAg) expression, which was not observed in response to CXCR5-CD8+ T cells. Anti-PD-1 and IL-21 treatment strengthen the function of immune cells involved in HBV clearance or the attack of cancer cells.
Rodriguez et al. studied cancer-associated fibroblasts to determine the effects of TLSs in a melanoma and colon adenocarcinoma mouse model [83]. Anti-PD-L1 and a combination of anti-PD-1 and anti-CTLA4 both increase the formation of TLSs in the tumor microenvironment, resulting in reduced tumor growth, and increased T cell recruitment to the tumor, as indicated by the formation of discrete T cell and B cell zones. They observed CXCL13 expression on cancer-associated fibroblasts, as assessed by immunofluorescence and real-time PCR. Finally, they found that immunotherapy increased the tumor-associated TLS size and number, which was associated with reduced cancer growth.
Balança et al. studied the role of CD4+ T cells in immunotherapy during the attack on cancer cells [95]. They applied single-cell RNA sequencing analysis to patient-derived samples of head and neck, cervical, and ovarian cancer to detect the expression of transcription factor TOX and the chemokine CXCL13. They found that anti-PD-1 therapy promoted the functional activity of exhausted CD4+ T cells, promoting dendritic cell maturation and CD8+ T cell proliferation. Exhausted CD4+ T cells also play important roles in regulating and coordinating immune cells in the attack on cancer cells after immunotherapy treatment.
Yang et al. analyzed the influence of CXCL13 on the ICI response in an ovarian cancer mouse model [117]. They identified different immune cell subsets in samples from patients with high-grade ovarian cancer that were able to secrete CXCL13. High levels of CXCL13 expression resulted in increased lymphocytes infiltration in the tumor, as assessed by immunofluorescence. Conclusively, anti-PD-1 combined with CXCL13 treatment is associated with a better treatment response than anti-PD-1 alone in a mouse model of ovarian cancer.
Zhang et al. combined the inhibitor of cyclin-dependent kinases 4 and 6 (CDK4/6i) with anti-PD-1 therapy and examined the synergistic effects in an ovarian cancer mouse model [119]. PCR and cytokine array methods were applied to detect cytokine and chemokine levels, which revealed that ID8 ovarian cancer cells secrete higher levels of CXCL10 and CXCL13 after CDK4/6i treatment resulting in increased lymphocytes infiltration in the tumor microenvironment. Combination treatment is associated with a better response and more active function among CD8+ and CD4+ T cells than anti-PD-1 or CDK4/6i alone, and the synergistic antitumor effects were dependent on the activities of B cells and CD8+ T cells.
B cells play important roles in the results of immunotherapy. Cabrita et al. reported that TLSs influence the immunotherapy response of melanoma, showing that the co-occurrence of CD8+ T cells and CD20+ B cells can predict the response to ICI therapy in patients with metastatic melanomas [118]. CXCL13, CXCR5, CD8+ T cells, and CD20+ B cells colocalized in melanoma samples obtained from patients using immunofluorescence staining, which revealed the TLS formation in these tumors. Moreover, gene expression associated with TLS formation was able to predict the clinical outcomes of immunotherapy. Without TLS formation, a dysfunctional molecular phenotype can be observed, with adverse effects on immunotherapy outcomes. TLSs play critical roles in influencing the response of the tumor microenvironment to ICI therapy, and therapeutic strategies that induce TLSs’ formation might improve the efficacy of immunotherapy.
CXCL13 has been identified as an exhaustion marker in previous research [120,121]. Studies have increasingly identified immunotherapy as being positively correlated with exhausted immune cells [122,123,124], including our research results for HCC [125]. The increased expression of exhaustion markers in the tumor microenvironment is an indicator of increased immune cell infiltration, which suggests a good response to immunotherapy. Bassez et al. identified several genes that were able to predict a better immune response to anti-PD-1 treatment associated with CXCL13 [126]. CXCL13 expression at high levels corresponds with T cells expansion after anti-PD-1 treatment for breast cancer, which is associated with a good response to immunotherapy.
Research continues into understanding why some subsets of patients respond to immunotherapy, but others do not respond. The heterogenicity of tumor cells and immune cells is a very important issue, which brings about different immunotherapy efficacy regardless of tumor types. The study divided cancer patients into two groups [126]. One group had expanding T cells, and the other had non-expanding T cells following anti-PD-1 therapy. The results demonstrated that CXCL13 expression on T cells is associated with the expansion of T cells, which implies that one subset of patients has a good response after immune checkpoint inhibitors treatment. The research sheds light on the exploration of the heterogenicity of immunotypes as well as associated genes for immunotherapy response.
We summarize the effects of CXCL13 and CXCR5 expression on preclinical models of the immunotherapy response. TLSs play critical roles in influencing the response to immune checkpoint blockade in the tumor microenvironment. Anti-PD-1 antibodies in the tumor microenvironment activate CD8+ T cells, and CD4+ T cells mediate CD8+ T cell proliferation and dendritic cell maturation (Figure 3). CXCL13 expression recruits additional immune cells to infiltrate the tumor environment to attack cancer cells following immunotherapy treatment.
5. CXCL13/CXCR5 Axis for ICI Response in Clinical Tumors
Immune checkpoint blockade can regulate tumor progression. Compared with chemotherapy, ICI therapy has a superior duration of response (20.4 months vs. 6.3 months) [127,128]. However, only 15–20% of patients benefit from immunotherapy [128] and the identification of biomarkers able to predict therapeutic response to ICI. Recently, some biomarkers were identified that were able to accurately predict the response of cancer patients to PD-1 blockade. Le et al. proposed that patients with mismatch repair deficiencies have a higher response rate to pembrolizumab treatment than patients with proficient mismatch repair processes [129]. In this section, we summarize clinical trials and research linking the CXCL13/CXCR5 axis with the ICI response in patients (Table 2).

Table 2.
Cancer immunotherapy approaches relative to the CXCL13/CXCR5 axis based on clinical data.
5.1. Breast Cancer
Gu-Trantien et al. analyzed CD4+ T cells in tumor tissues by profiling gene signatures. They identified TFH-related genes, such as CD200, CXCL13, ICOS, and PD-1, that were over-expressed in infiltrated tumors. They divided patients’ tumors into those with a high level of CD4+ tumor-infiltrating lymphocytes (TILs) and those with minimal infiltration [86]. Among patients with a high level of TILs, CXCL13 was the most highly overexpressed gene. IHC staining indicated that CXCL13 was not expressed in tumor cells but was detected intensively in TLSs and CD4+ TFH cells. Clinical correlations with CXCL13 were performed by investigating the disease-free survival (DFS) of 794 breast cancer patients, which revealed that CXCL13 expression was positively correlated with DFS in breast cancer, especially in the human epidermal growth factor 2 (HER2+) group. The authors also analyzed a cohort of 996 patients with breast cancer who received neoadjuvant chemotherapy, which showed that CXCL13 expression was highly correlated with a complete response in HER2+ patients.
Zhang et al. designed a clinical trial to identify key immune subpopulations associated with clinical outcomes for anti-PD-L1 blockade therapy in triple-negative breast cancer (TNBC) [131]. Twenty-two patients with late-stage TNBC were enrolled in this cohort, divided into two groups with equal numbers. Patients in one group were treated with paclitaxel, whereas the other group received paclitaxel plus atezolizumab. After 4 weeks, tumor biopsies and peripheral blood cells were collected from both groups. TILs were collected and analyzed by single-cell RNA-sequencing, single-cell ATAC-sequencing, and TCR-sequencing. In this cohort, the authors developed two indexes to associate immune subsets with clinical benefits in TNBC: the predictive index (Pi) and the therapeutic index (Ti). The Pi correlated the compositions of TIL subsets with changes in tumor volume, whereas the Ti correlated between TIL subsets dynamics with tumor volume changes. They identified B cells as being the most predictable subset for predicting the response to immune checkpoint blockade in the Pi analysis, whereas T cells were the most predictable subset to predict immune checkpoint blockade in the Ti analysis, indicating a functional change in T cells during ICI therapy. The authors clustered T cells using a high-resolution T cell map, which identified expanded CXCL13+CD8+ and CXCL13+CD4+ populations in patients who responded to paclitaxel plus atezolizumab but not patients who did not respond to treatment. Furthermore, the analysis of CXCL13+CD8+ T cells using a single-cell assay for transposase-accessible chromatin using sequencing (ATAC-seq) showed that CXCL13+CD8+ cells had more accessible chromatin regions in the IFNG, GZMK, and PDCD1 loci.
5.2. Bladder Cancer
Goswami et al. analyzed the combined data from two clinical trials, CheckMate275 and IMvigor210, to determine the correlations of the AT-rich interactive domain containing protein 1A (ARID1A) mutations and CXCL13 expression with the patient’s response to immune checkpoint blockade therapy [109]. CheckMate 275 is a phase 2 trial of nivolumab for the treatment of metastatic urothelial carcinoma (n = 265). IMvigor210 is a phase 2 trial of atezolizumab for the treatment of advanced or metastatic urothelial bladder cancer. They identified that an ARID1A mutation alone or CXCL13-high expression is correlated with favorable responses to ICI, and ARID1A mutation in combination with high levels of CXCL13 expression could predict the better response of patients to immune checkpoint blockade therapy than ARID1A mutation in combination with low levels of CXCL13 or high levels of CXCL13 in combination with ARID1A wild-type.
5.3. Non-Small Cell Lung Cancer
Thommen et al. classified CD8+ TILs derived from patients with NSCLC into three groups based on their PD-1 surface-level expression [34]. PD-1- were CD8+ TILs without detectable PD-1 expression; PD-1N were CD8+ TILs with PD-1 levels similar to those observed in healthy donors; PD-1T were CD8+ TILs PD-1 with considerably high expression levels. The reactive capacity of PD-1 TILs was confirmed by co-culturing PD-1T, PD-1N, and PD-1-, respectively, with autologous tumors isolated from eight patients. Among the eight patient-derived tumors, PD-1T TILs showed strong reactivity against the tumor cells in 6 of 8 cultures. Gene expression diversity among PD-1T, PD-1N, and PD-1- populations was identified by transcriptome analysis, which revealed that CXCL13 was one of the most upregulated genes in the PD-1T subset. The authors used bead-based immunoassays to quantify inflammatory cytokine and chemokine levels in sorted PD-1T TILs after 24h of culture, identifying that the levels of CXCL13 were the highest in the array. These data indicated that PD-1T TILs are likely to secrete CXCL13 into the tumor microenvironment. Of 21 stage IV NSCLC patients receiving anti-PD-1 therapy, 7 patients were identified as responders, whereas the other 14 patients were identified as non-responders. Responders were characterized by TIL subsets with a higher percentage of PD-1T TILs and total PD-1T TIL cell numbers. Those with PD-1T subsets > 1% of total cells were associated with longer median survival than those with PD-1T subsets < 1% of total cells following the administration of anti-PD-1 therapy. IHC and digital image analysis suggested that PD-1T TILs were localized in TLSs that formed both intratumorally and peritumorally. Intensive B cell marker staining and CD4+ TFH markers colocalized with PD-1T TILs, implying that CD8+PD-1T TILs might recruit CXCR5+ B cells and TFH cells into the tumor region and that B cells, as well as TFH cells, could reinforce the antitumor capacity of CD8+ T cells.
5.4. Hepatocellular Carcinoma
Liver cancer is one of the world’s most common cancers and the second leading cause of cancer deaths, and we also have some related studies in the past [125,134,135]. Hsu et al. established a platform to assess various biomarkers associated with prognostic efficacy for the immunotherapy response of patients with hepatocellular carcinoma (HCC) using Nanostring RNA analysis and multiplex immunofluorescence staining methods [125]. In this cohort, RNA was isolated from tumor samples before patients received anti-PD-1 or combination anti-PD-1 and anti-PD-L1 blockade therapy (n = 42). RNA was then hybridized to nCounter® probes for the 770 predefined genes featured in the PanCancer Immune Profiling Panel. They found that exhausted CD8+ T cells were enriched in responders and analyzed exhausted CD8+ T cells using transcriptome analysis. In this Immune Profiling Panel, nine genes expressed on exhausted T cells were positively correlated with a better response to anti-PD-1 monotherapy or combination anti-PD-1 and anti-PD-L1 therapy, including CXCL13 gene expression. In another study, the exploratory analysis identified distinct gene signatures associated with tumor response and resistance to anti-PD-1 monotherapy in HCC patients also showed that CXCL13 was positively correlated with a better response to anti-PD-1 therapy [136].
5.5. Pan-Cancers
Litchfield et al. collected exome/transcriptome data from 12 cohorts, including 1008 patients who received immune checkpoint blockade therapy for the treatment of urothelial cancer (n = 387), melanoma (n = 353), head and neck cancer (n = 107), non-small cell lung cancer (n = 76), renal cell carcinoma (n = 51), colorectal cancer (n = 20), and breast cancer (n = 14) [132]. Immune checkpoint blockade could be classified into (1) anti-PD-1 (n = 432), (2) anti-PD-L1 (n = 421), and anti-CTLA4 (n = 155). The authors collected raw fastq sequencing data from these 12 cohorts and reprocessed them using a uniform bioinformatics pipeline. Next, they obtained clinical response data for participants in each of the 12 cohorts and reclassified them into complete response (CR)/partial response (PR) group versus stable disease (SD)/progressive disease (PD). Using the CPI1000+ database v1.1, they identified 101 genes that were significantly upregulated in CR/PR versus SD/PD, among which CXCL13 was the most upregulated gene among responders (CR/PR), suggesting that CXCL13 expression may be correlated with a patient’s response to ICI therapy.
The incidence of ICI-induced mortality is estimated at about 0.3% to 1.3% [137], and the risk is lower than conventional therapy, such as 0–4% with targeted therapies [138] and about 15% with allogeneic hematopoietic stem cell transplantation [139]. Different doses [140,141] and the timing of administration and agents [142] may promote different levels of immune-related adverse events (irAEs) on patients receiving ICI. Common irAEs include headache, rash, pruritus, fatigue, pneumonitis, diarrhea, arthralgia, and endocrinopathies, which usually involve multiple organs [143,144,145]. Future research must address irAEs through more clinical trials, medical findings, and cutting-edge diagnosis tools, and the detailed mechanism causing these events will be better understood.
Two-thirds of CD8+ TILs express PD-1, and they mainly present an exhaustion situation [146]. ICI therapy is able to restore and enhance T cell function and effector cytokine production, such as TNF-α, IL-2, and IFN-γ [146,147,148]. Additionally, PD-1 inhibition could reverse the exhaustion status of specific T cells in tumor microenvironments [148].
We summarize the previous findings examining the association between CXCL13/CXCR5 expression and the clinical response to tumor immunotherapy. CXCL13 expression was able to predict the response to ICI by directly or indirectly influencing the responses of various cancer types. However, the mechanism through which CXCL13/CXCR5 signaling induces the response of various immune subsets to ICI treatment remains unknown, and determining the cellular subsets that serve as the major producers of the CXCL13/CXCR5 signal that induces immune cell infiltration in cancer requires additional study.
6. Conclusions and Perspectives
The CXCL13/CXCR5 signaling axis is able to recruit immune cells and enhance the capacity to attack cancers. On the other hand, the CXCL13/CXCR5 axis may induce tumor development and facilitate the downregulation of T cell immunity due to associated immunosuppression cells infiltrating within the tumor microenvironment, such as MDSC [149] and Treg [150]. CXCL13/CXCR5 signaling forms two opposite influences for cancer development. When two distinct pathways have similar strengths, cancers maintain homeostasis. However, anti-PD-1 is able to enhance the function of T cells, strengthening their anti-tumor ability. Additionally, MDSC and Treg express PD-L1 [151], and they may suppress T cell response by the PD-1/PD-L1 pathway. ICI therapy reduces immunosuppression and increases immune response, which is able to control tumor growth.
CXCL13 is a very important factor involved in the recruitment of CXCR5-producing immune cells to infiltrate the tumor microenvironment. CXCL13 expression was able to induce the establishment of TLS, which presents tumor antigens to T cells to determine consecutive T and B cell responses, resulting in the generation of effector T cells, plasma cells, and antibodies. In other words, the CXCL13/CXCR5 signal is able to turn “cold tumors” into “hot tumors”. The ability of immunotherapy indicates an improved response, which activates lymphocyte function to kill cancer cells. The validation of the CXCL13/CXCR5 signaling pathway components as useful biomarkers for cancer diagnosis or prognosis, particularly the response to ICI, requires further investigation. Exploring the regulation of this pathway could provide an avenue for the design or more effective immunotherapy-based treatment strategies for cancer.
Author Contributions
Conceptualization, C.H. and D.-L.O.; literature search, C.-H.H. and C.-Z.J.; writing—original draft preparation, C.-H.H. and C.-Z.J.; writing—review and editing, C.-H.H., C.-Z.J., L.-I.L., G.-S.L. and P.-Y.O.; supervision, L.-I.L., C.H. and D.-L.O.; funding acquisition, C.H. and D.-L.O. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the following research grants: NTU-109L901403, NTU-110L901404 (from Ministry of Education, Taiwan), MOST 106-2314-B-002-229-MY3, MOST 107-3017-F-002-002, MOST 107-2314-B-002-210-MY3, MOST 108-2314-B-002-075-MY3, MOST 108-3017-F-002-004, MOST 109-2634-F-002-043, 109-2314-B-002 -229 -MY3, MOST 110-2634-F-002-044 (from Ministry of Science and Technology, Taiwan), YongLin Chair Grant S-01, (from National Taiwan University), UN108-010, and UN109-051 (from National Taiwan University Hospital).
Acknowledgments
This work was financially supported by the Centers of Genomic and Precision Medicine, National Taiwan University, Taipei, Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Kokolus, K.M.; Obermajer, N.; Kalinski, P. Quantitative evaluation of tumor-specific T cells in tumors and lymphoid tissues. Methods Enzymol. 2019, 635, 149–166. [Google Scholar] [PubMed]
- Aerts, J.G.; Hegmans, J.P. Tumor-Specific Cytotoxic T Cells Are Crucial for Efficacy of Immunomodulatory Antibodies in Patients with Lung Cancer. Cancer Res. 2013, 73, 2381–2388. [Google Scholar] [CrossRef]
- Roybal, K.T.; Rupp, L.J.; Morsut, L.; Walker, W.J.; McNally, K.A.; Park, J.S.; Lim, W.A. Precision Tumor Recognition by T Cells with Combinatorial Antigen-Sensing Circuits. Cell 2016, 164, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Jacqueline, C.; Finn, O.J. Antibodies specific for disease-associated antigens (DAA) expressed in non-malignant diseases reveal potential new tumor-associated antigens (TAA) for immunotherapy or immunoprevention. Semin. Immunol. 2020, 47, 101394. [Google Scholar] [CrossRef]
- Gao, J.-Q.; Okada, N.; Mayumi, T.; Nakagawa, S. Immune Cell Recruitment and Cell-Based System for Cancer Therapy. Pharm. Res. 2008, 25, 752–768. [Google Scholar] [CrossRef] [PubMed]
- Leber, T. Uber die Entstehung der Entzundung und die entzundungerregeden Scadliekeiten. Fortschr. Med. 1888, 4, 460. [Google Scholar]
- McCutcheon, M. Chemotaxis in Leukocytes. Physiol. Rev. 1946, 26, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Hereld, D. Moving toward understanding eukaryotic chemotaxis. Eur. J. Cell Biol. 2006, 85, 905–913. [Google Scholar] [CrossRef]
- Bernardini, G.; Zabel, B.A. Editorial: The Role of Chemoattractants in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2671. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Bao, Z.; Tang, P.; Gong, W.; Yoshimura, T.; Wang, J.M. Chemokines in homeostasis and diseases. Cell. Mol. Immunol. 2018, 15, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Dangaj, D.; Bruand, M.; Grimm, A.J.; Ronet, C.; Barras, D.; Duttagupta, P.A.; Lanitis, E.; Duraiswamy, J.; Tanyi, J.L.; Benencia, F.; et al. Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors. Cancer Cell 2019, 35, 885–900.e10. [Google Scholar] [CrossRef]
- Molon, B.; Gri, G.; Bettella, M.; Gómez-Moutón, C.; Lanzavecchia, A.; Martínez-A, C.; Mañes, S.; Viola, A. T cell costimulation by chemokine receptors. Nat. Immunol. 2005, 6, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Sarukhan, A.; Bronte, V.; Molon, B. The pros and cons of chemokines in tumor immunology. Trends Immunol. 2012, 33, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Fein, M.R.; He, X.-Y.; Almeida, A.S.; Bružas, E.; Pommier, A.; Yan, R.; Eberhardt, A.; Fearon, D.T.; van Aelst, L.; Wilkinson, J.E.; et al. Cancer cell CCR2 orchestrates suppression of the adaptive immune response. J. Exp. Med. 2020, 217, e20181551. [Google Scholar] [CrossRef]
- Liu, Z.-Y.; Zheng, M.; Li, Y.-M.; Fan, X.-Y.; Wang, J.-C.; Li, Z.-C.; Yang, H.-J.; Yu, J.-M.; Cui, J.; Jiang, J.-L.; et al. RIP3 promotes colitis-associated colorectal cancer by controlling tumor cell proliferation and CXCL1-induced immune suppression. Theranostics 2019, 9, 3659–3673. [Google Scholar] [CrossRef]
- Franciszkiewicz, K.; Boissonnas, A.; Boutet, M.; Combadiere, C.; Mami-Chouaib, F. Role of Chemokines and Chemokine Receptors in Shaping the Effector Phase of the Antitumor Immune Response. Cancer Res. 2012, 72, 6325–6332. [Google Scholar] [CrossRef]
- Rossi, D.; Zlotnik, A. The Biology of Chemokines and their Receptors. Annu. Rev. Immunol. 2000, 18, 217–242. [Google Scholar] [CrossRef]
- Lacalle, R.A.; Blanco, R.; Carmona-Rodríguez, L.; Martín-Leal, A.; Mira, E.; Mañes, S. Chemokine Receptor Signaling and the Hallmarks of Cancer. Int. Rev. Cell Mol. Biol. 2017, 331, 181–244. [Google Scholar]
- Wang, Y.; Xu, P.; Qiu, L.; Zhang, M.; Huang, Y.; Zheng, J. CXCR7 Participates in CXCL12-mediated Cell Cycle and Proliferation Regulation in Mouse Neural Progenitor Cells. Curr. Mol. Med. 2016, 16, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Vlahakis, S.R.; Villasis-Keever, A.; Gomez, T.; Vanegas, M.; Vlahakis, N.; Paya, C.V. G Protein-Coupled Chemokine Receptors Induce Both Survival and Apoptotic Signaling Pathways. J. Immunol. 2002, 169, 5546–5554. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, S.H.; Baribaud, F.; Coughlan, C.M.; Sunshine, M.J.; Lee, V.M.Y.; Doms, R.W.; Littman, D.R.; Raper, J.A. The Chemokine Stromal Cell-Derived Factor-1 Promotes the Survival of Embryonic Retinal Ganglion Cells. J. Neurosci. 2003, 23, 4601–4612. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Filardi, E.; Nieto, C.; Domínguez-Soto, Á.; Barroso, R.; Sánchez-Mateos, P.; Puig-Kroger, A.; López-Bravo, M.; Joven, J.; Ardavín, C.; Rodríguez-Fernández, J.L.; et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: Identification of CCL2/CCR2-dependent gene expression profile. J. Immunol. 2014, 192, 3858–3867. [Google Scholar] [CrossRef]
- House, I.G.; Savas, P.; Lai, J.; Chen, A.X.Y.; Oliver, A.J.; Teo, Z.L.; Todd, K.L.; Henderson, M.A.; Giuffrida, L.; Petley, E.V.; et al. Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin. Cancer Res. 2020, 26, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Wang, B.; Yu, H.; Lin, J.; Xia, K.; Hou, W.; Yang, M.; Chen, J.; Yang, M.; Wang, X.; et al. Serum CCL27 predicts the response to Bacillus Calmette-Guerin immunotherapy in non-muscle-invasive bladder cancer. OncoImmunology 2020, 9, 1776060. [Google Scholar] [CrossRef] [PubMed]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Ansel, K.M.; Ngo, V.; Hyman, P.L.; Luther, S.; Forster, R.; Sedgwick, J.D.; Browning, J.; Lipp, M.; Cyster, J.G. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 2000, 406, 309–314. [Google Scholar] [CrossRef]
- Förster, R.; Mattis, A.E.; Kremmer, E.; Wolf, E.; Brem, G.; Lipp, M. A Putative Chemokine Receptor, BLR1, Directs B Cell Migration to Defined Lymphoid Organs and Specific Anatomic Compartments of the Spleen. Cell 1996, 87, 1037–1047. [Google Scholar] [CrossRef]
- del Molino del Barrio, I.; Kirby, J.; Ali, S. The Role of Chemokine and Glycosaminoglycan Interaction in Chemokine-Mediated Migration In Vitro and In Vivo. Methods Enzymol. 2016, 570, 309–333. [Google Scholar]
- Meijer, J.; Zeelenberg, I.S.; Sipos, B.; Roos, E. The CXCR5 Chemokine Receptor Is Expressed by Carcinoma Cells and Promotes Growth of Colon Carcinoma in the Liver. Cancer Res. 2006, 66, 9576–9582. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, X.; Guo, H.; Fu, L.; Pan, G.; Sun, Y. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway. Mol. Cell. Biochem. 2015, 400, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Qian, L.; Chen, X.; Ding, B. Prognostic significance of CXCL12, CXCR4, and CXCR7 in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 13217–13224. [Google Scholar]
- Thommen, D.S.; Koelzer, V.; Herzig, P.; Roller, A.; Trefny, M.; Dimeloe, S.; Kiialainen, A.; Hanhart, J.; Schill, C.; Hess, C.; et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018, 24, 994–1004. [Google Scholar] [CrossRef]
- Razis, E.; Kalogeras, K.T.; Kotsantis, I.; Koliou, G.-A.; Manousou, K.; Wirtz, R.; Veltrup, E.; Patsea, H.; Poulakaki, N.; Dionysopoulos, D.; et al. The Role of CXCL13 and CXCL9 in Early Breast Cancer. Clin. Breast Cancer 2020, 20, e36–e53. [Google Scholar] [CrossRef]
- Zhang, G.; Luo, X.; Zhang, W.; Chen, E.; Xu, J.; Wang, F.; Cao, G.; Ju, Z.; Jin, D.; Huang, X.; et al. CXCL-13 Regulates Resistance to 5-Fluorouracil in Colorectal Cancer. Cancer Res. Treat. 2020, 52, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ruan, H.; Song, Z.; Cao, Q.; Wang, K.; Bao, L.; Liu, D.; Tong, J.; Yang, H.; Chen, K.; et al. Identification of CXCL13 as a potential biomarker in clear cell renal cell carcinoma via comprehensive bioinformatics analysis. Biomed. Pharmacother. 2019, 118, 109264. [Google Scholar] [CrossRef]
- Jiao, F.; Sun, H.; Yang, Q.; Sun, H.; Wang, Z.; Liu, M.; Chen, J. Association of CXCL13 and Immune Cell Infiltration Signature in Clear Cell Renal Cell Carcinoma. Int. J. Med Sci. 2020, 17, 1610–1624. [Google Scholar] [CrossRef] [PubMed]
- Razis, E.; Kalogeras, K.T.; Kotoula, V.; Eleftheraki, A.; Nikitas, N.; Kronenwett, R.; Timotheadou, E.; Christodoulou, C.; Pectasides, D.; Gogas, H.; et al. Improved Outcome of High-Risk Early HER2 Positive Breast Cancer with High CXCL13-CXCR5 Messenger RNA Expression. Clin. Breast Cancer 2012, 12, 183–193. [Google Scholar] [CrossRef]
- Biswas, S.; Sengupta, S.; Chowdhury, S.R.; Jana, S.; Mandal, G.; Mandal, P.K.; Saha, N.; Malhotra, V.; Gupta, A.; Kuprash, D.V.; et al. CXCL13–CXCR5 co-expression regulates epithelial to mesenchymal transition of breast cancer cells during lymph node metastasis. Breast Cancer Res. Treat. 2014, 143, 265–276. [Google Scholar] [CrossRef]
- Qi, X.-W.; Xia, S.-H.; Yin, Y.; Jin, L.-F.; Pu, Y.; Hua, D.; Wu, H.-R. Expression features of CXCR5 and its ligand, CXCL13 associated with poor prognosis of advanced colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1916–1924. [Google Scholar] [PubMed]
- Singh, R.; Gupta, P.; Kloecker, G.H.; Singh, S.; Lillard, J.W., Jr. Expression and clinical significance of CXCR5/CXCL13 in human non-small cell lung carcinoma. Int. J. Oncol. 2014, 45, 2232–2240. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Blando, J.M.; Perez, C.J.; Abba, M.; Benavides, F.; Kazanietz, M.G. Protein Kinase C Epsilon Cooperates with PTEN Loss for Prostate Tumorigenesis through the CXCL13-CXCR5 Pathway. Cell Rep. 2017, 19, 375–388. [Google Scholar] [CrossRef]
- El-Haibi, C.P.; Singh, R.; Gupta, P.; Sharma, P.K.; Greenleaf, K.N.; Singh, S.; Lillard, J.W., Jr. Antibody Microarray Analysis of Signaling Networks Regulated by Cxcl13 and Cxcr5 in Prostate Cancer. J. Proteom. Bioinform. 2012, 5, 177–184. [Google Scholar] [CrossRef]
- Ammirante, M.; Shalapour, S.; Kang, Y.; Jamieson, C.A.M.; Karin, M. Tissue injury and hypoxia promote malignant progression of prostate cancer by inducing CXCL13 expression in tumor myofibroblasts. Proc. Natl. Acad. Sci. USA 2014, 111, 14776–14781. [Google Scholar] [CrossRef]
- Sambandam, Y.; Sundaram, K.; Liu, A.; Kirkwood, K.; Ries, W.L.; Reddy, S.V. CXCL13 activation of c-Myc induces RANK ligand expression in stromal/preosteoblast cells in the oral squamous cell carcinoma tumor-bone microenvironment. Oncogene 2012, 32, 97–105. [Google Scholar] [PubMed]
- Pandruvada, S.; Yuvaraj, S.; Liu, X.; Sundaram, K.; Shanmugarajan, S.; Ries, W.L.; Norris, J.S.; London, S.D.; Reddy, S.V. Role of CXC chemokine ligand 13 in oral squamous cell carcinoma associated osteolysis in athymic mice. Int. J. Cancer 2009, 126, 2319–2329. [Google Scholar] [CrossRef]
- Zheng, Z.; Cai, Y.; Chen, H.; Chen, Z.; Zhu, D.; Zhong, Q.; Xie, W. CXCL13/CXCR5 Axis Predicts Poor Prognosis and Promotes Progression Through PI3K/AKT/mTOR Pathway in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2018, 8, 682. [Google Scholar] [CrossRef]
- del Grosso, F.; Coco, S.; Scaruffi, P.; Stigliani, S.; Valdora, F.; Benelli, R.; Salvi, S.; Boccardo, S.; Truini, M.; Croce, M.; et al. Role of CXCL13-CXCR5 Crosstalk Between Malignant Neuroblastoma Cells and Schwannian Stromal Cells in Neuroblastic Tumors. Mol. Cancer Res. 2011, 9, 815–823. [Google Scholar] [CrossRef]
- Airoldi, I.; Cocco, C.; Morandi, F.; Prigione, I.; Pistoia, V. CXCR5 may be involved in the attraction of human metastatic neuroblastoma cells to the bone marrow. Cancer Immunol. Immunother. 2008, 57, 541–548. [Google Scholar] [CrossRef]
- Cha, Z.; Qian, G.; Zang, Y.; Gu, H.; Huang, Y.; Zhu, L.; Li, J.; Liu, Y.; Tu, X.; Song, H.; et al. Circulating CXCR5+CD4+ T cells assist in the survival and growth of primary diffuse large B cell lymphoma cells through interleukin 10 pathway. Exp. Cell Res. 2017, 350, 154–160. [Google Scholar] [CrossRef]
- Charbonneau, B.; Wang, A.H.; Maurer, M.; Asmann, Y.W.; Zent, C.S.; Link, B.; Ansell, S.M.; Weiner, G.; Ozsan, N.; Feldman, A.; et al. CXCR5 polymorphisms in non-Hodgkin lymphoma risk and prognosis. Cancer Immunol. Immunother. 2013, 62, 1475–1484. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Müller, G.; Lipp, M. Signal transduction by the chemokine receptor CXCR5: Structural requirements for G protein activation analyzed by chimeric CXCR1/CXCR5 molecules. Biol. Chem. 2001, 382, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- El-Haibi, C.P.; Sharma, P.; Singh, R.; Gupta, P.; Taub, D.D.; Singh, S.; Lillard, J.J.W. Differential G protein subunit expression by prostate cancer cells and their interaction with CXCR5. Mol. Cancer 2013, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Toyama, Y.; Kano, H.; Mase, Y.; Yokogawa, M.; Osawa, M.; Shimada, I. Dynamic regulation of GDP binding to G proteins revealed by magnetic field-dependent NMR relaxation analyses. Nat. Commun. 2017, 8, 14523. [Google Scholar] [CrossRef]
- Han, S.-B.; Moratz, C.; Huang, N.-N.; Kelsall, B.; Cho, H.; Shi, C.-S.; Schwartz, O.; Kehrl, J.H. Rgs1 and Gnai2 Regulate the Entrance of B Lymphocytes into Lymph Nodes and B Cell Motility within Lymph Node Follicles. Immunity 2005, 22, 343–354. [Google Scholar] [CrossRef]
- Hwang, I.-Y.; Park, C.; Kehrl, J.H. Impaired trafficking of Gnai2+/− and Gnai2−/− T lymphocytes: Implications for T cell movement within lymph nodes. J. Immunol. 2007, 179, 439–448. [Google Scholar] [CrossRef]
- Tybulewicz, V.L.J.; Henderson, R.B. Rho family GTPases and their regulators in lymphocytes. Nat. Rev. Immunol. 2009, 9, 630–644. [Google Scholar] [CrossRef]
- Bellamri, N.; Viel, R.; Morzadec, C.; Lecureur, V.; Joannes, A.; de Latour, B.; Llamas-Gutierrez, F.; Wollin, L.; Jouneau, S.; Vernhet, L. TNF-α and IL-10 Control CXCL13 Expression in Human Macrophages. J. Immunol. 2020, 204, 2492–2502. [Google Scholar] [CrossRef]
- Al-Kufaidy, R.; Vazquez-Tello, A.; BaHammam, A.S.; Al-Muhsen, S.; Hamid, Q.; Halwani, R. IL-17 enhances the migration of B cells during asthma by inducing CXCL13 chemokine production in structural lung cells. J. Allergy Clin. Immunol. 2017, 139, 696. [Google Scholar] [CrossRef]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.-W.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef]
- Franzoso, G.; Carlson, L.; Poljak, L.; Shores, E.W.; Epstein, S.; Leonardi, A.; Grinberg, A.; Tran, T.; Scharton-Kersten, T.; Anver, M. Mice deficient in nuclear factor (NF)-κB/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 1998, 187, 147–159. [Google Scholar] [CrossRef]
- Li, H.; Xia, J.Q.; Zhu, F.S.; Xi, Z.H.; Pan, C.Y.; Gu, L.M.; Tian, Y.Z. LPS promotes the expression of PD-L1 in gastric cancer cells through NF-κB activation. J. Cell. Biochem. 2018, 119, 9997–10004. [Google Scholar] [CrossRef]
- Gowrishankar, K.; Gunatilake, D.; Gallagher, S.J.; Tiffen, J.; Rizos, H.; Hersey, P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS ONE 2015, 10, e0123410. [Google Scholar] [CrossRef]
- Guo, R.; Li, Y.; Wang, Z.; Bai, H.; Duan, J.; Wang, S.; Wang, L.; Wang, J. Hypoxia-inducible factor-1α and nuclear factor-κB play important roles in regulating programmed cell death ligand 1 expression by epidermal growth factor receptor mutants in non-small-cell lung cancer cells. Cancer Sci. 2019, 110, 1665–1675. [Google Scholar] [CrossRef]
- Choi, J.; Diao, H.; Faliti, C.E.; Truong, J.; Rossi, M.; Bélanger, S.; Yu, B.; Goldrath, A.W.; Pipkin, M.E.; Crotty, S. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat. Immunol. 2020, 21, 777–789. [Google Scholar] [CrossRef]
- Miyazaki, M.; Rivera, R.R.; Miyazaki, K.; Lin, Y.; Agata, Y.; Murre, C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 2011, 12, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.A.; Chen, Y.; Ong, H.S.; Wu, D.; Man, K.; Deleage, C.; Minnich, M.; Meckiff, B.; Wei, Y.; Hou, Z.; et al. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 2016, 17, 1187–1196. [Google Scholar] [CrossRef]
- Celis, F.P.; Taborda, N.A.; Rugeles, M.T. Follicular CD8+ T Cells: Origin, Function and Importance during HIV Infection. Front. Immunol. 2017, 8, 1241. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.A.; Bélanger, S.; Omilusik, K.D.; Cho, S.; Scott-Browne, J.P.; Nance, J.P.; Goulding, J.; Lasorella, A.; Lu, L.-F.; Crotty, S. Id2 reinforces TH 1 differentiation and inhibits E2A to repress T FH differentiation. Nat. Immunol. 2016, 17, 834–843. [Google Scholar] [CrossRef]
- Johnston, R.J.; Poholek, A.C.; DiToro, D.; Yusuf, I.; Eto, D.; Barnett, B.; Dent, A.L.; Craft, J.; Crotty, S. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science 2009, 325, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, D. TCF-1 at the Tfh and Th1 Divergence. Trends Immunol. 2015, 36, 758–760. [Google Scholar] [CrossRef]
- Shao, P.; Li, F.; Wang, J.; Chen, X.; Liu, C.; Xue, H.-H. Cutting Edge: Tcf1 Instructs T Follicular Helper Cell Differentiation by Repressing Blimp1 in Response to Acute Viral Infection. J. Immunol. 2019, 203, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Shin, H.M.; Moseman, E.A.; Ji, Y.; Huang, B.; Harly, C.; Sen, J.M.; Berg, L.J.; Gattinoni, L.; McGavern, D.B.; et al. TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection. Cell Rep. 2015, 12, 2099–2110. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Gullicksrud, J.A.; Xing, S.; Zeng, Z.; Shan, Q.; Li, F.; Love, P.E.; Peng, W.; Xue, H.-H.; Crotty, S. LEF-1 and TCF-1 orchestrate T FH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 2015, 16, 980–990. [Google Scholar] [CrossRef]
- Mitkin, N.; Hook, C.D.; Schwartz, A.M.; Biswas, S.; Kochetkov, D.V.; Muratova, A.M.; Afanasyeva, M.; Kravchenko, J.E.; Bhattacharyya, A.; Kuprash, D.V. p53-dependent expression of CXCR5 chemokine receptor in MCF-7 breast cancer cells. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Alagesan, P.; Desai, N.K.; Pack, T.; Wu, J.-H.; Inoue, A.; Freedman, N.J.; Rajagopal, S. C-X-C Motif Chemokine Receptor 3 Splice Variants Differentially Activate Beta-Arrestins to Regulate Downstream Signaling Pathways. Mol. Pharmacol. 2017, 92, 136–150. [Google Scholar] [CrossRef]
- Thompson, B.D.; Jin, Y.; Wu, K.H.; Colvin, R.A.; Luster, A.D.; Birnbaumer, L.; Wu, M.X. Inhibition of Gαi2 activation by Gαi3 in CXCR3-mediated signaling. J. Biol. Chem. 2007, 282, 9547–9555. [Google Scholar] [CrossRef]
- Caggia, S.; Chunduri, H.; Millena, A.C.; Perkins, J.N.; Venugopal, S.V.; Vo, B.T.; Li, C.; Tu, Y.; Khan, S.A. Novel role of Giα2 in cell migration: Downstream of PI3-kinase–AKT and Rac1 in prostate cancer cells. J. Cell. Physiol. 2019, 234, 802–815. [Google Scholar] [CrossRef]
- Denecke, B.; Meyerdierks, A.; Böttger, E.C. RGS1 Is Expressed in Monocytes and Acts as a GTPase-activating Protein for G-protein-coupled Chemoattractant Receptors. J. Biol. Chem. 1999, 274, 26860–26868. [Google Scholar] [CrossRef]
- Legler, D.F.; Loetscher, M.; Roos, R.S.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. B Cell–attracting Chemokine 1, a Human CXC Chemokine Expressed in Lymphoid Tissues, Selectively Attracts B Lymphocytes via BLR1/CXCR5. J. Exp. Med. 1998, 187, 655–660. [Google Scholar] [CrossRef]
- Rouanne, M.; Arpaia, N.; Marabelle, A. CXCL13 shapes tertiary lymphoid structures and promotes response to immunotherapy in bladder cancer. Eur. J. Cancer 2021, 151, 245–248. [Google Scholar] [CrossRef]
- Rodriguez, A.B.; Peske, J.D.; Woods, A.N.; Leick, K.M.; Mauldin, I.S.; Meneveau, M.O.; Young, S.J.; Lindsay, R.S.; Melssen, M.M.; Cyranowski, S.; et al. Immune mechanisms orchestrate tertiary lymphoid structures in tumors via cancer-associated fibroblasts. Cell Rep. 2021, 36, 109422. [Google Scholar] [CrossRef]
- Schaerli, P.; Willimann, K.; Lang, A.B.; Lipp, M.; Loetscher, P.; Moser, B. Cxc Chemokine Receptor 5 Expression Defines Follicular Homing T Cells with B Cell Helper Function. J. Exp. Med. 2000, 192, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Gu-Trantien, C.; Migliori, E.; Buisseret, L.; de Wind, A.; Brohée, S.; Garaud, S.; Noël, G.; Chi, V.L.D.; Lodewyckx, J.-N.; Naveaux, C.; et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2017, 2, e91487. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; de Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef] [PubMed]
- Nakayamada, S.; Kanno, Y.; Takahashi, H.; Jankovic, D.; Lu, K.T.; Johnson, T.A.; Sun, H.-W.; Vahedi, G.; Hakim, O.; Handon, R.; et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity 2011, 35, 919–931. [Google Scholar] [CrossRef]
- Lönnberg, T.; Svensson, V.; James, K.R.; Fernandez-Ruiz, D.; Sebina, I.; Montandon, R.; Soon, M.S.; Fogg, L.G.; Nair, A.S.; Liligeto, U.; et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2017, 2, eaal2192. [Google Scholar] [CrossRef]
- Oestreich, K.J.; Huang, A.C.; Weinmann, A.S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 2011, 208, 1001–1013. [Google Scholar] [CrossRef]
- Lüthje, K.; Kallies, A.; Shimohakamada, Y.; Belz, G.; Light, A.; Tarlinton, D.; Nutt, S. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 2012, 13, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Pepper, M.; Pagán, A.J.; Igyártó, B.Z.; Taylor, J.J.; Jenkins, M.K. Opposing Signals from the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity 2011, 35, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Noël, G.; Fontsa, M.L.; Garaud, S.; de Silva, P.; de Wind, A.; Eynden, G.G.V.D.; Salgado, R.; Boisson, A.; Locy, H.; Thomas, N.; et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J. Clin. Investig. 2021, 131, e139905. [Google Scholar] [CrossRef] [PubMed]
- Kroenke, M.A.; Eto, D.; Locci, M.; Cho, M.; Davidson, T.; Haddad, E.K.; Crotty, S. Bcl6 and Maf Cooperate to Instruct Human Follicular Helper CD4 T Cell Differentiation. J. Immunol. 2012, 188, 3734–3744. [Google Scholar] [CrossRef]
- Denton, A.E.; Innocentin, S.; Carr, E.J.; Bradford, B.M.; Lafouresse, F.; Mabbott, N.A.; Mörbe, U.; Ludewig, B.; Groom, J.R.; Good-Jacobson, K.L.; et al. Type I interferon induces CXCL13 to support ectopic germinal center formation. J. Exp. Med. 2019, 216, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Balança, C.-C.; Salvioni, A.; Scarlata, C.-M.; Michelas, M.; Martinez-Gomez, C.; Gomez-Roca, C.; Sarradin, V.; Tosolini, M.; Valle, C.; Pont, F.; et al. PD-1 blockade restores helper activity of tumor-infiltrating, exhausted PD-1hiCD39+ CD4 T cells. JCI Insight 2021, 6, e142513. [Google Scholar] [CrossRef]
- Yoshitomi, H.; Kobayashi, S.; Miyagawa-Hayashino, A.; Okahata, A.; Doi, K.; Nishitani, K.; Murata, K.; Ito, H.; Tsuruyama, T.; Haga, H.; et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat. Commun. 2018, 9, 3762. [Google Scholar] [CrossRef]
- Workel, H.H.; Lubbers, J.M.; Arnold, R.; Prins, T.M.; van der Vlies, P.; de Lange, K.; Bosse, T.; van Gool, I.C.; Eggink, F.A.; Wouters, M.C.; et al. A Transcriptionally Distinct CXCL13+CD103+CD8+ T-cell Population Is Associated with B-cell Recruitment and Neoantigen Load in Human Cancer. Cancer Immunol. Res. 2019, 7, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, L.; Guo, L.; Chen, C.; Gu, S.; Zhou, Y.; Ye, G.; Li, X.; Wang, W.; Liao, X.; et al. CXCL13-mediated recruitment of intrahepatic CXCR5+CD8+ T cells favors viral control in chronic HBV infection. J. Hepatol. 2020, 72, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Le, K.S.; Amé-Thomas, P.; Tarte, K.; Gondois-Rey, F.; Granjeaud, S.; Orlanducci, F.; Foucher, E.D.; Broussais, F.; Bouabdallah, R.; Fest, T.; et al. CXCR5 and ICOS expression identifies a CD8 T-cell subset with T(FH) features in Hodgkin lymphomas. Blood Adv. 2018, 2, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Vermi, W.; Lonardi, S.; Bosisio, D.; Uguccioni, M.; Danelon, G.; Pileri, S.; Fletcher, C.; Sozzani, S.; Zorzi, F.; Arrigoni, G.; et al. Identification of CXCL13 as a new marker for follicular dendritic cell sarcoma. J. Pathol. 2008, 216, 356–364. [Google Scholar] [CrossRef]
- Vissers, J.L.M.; Hartgers, F.C.; Lindhout, E.; Figdor, C.G.; Adema, G.J. BLC (CXCL13) is expressed by different dendritic cell subsets in vitro and in vivo. Eur. J. Immunol. 2001, 31, 1544–1549. [Google Scholar] [CrossRef]
- Havenar-Daughton, C.; Lindqvist, M.; Heit, A.; Wu, J.E.; Reiss, S.M.; Kendric, K.; Bélanger, S.; Kasturi, S.P.; Landais, E.; Akondy, R.S.; et al. CXCL13 is a plasma biomarker of germinal center activity. Proc. Natl. Acad. Sci. USA 2016, 113, 2702–2707. [Google Scholar] [CrossRef]
- Feng, C.; Xu, Y.; Liu, Y.; Zhu, L.; Wang, L.; Cui, X.; Lu, J.; Zhang, Y.; Zhou, L.; Chen, M.; et al. Gene Expression Subtyping Reveals Immune alterations:TCGA Database for Prognosis in Ovarian Serous Cystadenocarcinoma. Front. Mol. Biosci. 2021, 8, 619027. [Google Scholar]
- Zanetti, C.; Kumar, R.; Ender, J.; Godavarthy, P.S.; Hartmann, M.; Hey, J.; Breuer, K.; Weissenberger, E.S.; Minciacchi, V.R.; Karantanou, C.; et al. The age of the bone marrow microenvironment influences B-cell acute lymphoblastic leukemia progression via CXCR5-CXCL13. Blood 2021, 138, 1870–1884. [Google Scholar] [CrossRef]
- Zhou, X.; Peng, M.; He, Y.; Peng, J.; Zhang, X.; Wang, C.; Xia, X.; Song, W. CXC Chemokines as Therapeutic Targets and Prognostic Biomarkers in Skin Cutaneous Melanoma Microenvironment. Front. Oncol. 2021, 11, 619003. [Google Scholar] [CrossRef]
- Lv, Y.; Lv, D.; Lv, X.; Xing, P.; Zhang, J.; Zhang, Y. Immune Cell Infiltration-Based Characterization of Triple-Negative Breast Cancer Predicts Prognosis and Chemotherapy Response Markers. Front. Genet. 2021, 12, 616469. [Google Scholar] [CrossRef]
- Li, N.; Li, B.; Zhan, X. Comprehensive Analysis of Tumor Microenvironment Identified Prognostic Immune-Related Gene Signature in Ovarian Cancer. Front. Genet. 2021, 12, 616073. [Google Scholar] [CrossRef]
- Li, Y.; Wu, T.; Gong, S.; Zhou, H.; Yu, L.; Liang, M.; Shi, R.; Wu, Z.; Zhang, J.; Li, S. Analysis of the Prognosis and Therapeutic Value of the CXC Chemokine Family in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 10, 570736. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Chen, Y.; Anandhan, S.; Szabo, P.M.; Basu, S.; Blando, J.M.; Liu, W.; Zhang, J.; Natarajan, S.M.; Xiong, L.; et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci. Transl. Med. 2020, 12, eabc4220. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Hamanishi, J.; Ukita, M.; Yamanoi, K.; Takamatsu, S.; Abiko, K.; Murakami, R.; Miyamoto, T.; Suzuki, H.; Ueda, A.; et al. Tertiary lymphoid structures are associated with favorable survival outcomes in patients with endometrial cancer. Cancer Immunol. Immunother. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dieu-Nosjean, M.-C. Tumor-Associated Tertiary Lymphoid Structures: A Cancer Biomarker and a Target for Next-generation Immunotherapy. Adv. Exp. Med. Biol. 2021, 1329, 51–68. [Google Scholar]
- Delvecchio, F.R.; Fincham, R.E.; Spear, S.; Clear, A.; Roy-Luzarraga, M.; Balkwill, F.R.; Gribben, J.G.; Bombardieri, M.; Hodivala-Dilke, K.; Capasso, M.; et al. Pancreatic Cancer Chemotherapy Is Potentiated by Induction of Tertiary Lymphoid Structures in Mice. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1543–1565. [Google Scholar] [CrossRef]
- Wennhold, K.; Thelen, M.; Lehmann, J.; Schran, S.; Preugszat, E.; Garcia-Marquez, M.; Lechner, A.; Shimabukuro-Vornhagen, A.; Ercanoglu, M.S.; Klein, F.; et al. CD86+ Antigen-Presenting B Cells Are Increased in Cancer, Localize in Tertiary Lymphoid Structures, and Induce Specific T-cell Responses. Cancer Immunol. Res. 2021, 9, 1098–1108. [Google Scholar] [CrossRef]
- Nebhan, C.A.; Cortellini, A.; Ma, W.; Ganta, T.; Song, H.; Ye, F.; Irlmeier, R.; Debnath, N.; Saeed, A.; Radford, M.; et al. Clinical Outcomes and Toxic Effects of Single-Agent Immune Checkpoint Inhibitors Among Patients Aged 80 Years or Older with Cancer: A Multicenter International Cohort Study. JAMA Oncol. 2021, 7, 1856–1861. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lu, J.; Zhang, G.; Wang, Y.; He, M.; Xu, Q.; Xu, C.; Liu, H. CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J. Immunother. Cancer 2021, 9, e001136. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Larsen, M.S.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Li, J.; Jiang, K.; Wang, R.; Ge, J.L.; Yang, H.; Liu, S.J.; Jia, L.T.; Wang, L.; Chen, B.L. CDK4/6 inhibition promotes immune infiltration in ovarian cancer and synergizes with PD-1 blockade in a B cell-dependent manner. Theranostics 2020, 10, 10619–10633. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, X.; Liu, R.; Chen, Y.; Ren, C.; Du, S. TOX correlates with prognosis, immune infiltration, and T cells exhaustion in lung adenocarcinoma. Cancer Med. 2020, 9, 6694–6709. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Liao, P.; Yan, L.-Y.; Zhao, Q.-Y.; Xie, Z.-Y.; Dong, J.; Sun, H.-T. Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 2021, 21, 416. [Google Scholar] [CrossRef] [PubMed]
- Terranova-Barberio, M.; Pawlowska, N.; Dhawan, M.; Moasser, M.; Chien, A.J.; Melisko, M.E.; Rugo, H.; Rahimi, R.; Deal, T.; Daud, A.; et al. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat. Commun. 2020, 11, 3584. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.I.; Loo, K.; Pauli, M.L.; Sanchez-Rodriguez, R.; Sandoval, P.M.; Taravati, K.; Tsai, K.; Nosrati, A.; Nardo, L.; Alvarado, M.D.; et al. Tumor immune profiling predicts response to anti–PD-1 therapy in human melanoma. J. Clin. Investig. 2016, 126, 3447–3452. [Google Scholar] [CrossRef]
- Tian, S.; Wang, F.; Zhang, R.; Chen, G. Global Pattern of CD8+ T-Cell Infiltration and Exhaustion in Colorectal Cancer Predicts Cancer Immunotherapy Response. Front. Pharmacol. 2021, 12, 715721. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-L.; Ou, D.-L.; Bai, L.-Y.; Chen, C.-W.; Lin, L.; Huang, S.-F.; Cheng, A.-L.; Jeng, Y.-M.; Hsu, C. Exploring Markers of Exhausted CD8 T Cells to Predict Response to Immune Checkpoint Inhibitor Therapy for Hepatocellular Carcinoma. Liver Cancer 2021, 10, 1–14. [Google Scholar] [CrossRef]
- Bassez, A.; Vos, H.; van Dyck, L.; Floris, G.; Arijs, I.; Desmedt, C.; Boeckx, B.; Bempt, M.V.; Nevelsteen, I.; Lambein, K.; et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 2021, 27, 820–832. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Friedlaender, A.; Kim, C.; Addeo, A. Rethinking the Optimal Duration of Immune Checkpoint Inhibitors in Non-small Cell Lung Cancer Throughout the COVID-19 Pandemic. Front. Oncol. 2020, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Chen, X.; Xu, R.; He, D.; Zhang, Y.; Chen, H.; Zhu, Y.; Cheng, Y.; Liu, R.; Zhu, R.; Gong, L.; et al. CD8+ T effector and immune checkpoint signatures predict prognosis and responsiveness to immunotherapy in bladder cancer. Oncogene 2021, 40, 6223–6234. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Mo, H.; Hu, X.; Gao, R.; Zhao, Y.; Liu, B.; Niu, L.; Sun, X.; Yu, X.; et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 2021, 39, 1578–1593.e8. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Reading, J.L.; Puttick, C.; Thakkar, K.; Abbosh, C.; Bentham, R.; Watkins, T.B.K.; Rosenthal, R.; Biswas, D.; Rowan, A.; et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021, 184, 596–614.e14. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Lin, L.-I.; Cheng, Y.-C.; Feng, Z.-R.; Shao, Y.-Y.; Cheng, A.-L.; Ou, D.-L. Cyclin E1 Inhibition can Overcome Sorafenib Resistance in Hepatocellular Carcinoma Cells Through Mcl-1 Suppression. Clin. Cancer Res. 2016, 22, 2555–2564. [Google Scholar] [CrossRef]
- Lu, J.-W.; Ho, Y.-J.; Yang, Y.-J.; Liao, H.-A.; Ciou, S.-C.; Lin, L.-I.; Ou, D.-L. Zebrafish as a disease model for studying human hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 12042–12058. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Edeline, J.; Masi, G.; Ma, Y.T.; Wang, W.; Wege, H.; Fei, C.; Ling, C.; Ma, X.; Zhang, P.; et al. 360 Tumor-immune signatures associated with response or resistance to tislelizumab in patients with previously treated advanced hepatocellular carcinoma (HCC). J. Immunother. Cancer 2021, 9, A387. [Google Scholar] [CrossRef]
- Wang, D.Y.; Salem, J.E.; Cohen, J.V.; Chandra, S.; Menzer, C.; Ye, F.; Zhao, S.; Das, S.; Beckermann, K.E.; Ha, L.; et al. Fatal Toxic Effects Associated with Immune Checkpoint Inhibitors: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 1721–1728. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Arén Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Gooley, T.A.; Chien, J.W.; Pergam, S.; Hingorani, S.; Sorror, M.L.; Boeckh, M.; Martin, P.J.; Sandmaier, B.M.; Marr, K.A.; Appelbaum, F.R.; et al. Reduced Mortality after Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2010, 363, 2091–2101. [Google Scholar] [CrossRef]
- Eggermont, A.M.; Chiarion-Sileni, V.; Grob, J.-J.; Dummer, R.; Wolchok, J.D.; Schmidt, H.; Hamid, O.; Robert, C.; Ascierto, P.A.; Richards, J.M.; et al. Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy. N. Engl. J. Med. 2016, 375, 1845–1855. [Google Scholar] [CrossRef]
- Weber, J.S.; Kähler, K.C.; Hauschild, A. Management of Immune-Related Adverse Events and Kinetics of Response with Ipilimumab. J. Clin. Oncol. 2012, 30, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Chen, Y.; Du, X.-J.; Liu, J.-Q.; Huang, C.-L.; Chen, L.; Zhou, G.-Q.; Li, W.-F.; Mao, Y.-P.; Hsu, C.; et al. Comparative safety of immune checkpoint inhibitors in cancer: Systematic review and network meta-analysis. BMJ 2018, 363, k4226. [Google Scholar] [CrossRef] [PubMed]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Eggermont, A.M.M.; Blank, C.U.; Mandalà, M.; Long, G.V.; Atkinson, V.; Dalle, S.; Haydon, A.; Lichinitser, M.; Khattak, A.; Carlino, M.S.; et al. Adjuvant Pembrolizumab versus Placebo in Resected Stage III Melanoma. N. Engl. J. Med. 2018, 378, 1789–1801. [Google Scholar] [CrossRef]
- Duraiswamy, J.; Kaluza, K.M.; Freeman, G.J.; Coukos, G. Dual Blockade of PD-1 and CTLA-4 Combined with Tumor Vaccine Effectively Restores T-Cell Rejection Function in Tumors. Cancer Res. 2013, 73, 3591–3603. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Thudium, K.B.; Han, M.; Wang, X.-T.; Huang, H.; Feingersh, D.; Garcia, C.; Wu, Y.; Kuhne, M.; Srinivasan, M.; et al. In Vitro Characterization of the Anti-PD-1 Antibody Nivolumab, BMS-936558, and In Vivo Toxicology in Non-Human Primates. Cancer Immunol. Res. 2014, 2, 846–856. [Google Scholar] [CrossRef]
- Balança, C.-C.; Scarlata, C.-M.; Michelas, M.; Devaud, C.; Sarradin, V.; Franchet, C.; Gomez, C.M.; Gomez-Roca, C.; Tosolini, M.; Heaugwane, D.; et al. Dual Relief of T-lymphocyte Proliferation and Effector Function Underlies Response to PD-1 Blockade in Epithelial Malignancies. Cancer Immunol. Res. 2020, 8, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shen, J.; Zhang, G.; Chen, X.; Wu, J.; Chen, W. CD40 controls CXCR5-induced recruitment of myeloid-derived suppressor cells to gastric cancer. Oncotarget 2015, 6, 38901–38911. [Google Scholar] [CrossRef]
- Chen, X.; Takemoto, Y.; Deng, H.; Middelhoff, M.; Friedman, R.A.; Chu, T.H.; Churchill, M.J.; Ma, Y.; Nagar, K.K.; Tailor, Y.H.; et al. Histidine decarboxylase (HDC)-expressing granulocytic myeloid cells induce and recruit Foxp3+ regulatory T cells in murine colon cancer. OncoImmunology 2017, 6, e1290034. [Google Scholar] [CrossRef]
- Amarnath, S.; Costanzo, C.M.; Mariotti, J.; Ullman, J.L.; Telford, W.G.; Kapoor, V.; Riley, J.L.; Levine, B.L.; June, C.H.; Fong, T.; et al. Regulatory T Cells and Human Myeloid Dendritic Cells Promote Tolerance via Programmed Death Ligand-1. PLoS Biol. 2010, 8, e1000302. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).