Toward Tumor Fight and Tumor Microenvironment Remodeling: PBA Induces Cell Cycle Arrest and Reduces Tumor Hybrid Cells’ Pluripotency in Bladder Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Reagents
2.2. Cell Viability Assays
2.3. Cell Cycle Analysis
2.4. Western Blot and Immunohistochemistry
2.5. Quantitative PCR with Reverse Transcription (RT-qPCR)
2.6. Human Macrophage Isolation and Co-Cultures
2.7. Lymphocyte Polarization and Intracellular Cytokine Staining
2.8. Vital Colorant Assays
2.9. Cell Sorting and Flow Cytometry
2.10. Soluble Cytokine Quantification
2.11. Statistical Analysis
3. Results
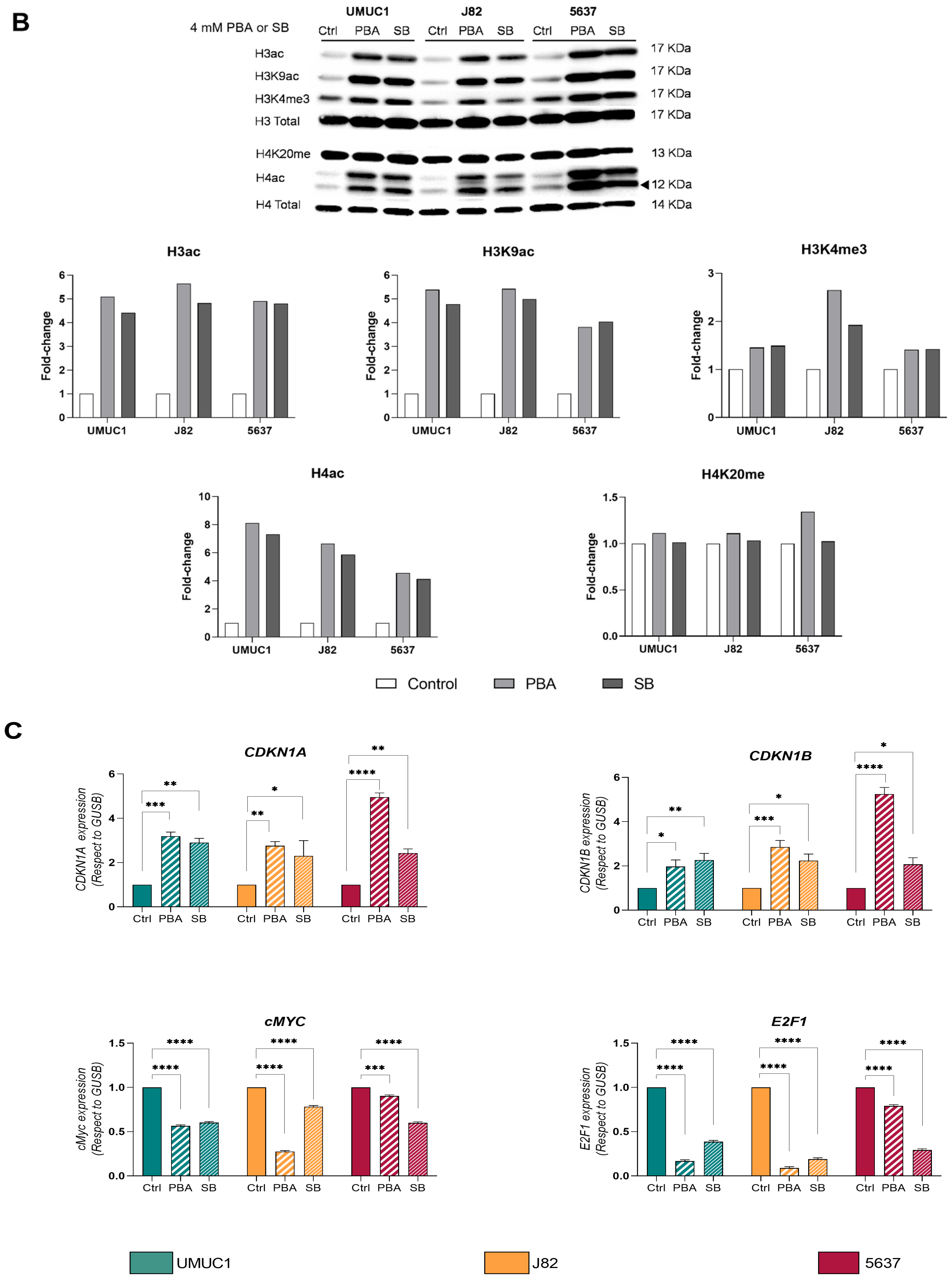
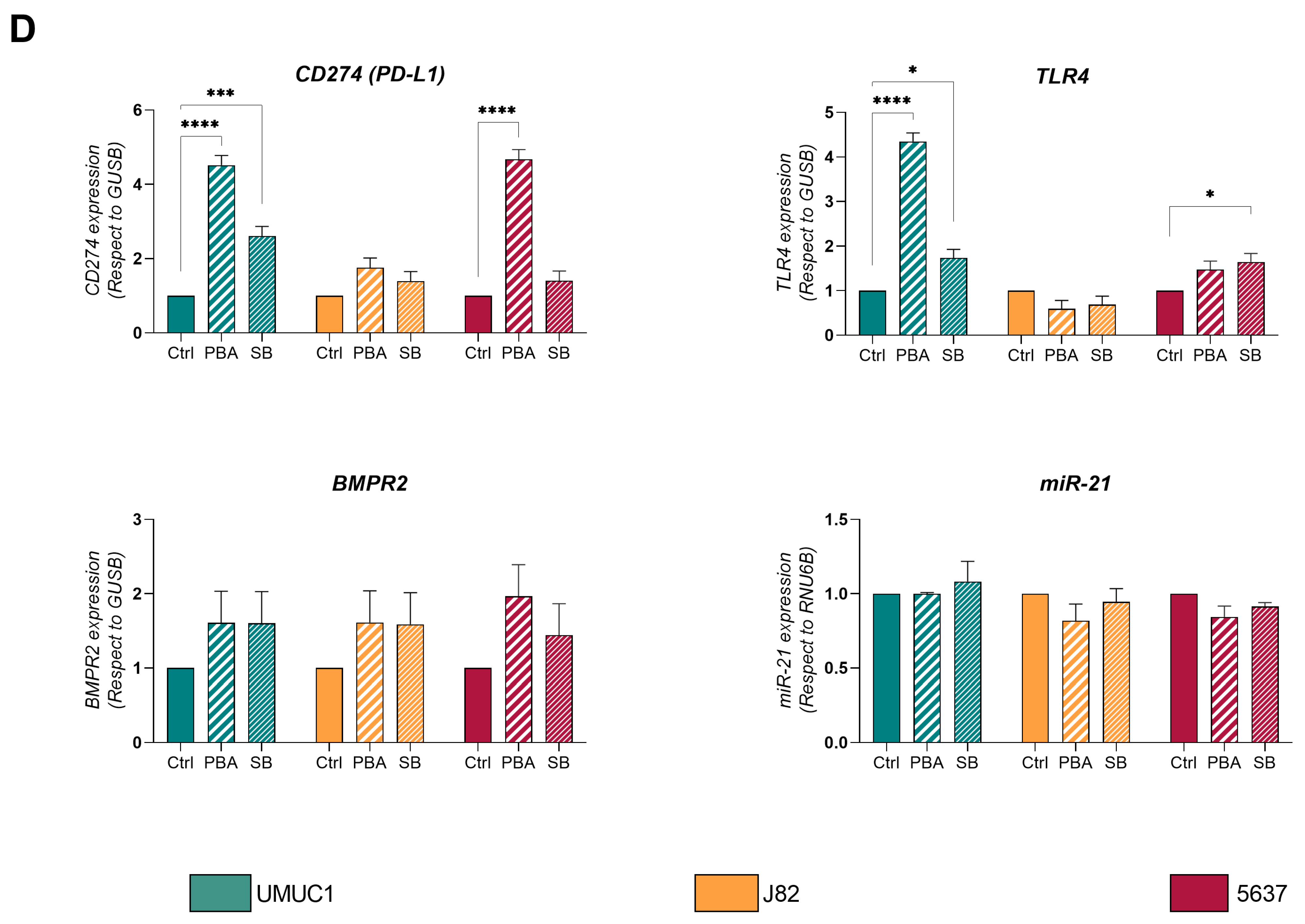
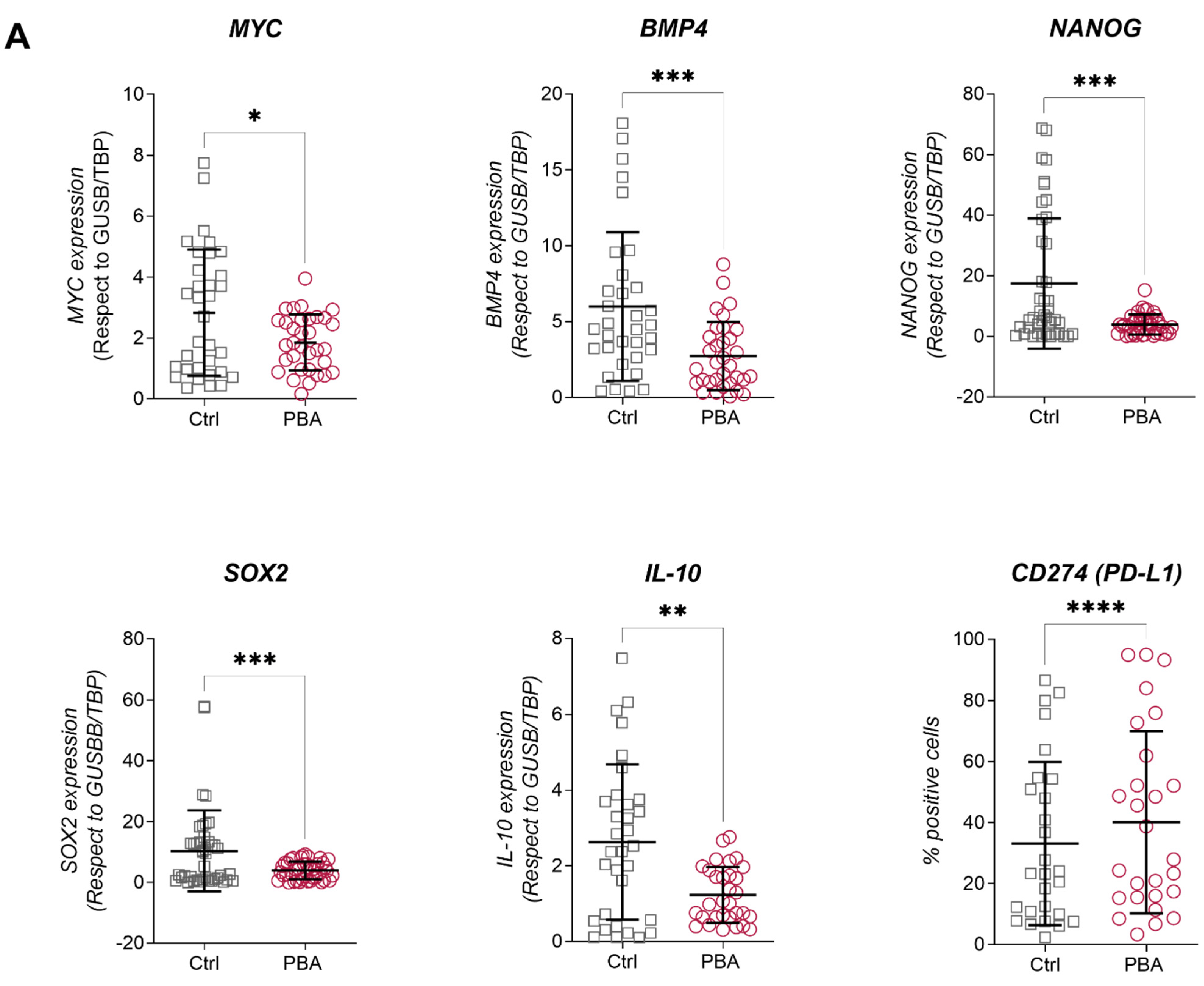
3.1. PBA and SB Induce Cell Cycle Arrest in BC Cell Lines and Impacts Their Immune Profile
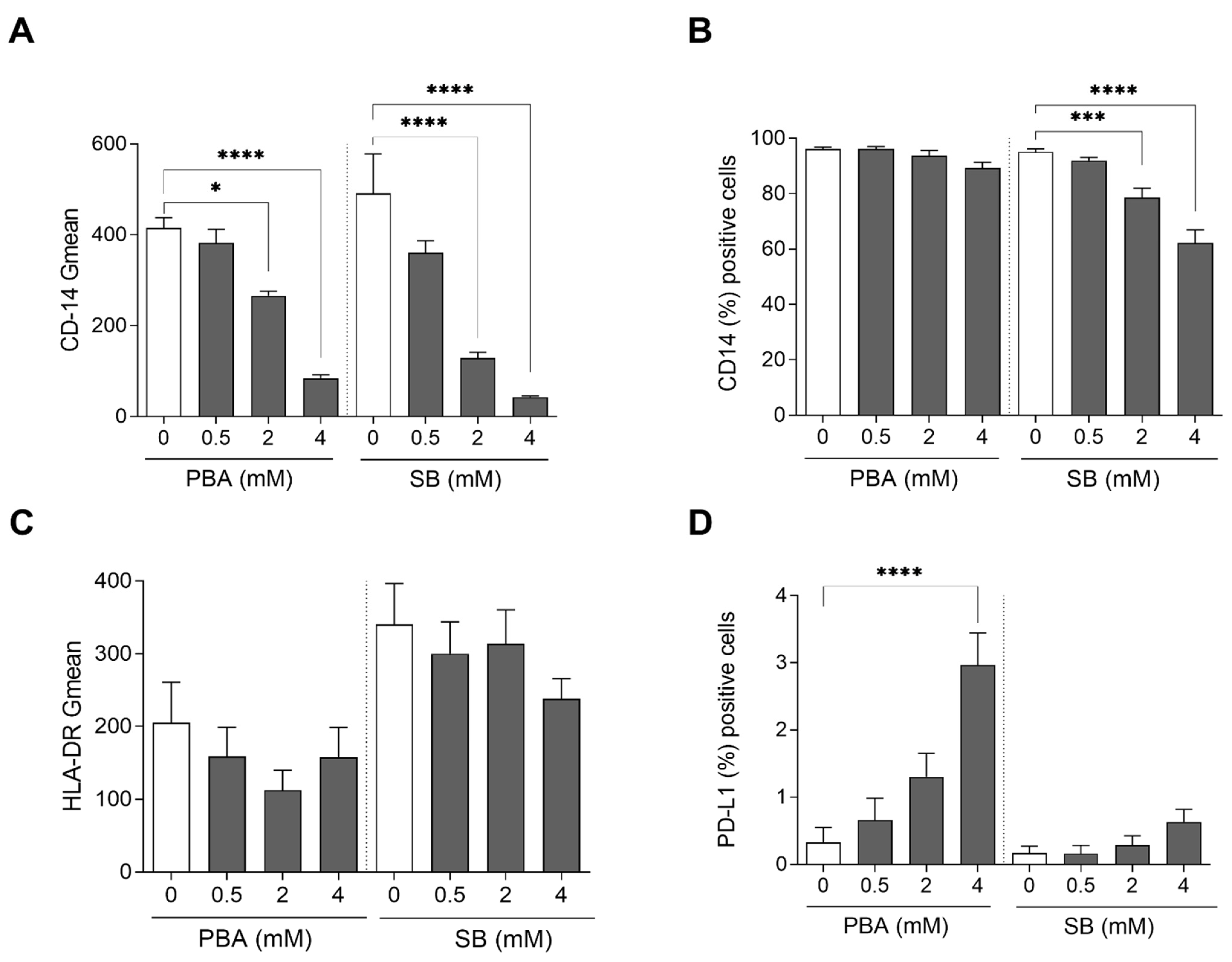
3.2. Butyrate Derivatives Increase PD-L1 Expression, Induce Non-Classical Phenotype on Monocytes and Reduce M2 Polarization in Human Macrophages
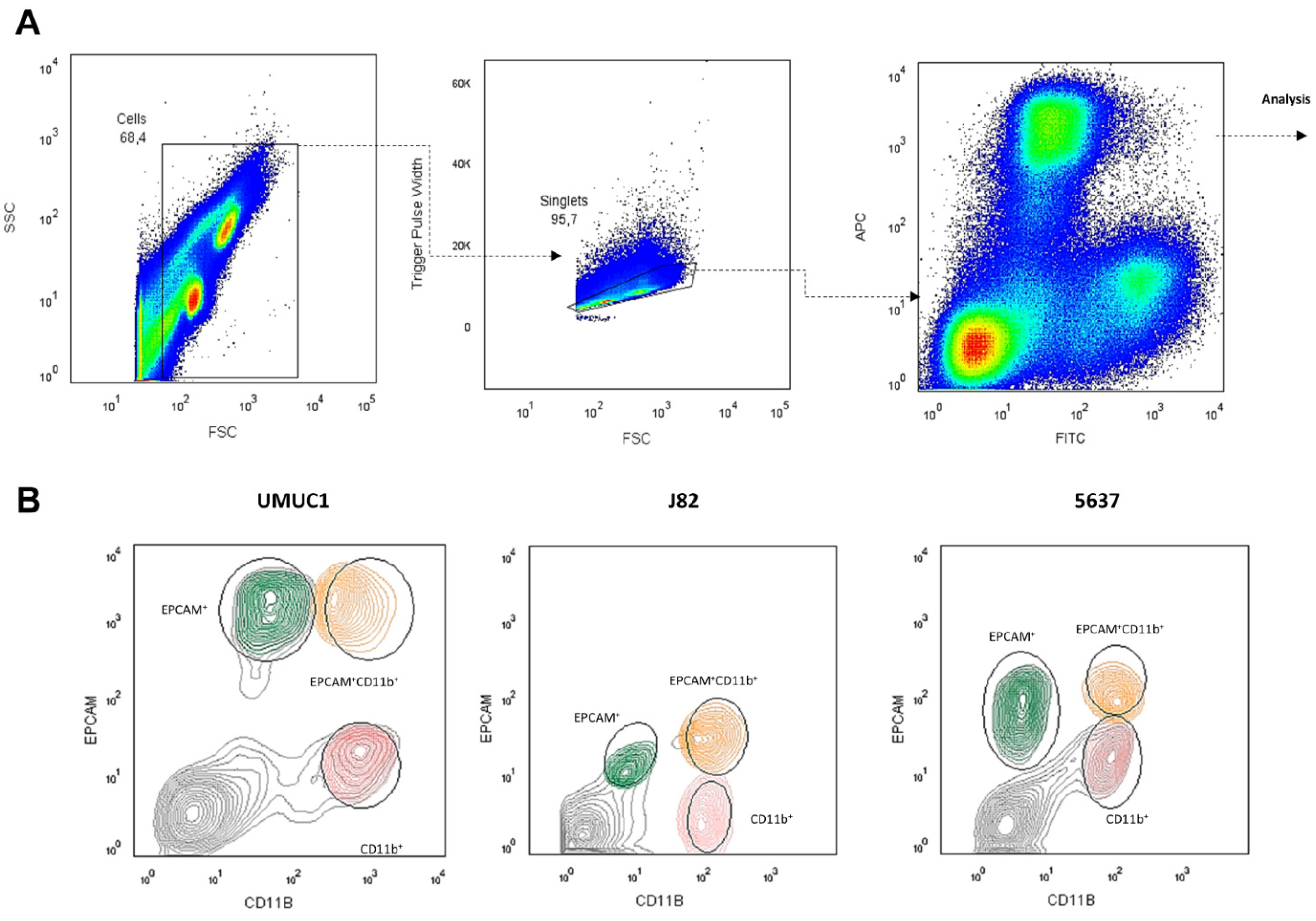
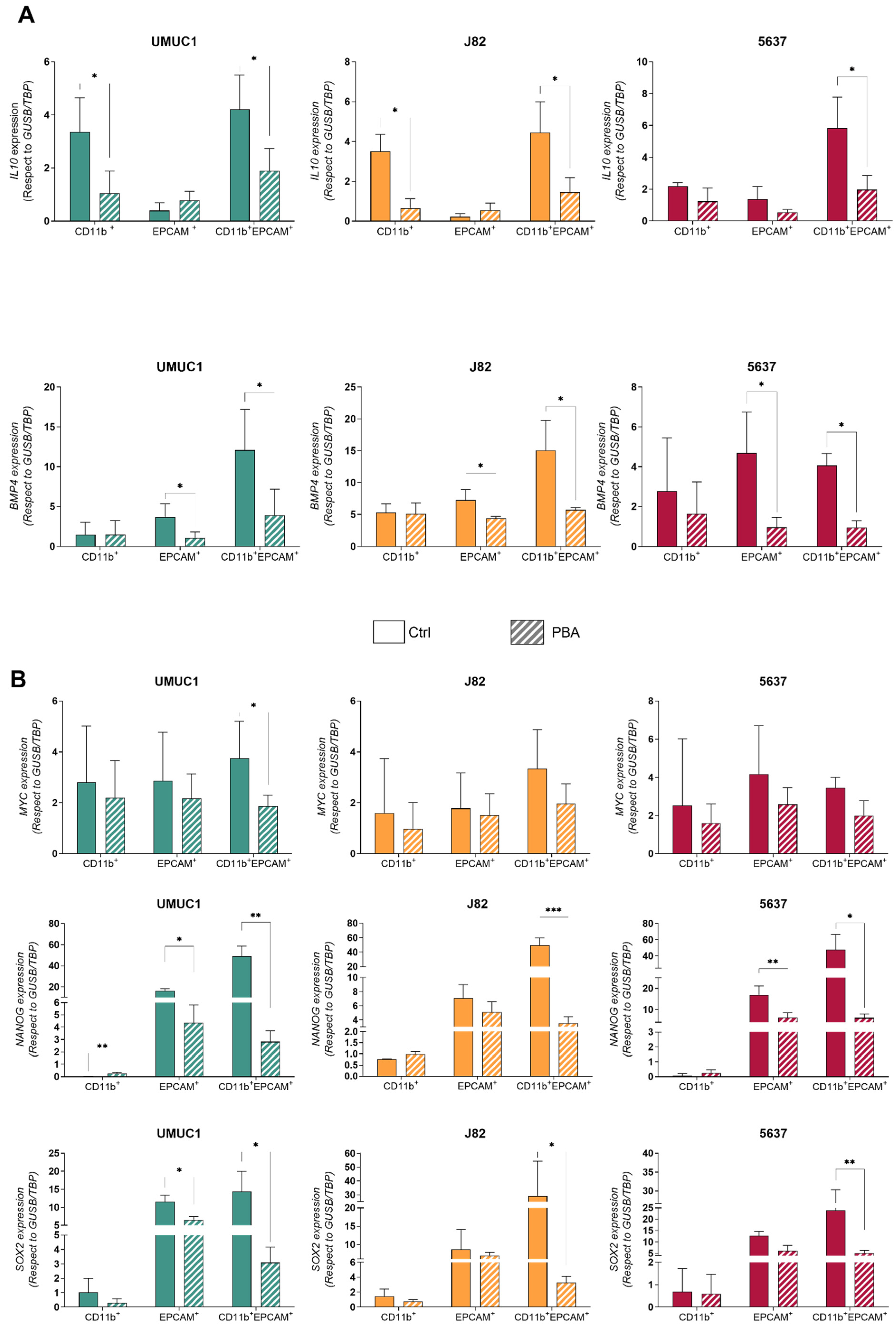
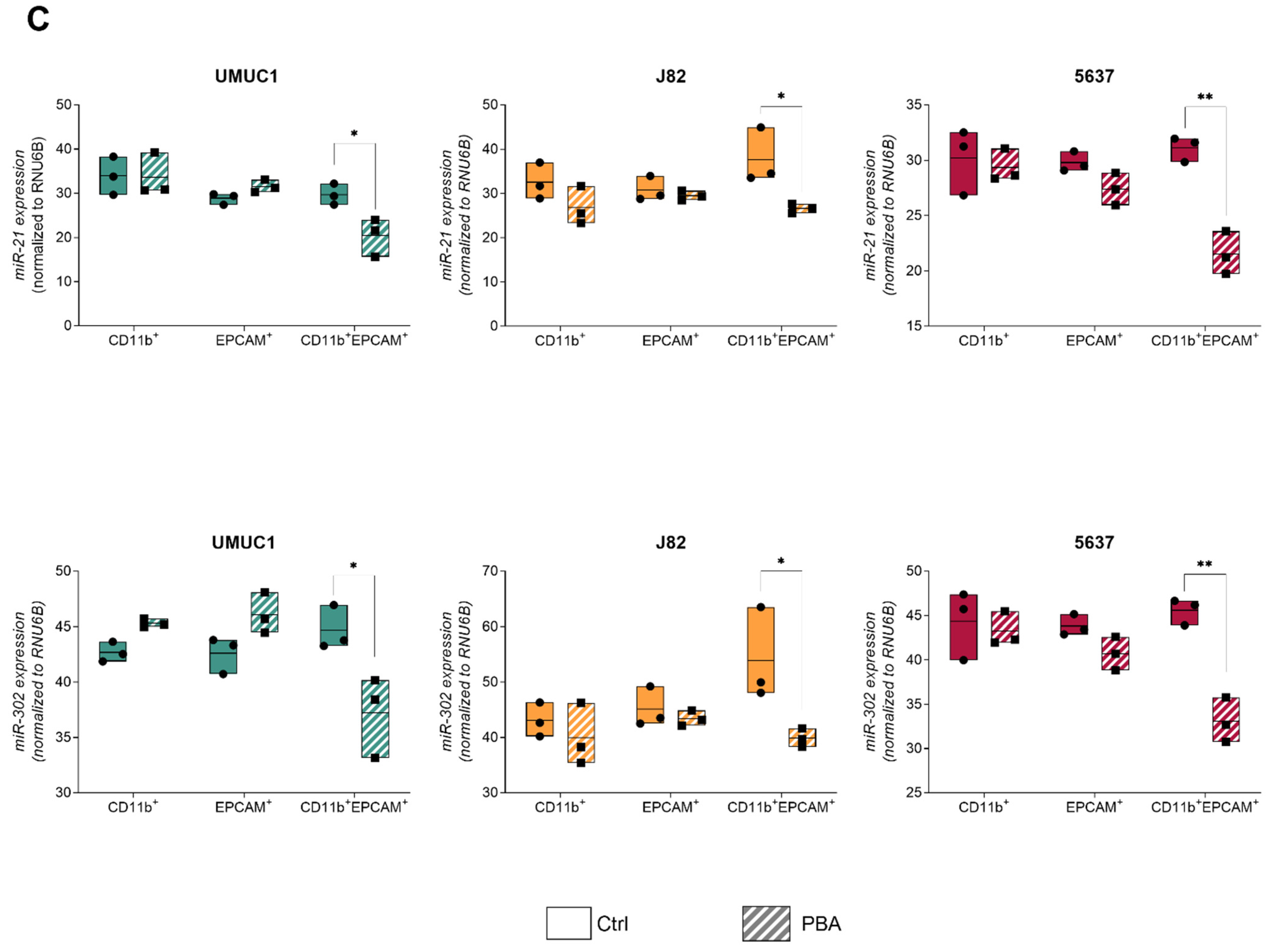
3.3. Macrophage–BC Cell Co-Cultures Generate a THC Population with Enhanced PD-L1 and Pluripotency Genes Expression
3.4. THC Population Is More Sensitive to PBA Treatment Than Parental Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babjuk, M.; Oosterlinck, W.; Sylvester, R.; Kaasinen, E.; Böhle, A.; Palou-Redorta, J. Guía Clínica Del Carcinoma Urotelial No Músculo Invasivo de La Asociación Europea de Urología. Actas Urol. Esp. 2009, 33, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Morales, A.; Eidinger, D.; Bruce, A. Intracavitary Bacillus Calmette-Guerin in the Treatment of Superficial Bladder Tumors. J. Urol. 1976, 116, 180–182. [Google Scholar] [CrossRef]
- Miyake, M.; Tatsumi, Y.; Gotoh, D.; Ohnishi, S.; Owari, T.; Iida, K.; Ohnishi, K.; Hori, S.; Morizawa, Y.; Itami, Y.; et al. Regulatory T Cells and Tumor-Associated Macrophages in the Tumor Microenvironment in Non-Muscle Invasive Bladder Cancer Treated with Intravesical Bacille Calmette-Guérin: A Long-Term Follow-Up Study of a Japanese Cohort. Int. J. Mol. Sci. 2017, 18, 2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.J.; Nguyen, P.H.D.; Wasser, M.; Kumar, P.; Lee, Y.H.; Nasir, N.J.M.; Chua, C.; Lai, L.; Hazirah, S.N.; Loh, J.J.H.; et al. Immunological Hallmarks for Clinical Response to BCG in Bladder Cancer. Front. Immunol. 2020, 11, 615091. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, L.A.; Montalbán-Hernández, K.; Avendaño-Ortiz, J.; Marín, E.; Lozano, R.; Toledano, V.; Sánchez-Maroto, L.; Terrón, V.; Valentín, J.; Pulido, E.; et al. Tumor Stem Cells Fuse with Monocytes to Form Highly Invasive Tumor-Hybrid Cells. Oncoimmunology 2020, 9, 1773204. [Google Scholar] [CrossRef]
- Manjunath, Y.; Porciani, D.; Mitchem, J.B.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Burke, D.H.; Li, G.; Kaifi, J.T. Tumor-Cell–Macrophage Fusion Cells as Liquid Biomarkers and Tumor Enhancers in Cancer. Int. J. Mol. Sci. 2020, 21, 1872. [Google Scholar] [CrossRef] [Green Version]
- LaBerge, G.; Duvall, E.; Grasmick, Z.; Haedicke, K.; Galan, A.; Pawelek, J. A Melanoma Patient with Macrophage-Cancer Cell Hybrids in the Primary Tumor, a Lymph Node Metastasis and a Brain Metastasis. Cancer Genet. 2021, 256, 162–164. [Google Scholar] [CrossRef]
- Pawelek, J.; Chakraborty, A.; Lazova, R.; Yilmaz, Y.; Cooper, D.; Brash, D.; Handerson, T. Co-Opting Macrophage Traits in Cancer Progression: A Consequence of Tumor Cell Fusion? Contrib. Microbiol. 2006, 13, 138–155. [Google Scholar] [CrossRef]
- Peng, X.-C.; Zhang, M.; Meng, Y.-Y.; Liang, Y.-F.; Wang, Y.-Y.; Liu, X.-Q.; Cai, W.-Q.; Zhou, Y.; Wang, X.-W.; Ma, Z.-W.; et al. Cell-cell Fusion as an Important Mechanism of Tumor Metastasis (Review). Oncol. Rep. 2021, 46, 145. [Google Scholar] [CrossRef] [PubMed]
- Maniecki, M.B.; Etzerodt, A.; Ulhøi, B.P.; Steiniche, T.; Borre, M.; Dyrskjøt, L.; Ørntoft, T.F.; Moestrup, S.K.; Møller, H.J. Tumor-Promoting Macrophages Induce the Expression of the Macrophage-Specific Receptor CD163 in Malignant Cells. Int. J. Cancer 2012, 131, 2320–2331. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.M.G.; Rubio, C.; Martínez-Fernández, M.; Segovia, C.; López-Calderón, F.; Garin, M.I.; Teijeira, A.; Munera-Maravilla, E.; Varas, A.; Sacedón, R.; et al. BMP4 Induces M2 Macrophage Polarization and Favors Tumor Progression in Bladder Cancer. Clin. Cancer Res. 2017, 23, 7388–7399. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, H.; Tanaka, M.; Tanaka, A.; Tsunemi, A.; Yamamoto, H. Predominance of M2-polarized Macrophages in Bladder Cancer Affects Angiogenesis, Tumor Grade and Invasiveness. Oncol. Lett. 2016, 11, 3403–3408. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Louie, J.; Weisman, A.; Sheu-Gruttadauria, J.; Davis-Dusenbery, B.; Lagna, G.; Hata, A. Inhibition of MicroRNA-302 (MiR-302) by Bone Morphogenetic Protein 4 (BMP4) Facilitates the BMP Signaling Pathway. J. Biol. Chem. 2012, 287, 38656–38664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolden, J.E.; Peart, M.J.; Johnstone, R. Anticancer Activities of Histone Deacetylase Inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Carew, J.S.; Giles, F.J.; Nawrocki, S.T. Histone Deacetylase Inhibitors: Mechanisms of Cell Death and Promise in Combination Cancer Therapy. Cancer Lett. 2008, 269, 7–17. [Google Scholar] [CrossRef]
- Chun, P. Histone Deacetylase Inhibitors in Hematological Malignancies and Solid Tumors. Arch. Pharm. Res. 2015, 38, 933–949. [Google Scholar] [CrossRef]
- Ferrarelli, L.K. HDAC Inhibitors in Solid Tumors and Blood Cancers. Sci. Signal. 2016, 9, ec216. [Google Scholar] [CrossRef]
- Mishra, A.; La Perle, K.; Kwiatkowski, S.; Sullivan, L.A.; Sams, G.H.; Johns, J.; Curphey, D.P.; Wen, J.; McConnell, K.; Qi, J.; et al. Mechanism, Consequences, and Therapeutic Targeting of Abnormal IL15 Signaling in Cutaneous T-Cell Lymphoma. Cancer Discov. 2016, 6, 986–1005. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Qu, J.; Jin, N.; Xu, J.; Lin, C.; Chen, Y.; Yang, X.; He, X.; Tang, S.; Lan, X.; et al. Feedback Activation of Leukemia Inhibitory Factor Receptor Limits Response to Histone Deacetylase Inhibitors in Breast Cancer. Cancer Cell 2016, 30, 459–473. [Google Scholar] [CrossRef] [Green Version]
- Ariffin, J.K.; Das Gupta, K.; Kapetanovic, R.; Iyer, A.; Reid, R.C.; Fairlie, D.P.; Sweet, M.J. Histone Deacetylase Inhibitors Promote Mitochondrial Reactive Oxygen Species Production and Bacterial Clearance by Human Macrophages. Antimicrob. Agents Chemother. 2016, 60, 1521–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, X.; Chen, Q.; Jiao, F.; Shi, C.; Pei, M.; Wang, L.; Gong, Z. Histone Deacetylase 6 Regulates the Activation of M1 Macrophages by the Glycolytic Pathway During Acute Liver Failure. J. Inflamm. Res. 2021, 14, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Das Gupta, K.; Shakespear, M.; Iyer, A.; Fairlie, D.; Sweet, M.J. Histone Deacetylases in Monocyte/Macrophage Development, Activation and Metabolism: Refining HDAC Targets for Inflammatory and Infectious Diseases. Clin. Transl. Immunol. 2016, 5, e62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luque-Martin, R.; van den Bossche, J.; Furze, R.C.; Neele, A.E.; van der Velden, S.; Gijbels, M.J.J.; van Roomen, C.P.P.A.; Bernard, S.G.; de Jonge, W.J.; Rioja, I.; et al. Targeting Histone Deacetylases in Myeloid Cells Inhibits Their Maturation and Inflammatory Function With Limited Effects on Atherosclerosis. Front. Pharmacol. 2019, 10, 1242. [Google Scholar] [CrossRef] [PubMed]
- Segain, J.-P.; De La Blétière, D.R.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottiere, H.; Galmiche, J.-P. Butyrate Inhibits Inflammatory Responses through NFkappaB Inhibition: Implications for Crohn’s Disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Shu, D.; Zheng, M.; Wang, J.; Luo, C.; Wang, Y.; Guo, F.; Zou, X.; Lv, X.; Li, Y.; et al. Microbial Metabolite Butyrate Facilitates M2 Macrophage Polarization and Function. Sci. Rep. 2016, 6, 24838. [Google Scholar] [CrossRef]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Rowe, M.; Ren, M.; Hong, J.-S.; Chen, P.-S.; Chuang, D.-M. Histone Deacetylase Inhibitors Exhibit Anti-Inflammatory and Neuroprotective Effects in a Rat Permanent Ischemic Model of Stroke: Multiple Mechanisms of Action. J. Pharmacol. Exp. Ther. 2007, 321, 892–901. [Google Scholar] [CrossRef]
- Cacabelos, R.; Torrellas, C. Pharmacoepigenomics. Med. Epigenetics 2016, 585–617. [Google Scholar] [CrossRef]
- Earl, J.; Rico, D.; Carrillo-de-Santa-Pau, E.; Rodríguez-Santiago, B.; Méndez-Pertuz, M.; Auer, H.; Gómez, G.; Grossman, H.B.; Pisano, D.G.; Schulz, W.A.; et al. The UBC-40 Urothelial Bladder Cancer Cell Line Index: A Genomic Resource for Functional Studies. BMC Genom. 2015, 16, 403. [Google Scholar] [CrossRef]
- Toledano, V.; Hernández-Jiménez, E.; Cubillos-Zapata, C.; Flandez, M.; Álvarez, E.; Varela-Serrano, A.; Cantero, R.; Valles, G.; García-Rio, F.; López-Collazo, E. Galactomannan Downregulates the Inflammation Responses in Human Macrophages via NF κ B2/P100. Mediat. Inflamm. 2015, 2015, 942517. [Google Scholar] [CrossRef] [Green Version]
- Povo-Retana, A.; Mojena, M.; Stremtan, A.B.; Fernández-García, V.B.; Gómez-Sáez, A.; Nuevo-Tapioles, C.; Molina-Guijarro, J.M.; Avendaño-Ortiz, J.; Cuezva, J.M.; López-Collazo, E.; et al. Specific Effects of Trabectedin and Lurbinectedin on Human Macrophage Function and Fate—Novel Insights. Cancers 2020, 12, 3060. [Google Scholar] [CrossRef] [PubMed]
- Gast, C.E.; Silk, A.D.; Zarour, L.; Riegler, L.; Burkhart, J.G.; Gustafson, K.T.; Parappilly, M.S.; Roh-Johnson, M.; Goodman, J.R.; Olson, B.; et al. Cell Fusion Potentiates Tumor Heterogeneity and Reveals Circulating Hybrid Cells That Correlate with Stage and Survival. Sci. Adv. 2018, 4, eaat7828. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Fang, D.; Zhang, H.; Xue, J.; Wangchuk, D.; Du, J.; Jiang, L. Sodium Butyrate Selectively Kills Cancer Cells and Inhibits Migration in Colorectal Cancer by Targeting Thioredoxin-1. OncoTargets Ther. 2020, 13, 4691–4704. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; di Costanzo, M.; Leone, L. The Epigenetic Effects of Butyrate: Potential Therapeutic Implications for Clinical Practice. Clin. Epigenetics 2012, 4, 4. [Google Scholar] [CrossRef]
- Zeitlin, P.L.; Diener-West, M.; Rubenstein, R.C.; Boyle, M.P.; Lee, C.K.K.; Brass-Ernst, L. Evidence of CFTR Function in Cystic Fibrosis after Systemic Administration of 4-Phenylbutyrate. Mol. Ther. 2002, 6, 119–126. [Google Scholar] [CrossRef]
- Li, X.N.; Parikh, S.; Shu, Q.; Jung, H.L.; Chow, C.W.; Perlaky, L.; Leung, H.C.E.; Su, J.; Blaney, S.; Lau, C.C. Phenylbutyrate and Phenylacetate Induce Differentiation and Inhibit Proliferation of Human Medulloblastoma Cells. Clin. Cancer Res. 2004, 10, 1150–1159. [Google Scholar] [CrossRef] [Green Version]
- Martinez, V.G.; Munera-Maravilla, E.; Bernardini, A.; Rubio, C.; Suarez-Cabrera, C.; Segovia, C.; Lodewijk, I.; Dueñas, M.; Martínez-Fernández, M.; Paramio, J.M. Epigenetics of Bladder Cancer: Where Biomarkers and Therapeutic Targets Meet. Front. Genet. 2019, 10, 1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Zheng, X. LSC—2017—Re-Education of Tumor-Associated Macrophages by Modulating Histone Deacetylases in Lung Cancer. Eur. Respir. J. 2017, 50, PA3297. [Google Scholar] [CrossRef]
- Larionova, I.; Cherdyntseva, N.; Liu, T.; Patysheva, M.; Rakina, M.; Kzhyshkowska, J. Interaction of Tumor-Associated Macrophages and Cancer Chemotherapy. Oncoimmunology 2019, 8, e1596004. [Google Scholar] [CrossRef] [Green Version]
- Sumitomo, R.; Hirai, T.; Fujita, M.; Murakami, H.; Otake, Y.; Huang, C. M2 Tumor-Associated Macrophages Promote Tumor Progression in Non-Small-Cell Lung Cancer. Exp. Ther. Med. 2019, 18, 4490–4498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Xu, J.; Lan, H. Tumor-Associated Macrophages in Tumor Metastasis: Biological Roles and Clinical Therapeutic Applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumor-Associated Macrophages as Treatment Targets in Oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Van Dalen, F.J.; Van Stevendaal, M.H.M.E.; Fennemann, F.L.; Verdoes, M.; Ilina, O. Molecular Repolarisation of Tumour-Associated Macrophages. Molecules 2018, 24, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.-Q.; Xu, R.; Li, X.-F.; Zhao, X.-K.; Qian, B.-Z. Prognostic Roles of Tumor Associated Macrophages in Bladder Cancer: A System Review and Meta-Analysis. Oncotarget 2018, 9, 25294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, H.; Nishimura, K.; Tsujimura, A.; Nakai, Y.; Nakayama, M.; Aozasa, K.; Okuyama, A.; Nonomura, N. Increased Infiltration of Tumor Associated Macrophages Is Associated with Poor Prognosis of Bladder Carcinoma in Situ after Intravesical Bacillus Calmette-Guerin Instillation. J. Urol. 2009, 181, 1894–1900. [Google Scholar] [CrossRef]
- Wang, L.-A.; Yang, B.; Tang, T.; Yang, Y.; Zhang, D.; Xiao, H.; Xu, J.; Wang, L.; Lin, L.; Jiang, J. Correlation of APE1 with VEGFA and CD163 + Macrophage Infiltration in Bladder Cancer and Their Prognostic Significance. Oncol. Lett. 2020, 20, 2881–2887. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, L.; Tian, J.; Man, H.; Li, P.; Shan, B. High Expression of B7-H3 and CD163 in Cancer Tissues Indicates Malignant Clinicopathological Status and Poor Prognosis of Patients with Urothelial Cell Carcinoma of the Bladder. Oncol. Lett. 2018, 15, 6519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garvin, S.; Oda, H.; Arnesson, L.-G.; Lindström, A.; Shabo, I. Tumor Cell Expression of CD163 Is Associated to Postoperative Radiotherapy and Poor Prognosis in Patients with Breast Cancer Treated with Breast-Conserving Surgery. J. Cancer Res. Clin. Oncol. 2018, 144, 1253. [Google Scholar] [CrossRef] [Green Version]
- Shabo, I.; Stål, O.; Olsson, H.; Doré, S.; Svanvik, J. Breast Cancer Expression of CD163, a Macrophage Scavenger Receptor, Is Related to Early Distant Recurrence and Reduced Patient Survival. Int. J. Cancer 2008, 123, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Saed, G.M.; Fletcher, N.M.; Diamond, M.P.; Morris, R.T.; Gomez-Lopez, N.; Memaj, I. Novel Expression of CD11b in Epithelial Ovarian Cancer: Potential Therapeutic Target. Gynecol. Oncol. 2018, 148, 567–575. [Google Scholar] [CrossRef]
- Huysentruyt, L.C.; Mukherjee, P.; Banerjee, D.; Shelton, L.M.; Seyfried, T.N. Metastatic Cancer Cells with Macrophage Properties: Evidence from a New Murine Tumor Model. Int. J. Cancer 2008, 123, 73–84. [Google Scholar] [CrossRef]
- Martínez-Fernández, M.; Dueñas, M.; Feber, A.; Segovia, C.; García-Escudero, R.; Rubio, C.; López-Calderón, F.F.; Díaz-García, C.; Villacampa, F.; Duarte, J.; et al. A Polycomb-Mir200 Loop Regulates Clinical Outcome in Bladder Cancer. Oncotarget 2015, 6, 42258–42275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornachea, O.; Santos, M.; Martínez-Cruz, A.B.; García-Escudero, R.; Dueñas, M.; Costa, C.; Segrelles, C.; Lorz, C.; Buitrago, A.; Saiz-Ladera, C.; et al. EMT and Induction of MiR-21 Mediate Metastasis Development in Trp53-Deficient Tumours. Sci. Rep. 2012, 2, 434. [Google Scholar] [CrossRef]
- Woods, D.M.; Sodré, A.L.; Villagra, A.; Sarnaik, A.; Sotomayor, E.M.; Weber, J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol. Res. 2015, 3, 1375. [Google Scholar] [CrossRef] [Green Version]
- Rubio, C.; Munera-Maravilla, E.; Lodewijk, I.; Suárez, C.; Karaivanova, V.; Ruiz-Palomares, R.; Paramio, J.M.; Dueñas, M. Macrophage Polarization as a Novel Weapon in Conditioning Tumor Microenvironment for Bladder Cancer: Can We Turn Demons into Gods? Clin. Transl. Oncol. 2019, 21, 391–403. [Google Scholar] [CrossRef]
- Jenke, R.; Reßing, N.; Hansen, F.K.; Aigner, A.; Büch, T. Anticancer Therapy with HDAC Inhibitors: Mechanism-Based Combination Strategies and Future Perspectives. Cancers 2021, 13, 634. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubio, C.; Avendaño-Ortiz, J.; Ruiz-Palomares, R.; Karaivanova, V.; Alberquilla, O.; Sánchez-Domínguez, R.; Casalvilla-Dueñas, J.C.; Montalbán-Hernández, K.; Lodewijk, I.; Rodríguez-Izquierdo, M.; et al. Toward Tumor Fight and Tumor Microenvironment Remodeling: PBA Induces Cell Cycle Arrest and Reduces Tumor Hybrid Cells’ Pluripotency in Bladder Cancer. Cancers 2022, 14, 287. https://doi.org/10.3390/cancers14020287
Rubio C, Avendaño-Ortiz J, Ruiz-Palomares R, Karaivanova V, Alberquilla O, Sánchez-Domínguez R, Casalvilla-Dueñas JC, Montalbán-Hernández K, Lodewijk I, Rodríguez-Izquierdo M, et al. Toward Tumor Fight and Tumor Microenvironment Remodeling: PBA Induces Cell Cycle Arrest and Reduces Tumor Hybrid Cells’ Pluripotency in Bladder Cancer. Cancers. 2022; 14(2):287. https://doi.org/10.3390/cancers14020287
Chicago/Turabian StyleRubio, Carolina, José Avendaño-Ortiz, Raquel Ruiz-Palomares, Viktoriya Karaivanova, Omaira Alberquilla, Rebeca Sánchez-Domínguez, José Carlos Casalvilla-Dueñas, Karla Montalbán-Hernández, Iris Lodewijk, Marta Rodríguez-Izquierdo, and et al. 2022. "Toward Tumor Fight and Tumor Microenvironment Remodeling: PBA Induces Cell Cycle Arrest and Reduces Tumor Hybrid Cells’ Pluripotency in Bladder Cancer" Cancers 14, no. 2: 287. https://doi.org/10.3390/cancers14020287
APA StyleRubio, C., Avendaño-Ortiz, J., Ruiz-Palomares, R., Karaivanova, V., Alberquilla, O., Sánchez-Domínguez, R., Casalvilla-Dueñas, J. C., Montalbán-Hernández, K., Lodewijk, I., Rodríguez-Izquierdo, M., Munera-Maravilla, E., Nunes, S. P., Suárez-Cabrera, C., Pérez-Crespo, M., Martínez, V. G., Morales, L., Pérez-Escavy, M., Alonso-Sánchez, M., Lozano-Rodríguez, R., ... Dueñas, M. (2022). Toward Tumor Fight and Tumor Microenvironment Remodeling: PBA Induces Cell Cycle Arrest and Reduces Tumor Hybrid Cells’ Pluripotency in Bladder Cancer. Cancers, 14(2), 287. https://doi.org/10.3390/cancers14020287









