Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma?
Abstract
Simple Summary
Abstract
1. Introduction
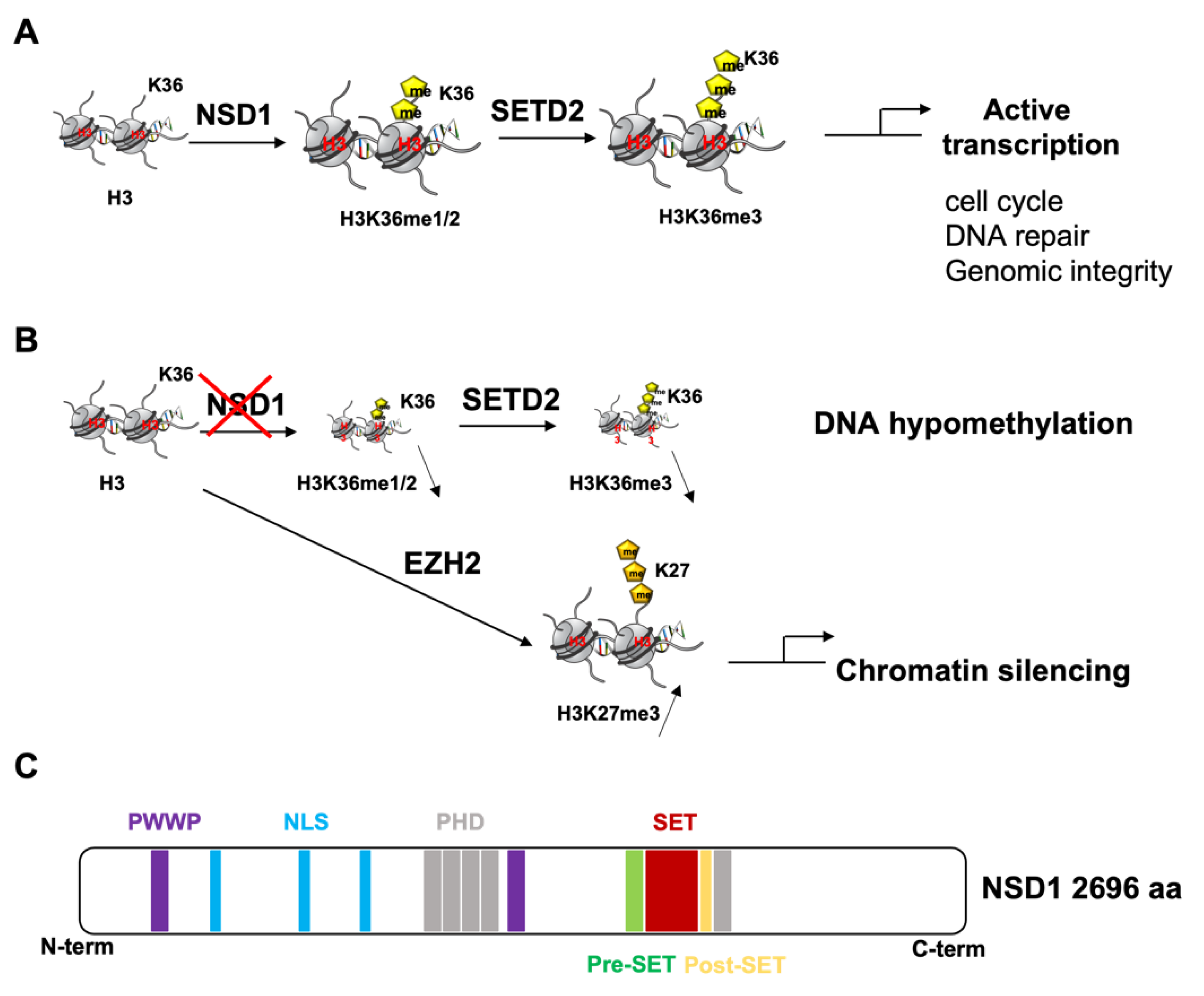
2. NSD1 Structure and Function
2.1. Physiological Function
2.2. NSD1′s Role in Congenital Developmental Disorders
2.3. NSD1′s Role in Cancer
2.3.1. Anti-Tumoral Role of NSD1
2.3.2. Pro-Tumoral Role of NSD1
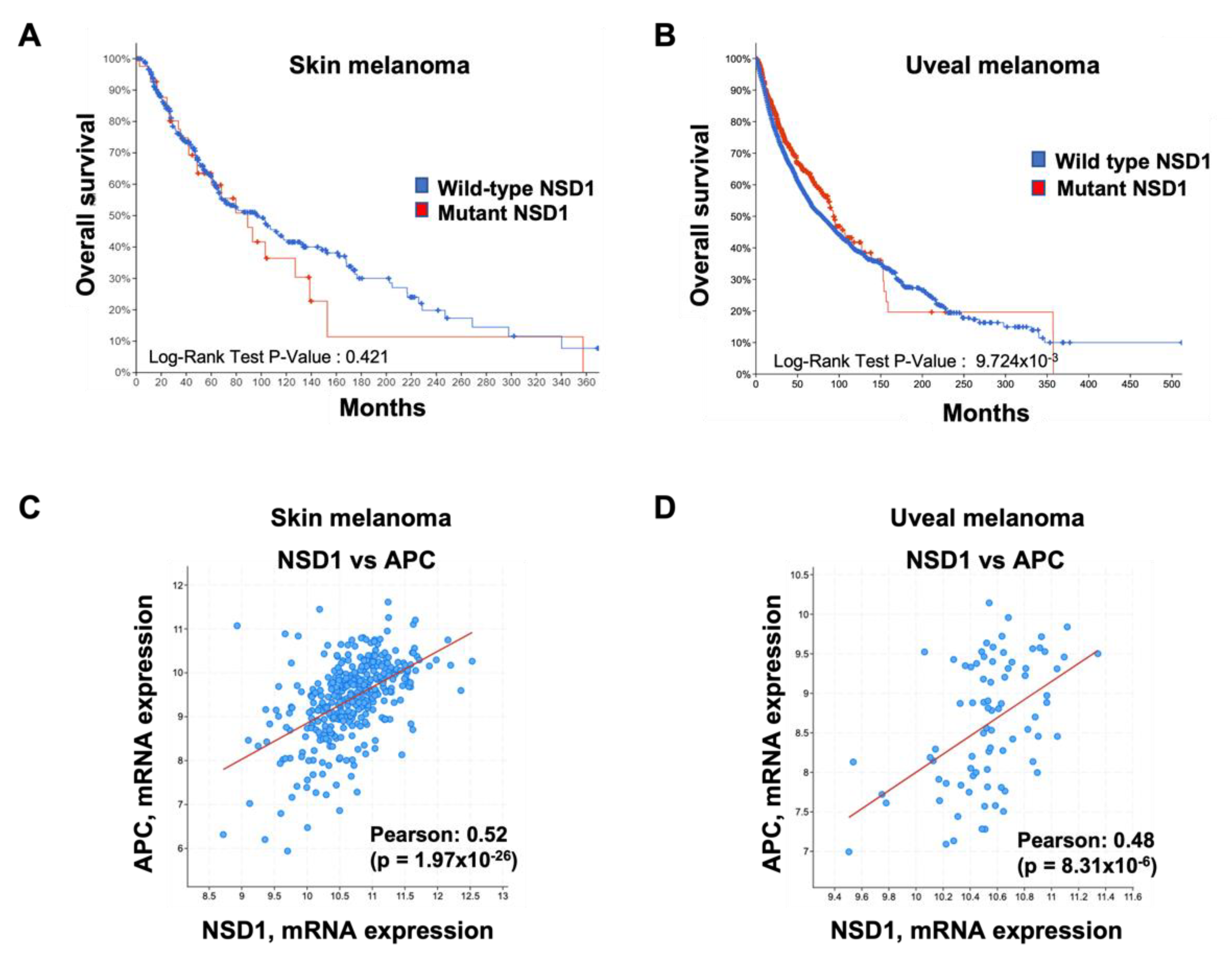
3. NSD1 in Melanoma
4. Therapeutic Opportunities in the NSD1 Signaling Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Qureshi, I.A.; Mehler, M.F. Epigenetic Mechanisms Underlying Nervous System Diseases. In Handbook of Clinical Neurology; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 147, pp. 43–58. [Google Scholar] [CrossRef]
- Strub, T.; Ballotti, R.; Bertolotto, C. The “ART” of Epigenetics in Melanoma: From Histone “Alterations, to Resistance and Therapies”. Theranostics 2020, 10, 1777–1797. [Google Scholar] [CrossRef]
- Klein, B.J.; Krajewski, K.; Restrepo, S.; Lewis, P.W.; Strahl, B.D.; Kutateladze, T.G. Recognition of Cancer Mutations in Histone H3K36 by Epigenetic Writers and Readers. Epigenetics 2018, 13, 683–692. [Google Scholar] [CrossRef]
- Lachner, M.; O’Sullivan, R.; Jenuwein, T. An Epigenetic Road Map for Histone Lysine Methylation. J. Cell Sci. 2003, 116, 2117–2124. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.; Schones, D.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Zhang, X.; Wen, H.; Shi, X. Lysine Methylation: Beyond Histones. Acta Biochim. Biophys. Sin. 2012, 44, 14–27. [Google Scholar] [CrossRef]
- Sankaran, S.M.; Wilkinson, A.W.; Elias, J.E.; Gozani, O. A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin. J. Biol. Chem. 2016, 291, 8465–8474. [Google Scholar] [CrossRef]
- Qiao, Q.; Li, Y.; Chen, Z.; Wang, M.; Reinberg, D.; Xu, R.M. The Structure of NSD1 Reveals an Autoregulatory Mechanism Underlying Histone H3K36 Methylation. J. Biol. Chem. 2011, 286, 8361–8368. [Google Scholar] [CrossRef]
- Bennett, R.L.; Swaroop, A.; Troche, C.; Licht, J.D. The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026708. [Google Scholar] [CrossRef]
- Wang, X.; Yeh, S.; Wu, G.; Hsu, C.L.; Wang, L.; Chiang, T.; Yang, Y.; Guo, Y.; Chang, C. Identification and Characterization of a Novel Androgen Receptor Coregulator ARA267-α in Prostate Cancer Cells. J. Biol. Chem. 2001, 276, 40417–40423. [Google Scholar] [CrossRef]
- Dingwall, C.; Laskey, R.A. Nuclear Targeting Sequences—A Consensus? Trends Biochem. Sci. 1991, 16, 478–481. [Google Scholar] [CrossRef]
- Rao, B.; Shibata, Y.; Strahl, B.D.; Lieb, J.D. Dimethylation of Histone H3 at Lysine 36 Demarcates Regulatory and Nonregulatory Chromatin Genome-Wide. Mol. Cell. Biol. 2005, 25, 9447. [Google Scholar] [CrossRef]
- Wagner, E.J.; Carpenter, P.B. Understanding the Language of Lys36 Methylation at Histone H3. Nat. Rev. Mol. Cell Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef]
- McDaniel, S.L.; Strahl, B.D. Shaping the Cellular Landscape with Set2/SETD2 Methylation. Cell. Mol. Life Sci. 2017, 74, 3317–3334. [Google Scholar] [CrossRef]
- Li, J.; Duns, G.; Westers, H.; Sijmons, R.; van den Berg, A.; Kok, K. SETD2: An Epigenetic Modifier with Tumor Suppressor Functionality. Oncotarget 2016, 7, 50719–50734. [Google Scholar] [CrossRef]
- Lucio-Eterovic, A.K.; Singh, M.M.; Gardner, J.E.; Veerappan, C.S.; Rice, J.C.; Carpenter, P.B. Role for the Nuclear Receptor-Binding SET Domain Protein 1 (NSD1) Methyltransferase in Coordinating Lysine 36 Methylation at Histone 3with RNApolymerase II Function. Proc. Natl. Acad. Sci. USA 2010, 107, 16952–16957. [Google Scholar] [CrossRef]
- Kurotaki, N.; Harada, N.; Yoshiura, K.I.; Sugano, S.; Niikawa, N.; Matsumoto, N. Molecular Characterization of NSD1, a Human Homologue of the Mouse Nsd1 Gene. Gene 2001, 279, 197–204. [Google Scholar] [CrossRef]
- Rayasam, G.V.; Wendling, O.; Angrand, P.-O.; Mark, M.; Niederreither, K.; Song, L.; Lerouge, T.; Hager, G.L.; Chambon, P.; Losson, R. NSD1 Is Essential for Early Post-Implantation Development and Has a Catalytically Active SET Domain. EMBO J. 2003, 22, 3153–3163. [Google Scholar] [CrossRef]
- Tamaru, H.; Selker, E.U. A Histone H3 Methyltransferase Controls DNA Methylation in Neurospora Crassa. Nature 2001, 414, 277–283. [Google Scholar] [CrossRef]
- Choufani, S.; Cytrynbaum, C.; Chung, B.H.Y.; Turinsky, A.L.; Grafodatskaya, D.; Chen, Y.A.; Cohen, A.S.A.; Dupuis, L.; Butcher, D.T.; Siu, M.T.; et al. NSD1 Mutations Generate a Genome-Wide DNA Methylation Signature. Nat. Commun. 2015, 6, 10207. [Google Scholar] [CrossRef]
- Papillon-Cavanagh, S.; Lu, C.; Gayden, T.; Mikael, L.G.; Bechet, D.; Karamboulas, C.; Ailles, L.; Karamchandani, J.; Marchione, D.M.; Garcia, B.A.; et al. Impaired H3K36 Methylation Defines a Subset of Head and Neck Squamous Cell Carcinomas. Nat. Genet. 2017, 49, 180–185. [Google Scholar] [CrossRef]
- Weinberg, D.N.; Papillon-Cavanagh, S.; Chen, H.; Yue, Y.; Chen, X.; Rajagopalan, K.N.; Horth, C.; McGuire, J.T.; Xu, X.; Nikbakht, H.; et al. The Histone Mark H3K36me2 Recruits DNMT3A and Shapes the Intergenic DNA Methylation Landscape. Nature 2019, 573, 281–286. [Google Scholar] [CrossRef]
- Vougiouklakis, T.; Hamamoto, R.; Nakamura, Y.; Saloura, V. The NSD Family of Protein Methyltransferases in Human Cancer. Epigenomics 2015, 7, 863–874. [Google Scholar] [CrossRef]
- Sotos, J.F.; Dodge, P.R.; Muirhead, D.; Crawford, J.D.; Talbot, N.B. Cerebral Gigantism in Childhood—A Syndrome of Excessively Rapid Growth with Acromegalic Features and a Nonprogressive Neurologic Disorder. N. Engl. J. Med. 1964, 271, 109–116. [Google Scholar] [CrossRef]
- Kurotaki, N.; Imaizumi, K.; Harada, N.; Masuno, M.; Kondoh, T.; Nagai, T.; Ohashi, H.; Naritomi, K.; Tsukahara, M.; Makita, Y.; et al. Haploinsufficiency of NSD1 Causes Sotos Syndrome. Nat. Genet. 2002, 30, 365–366. [Google Scholar] [CrossRef]
- Kurotaki, N.; Stankiewicz, P.; Wakui, K.; Niikawa, N.; Lupski, J.R. Sotos Syndrome Common Deletion Is Mediated by Directly Oriented Subunits within Inverted Sos-REP Low-Copy Repeats. Hum. Mol. Genet. 2005, 14, 535–542. [Google Scholar] [CrossRef][Green Version]
- Rahman, N. Mechanisms Predisposing to Childhood Overgrowth and Cancer. Curr. Opin. Genet. Dev. 2005, 15, 227–233. [Google Scholar] [CrossRef]
- Douglas, J.; Hanks, S.; Temple, I.K.; Davies, S.; Murray, A.; Upadhyaya, M.; Tomkins, S.; Hughes, H.E.; Cole, T.R.P.; Rahman, N. NSD1 Mutations Are the Major Cause of Sotos Syndrome and Occur in Some Cases of Weaver Syndrome but Are Rare in Other Overgrowth Phenotypes. Am. J. Hum. Genet. 2003, 72, 132–143. [Google Scholar] [CrossRef]
- Sachwitz, J.; Meyer, R.; Fekete, G.; Spranger, S.; Matulevičienė, A.; Kučinskas, V.; Bach, A.; Luczay, A.; Brüchle, N.O.; Eggermann, K.; et al. NSD1 Duplication in Silver–Russell Syndrome (SRS): Molecular Karyotyping in Patients with SRS Features. Clin. Genet. 2017, 91, 73–78. [Google Scholar] [CrossRef]
- Pasillas, M.P.; Shah, M.; Kamps, M.P. NSD1 PHD Domains Bind Methylated H3K4 and H3K9 Using Interactions Disrupted by Point Mutations in Human Sotos Syndrome. Hum. Mutat. 2011, 32, 292–298. [Google Scholar] [CrossRef]
- Brennan, K.; Zheng, H.; Fahrner, J.A.; Shin, J.H.; Gentles, A.J.; Schaefer, B.; Sunwoo, J.B.; Bernstein, J.A.; Gevaert, O. NSD1 Mutations Deregulate Transcription and DNA Methylation of Bivalent Developmental Genes in Sotos Syndrome. Hum. Mol. Genet. 2022, 31, 2164–2184. [Google Scholar] [CrossRef]
- Fang, Y.; Tang, Y.; Zhang, Y.; Pan, Y.; Jia, J.; Sun, Z.; Zeng, W.; Chen, J.; Yuan, Y.; Fang, D. The H3K36me2 Methyltransferase NSD1 Modulates H3K27ac at Active Enhancers to Safeguard Gene Expression. Nucleic Acids Res. 2021, 49, 6281–6295. [Google Scholar] [CrossRef]
- Streubel, G.; Watson, A.; Jammula, S.G.; Scelfo, A.; Fitzpatrick, D.J.; Oliviero, G.; McCole, R.; Conway, E.; Glancy, E.; Negri, G.L.; et al. The H3K36me2 Methyltransferase Nsd1 Demarcates PRC2-Mediated H3K27me2 and H3K27me3 Domains in Embryonic Stem Cells. Mol. Cell 2018, 70, 371–379.e5. [Google Scholar] [CrossRef]
- Jani, K.S.; Jain, S.U.; Ge, E.J.; Diehl, K.L.; Lundgren, S.M.; Müller, M.M.; Lewis, P.W.; Muir, T.W. Histone H3 Tail Binds a Unique Sensing Pocket in EZH2 to Activate the PRC2 Methyltransferase. Proc. Natl. Acad. Sci. USA 2019, 116, 8295–8300. [Google Scholar] [CrossRef]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 Methylation Antagonizes PRC2-Mediated H3K27 Methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef]
- Cedar, H.; Bergman, Y. Linking DNA Methylation and Histone Modification: Patterns and Paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Martin-Herranz, D.E.; Aref-Eshghi, E.; Bonder, M.J.; Stubbs, T.M.; Choufani, S.; Weksberg, R.; Stegle, O.; Sadikovic, B.; Reik, W.; Thornton, J.M. Screening for Genes That Accelerate the Epigenetic Aging Clock in Humans Reveals a Role for the H3K36 Methyltransferase NSD1. Genome Biol. 2019, 20, 146. [Google Scholar] [CrossRef]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, A.; Kuh, D.; Paul, D.S.; Rakyan, V.K.; Leslie, R.D.; Zheng, S.C.; Widschwendter, M.; Beck, S.; Teschendorff, A.E. Correlation of an Epigenetic Mitotic Clock with Cancer Risk. Genome Biol. 2016, 17, 205. [Google Scholar] [CrossRef]
- Almuriekhi, M.; Shintani, T.; Fahiminiya, S.; Fujikawa, A.; Kuboyama, K.; Takeuchi, Y.; Nawaz, Z.; Nadaf, J.; Kamel, H.; Kitam, A.K.; et al. Loss-of-Function Mutation in APC2 Causes Sotos Syndrome Features. Cell Rep. 2015, 10, 1585–1598. [Google Scholar] [CrossRef]
- Harris, J.R.; Fahrner, J.A. Disrupted Epigenetics in the Sotos Syndrome Neurobehavioral Phenotype. Curr. Opin. Psychiatry 2019, 32, 55–59. [Google Scholar] [CrossRef]
- Visser, R.; Landman, E.B.M.; Goeman, J.; Wit, J.M.; Karperien, M. Sotos Syndrome Is Associated with Deregulation of the MAPK/ERK-Signaling Pathway. PLoS ONE 2012, 7, e49229. [Google Scholar] [CrossRef]
- Leventopoulos, G.; Kitsiou-Tzeli, S.; Kritikos, K.; Psoni, S.; Mavrou, A.; Kanavakis, E.; Fryssira, H. A Clinical Study of Sotos Syndrome Patients With Review of the Literature. Pediatr. Neurol. 2009, 40, 357–364. [Google Scholar] [CrossRef]
- Hill, D.E.; Roberts, C.C.; Inwards, C.Y.; Sim, F.H. Childhood Soft-Tissue Sarcoma Associated with Sotos Syndrome. Radiol. Case Rep. 2010, 5, 384. [Google Scholar] [CrossRef][Green Version]
- Corsello, G.; Giuffrb, M.; Carcione, A.; Cuzto, M.L.; Piccione, M.; Ziino, O. Lymphoproliferative Disorders in Sotos Syndrome: Observation of Two Cases. Am. J. Med. Genet. 1996, 64, 588–593. [Google Scholar] [CrossRef]
- Al-Mulla, N.; Belgaumi, A.F.; Teebi, A. Cancer in Sotos Syndrome: Report of a Patient with Acute Myelocytic Leukemia and Review of the Literature. J. Pediatr. Hematol. Oncol. 2004, 26, 204–208. [Google Scholar] [CrossRef]
- Hersh, J.H.; Cole, T.R.P.; Bloom, A.S.; Bertolone, S.J.; Hughes, H.E. Risk of Malignancy in Sotos Syndrome. J. Pediatr. 1992, 120, 572–574. [Google Scholar] [CrossRef]
- Baylin, S.B. DNA Methylation and Gene Silencing in Cancer. Nat. Clin. Pract. Oncol. 2005, 2, S4–S11. [Google Scholar] [CrossRef]
- Hake, S.B.; Xiao, A.; Allis, C.D. Linking the Epigenetic “language” of Covalent Histone Modifications to Cancer. Br. J. Cancer 2004, 90, 761–769. [Google Scholar] [CrossRef]
- Tauchmann, S.; Schwaller, J.; Jeltsch, A.; Dhayalan, A. NSD1: A Lysine Methyltransferase between Developmental Disorders and Cancer. Life 2021, 11, 877. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Rahman, N. The NSD1 and EZH2 Overgrowth Genes, Similarities and Differences. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163, 86–91. [Google Scholar] [CrossRef]
- Berdasco, M.; Ropero, S.; Setien, F.; Fraga, M.F.; Lapunzina, P.; Losson, R.; Alaminos, M.; Cheung, N.K.; Rahman, N.; Esteller, M. Epigenetic Inactivation of the Sotos Overgrowth Syndrome Gene Histone Methyltransferase NSD1 in Human Neuroblastoma and Glioma. Proc. Natl. Acad. Sci. USA 2009, 106, 21830–21835. [Google Scholar] [CrossRef]
- Leonards, K.; Almosailleakh, M.; Tauchmann, S.; Bagger, F.O.; Thirant, C.; Juge, S.; Bock, T.; Méreau, H.; Bezerra, M.F.; Tzankov, A.; et al. Nuclear Interacting SET Domain Protein 1 Inactivation Impairs GATA1-Regulated Erythroid Differentiation and Causes Erythroleukemia. Nat. Commun. 2020, 11, 2807. [Google Scholar] [CrossRef]
- Brumbaugh, J.; Kim, I.S.; Ji, F.; Huebner, A.J.; Di Stefano, B.; Schwarz, B.A.; Charlton, J.; Coffey, A.; Choi, J.; Walsh, R.M.; et al. Inducible Histone K-to-M Mutations Are Dynamic Tools to Probe the Physiological Role of Site-Specific Histone Methylation in Vitro and in Vivo. Nat. Cell Biol. 2019, 21, 1449–1461. [Google Scholar] [CrossRef]
- Brennan, K.; Shin, J.H.; Tay, J.K.; Prunello, M.; Gentles, A.J.; Sunwoo, J.B.; Gevaert, O. NSD1 Inactivation Defines an Immune Cold, DNA Hypomethylated Subtype in Squamous Cell Carcinoma. Sci. Rep. 2017, 7, 17064. [Google Scholar] [CrossRef]
- Farhangdoost, N.; Horth, C.; Hu, B.; Bareke, E.; Chen, X.; Li, Y.; Coradin, M.; Garcia, B.; Lu, C.; Majewski, J. Epigenome Dysregulation Resulting from NSD1 Mutation in Head and Neck Squamous Cell Carcinoma. bioRxiv 2020. [Google Scholar] [CrossRef]
- Farhangdoost, N.; Horth, C.; Hu, B.; Bareke, E.; Chen, X.; Li, Y.; Coradin, M.; Garcia, B.A.; Lu, C.; Majewski, J. Chromatin Dysregulation Associated with NSD1 Mutation in Head and Neck Squamous Cell Carcinoma. Cell Rep. 2021, 34, 108769. [Google Scholar] [CrossRef]
- Gameiro, S.F.; Ghasemi, F.; Zeng, P.Y.F.; Mundi, N.; Howlett, C.J.; Plantinga, P.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. Low Expression of NSD1, NSD2, and NSD3 Define a Subset of Human Papillomavirus-Positive Oral Squamous Carcinomas with Unfavorable Prognosis. Infect. Agents Cancer 2021, 16, 13. [Google Scholar] [CrossRef]
- Pan, C.; Izreig, S.; Yarbrough, W.G.; Issaeva, N. NSD1 Mutations by HPV Status in Head and Neck Cancer: Differences in Survival and Response to DNA-Damaging Agents. Cancers Head Neck 2019, 4, 3. [Google Scholar] [CrossRef]
- Bambury, R.M.; Jordan, E.; Zabor, E.C.; Bouvier, N.; Al-Ahmadie, H.; Boyd, M.E.; Mullane, S.A.; Cha, E.K.; Ostrovnaya, I.; Hyman, D.M.; et al. Association of Somatic Mutations in DNA Damage Repair (DDR) Genes with Efficacy of Platinum-Based Chemotherapy in Advanced Urothelial Carcinoma. J. Clin. Oncol. 2015, 33, 4532. [Google Scholar] [CrossRef]
- Ceccaldi, R.; O’Connor, K.W.; Mouw, K.W.; Li, A.Y.; Matulonis, U.A.; D’Andrea, A.D.; Konstantinopoulos, P.A. A Unique Subset of Epithelial Ovarian Cancers with Platinum Sensitivity and PARP Inhibitor Resistance. Cancer Res. 2015, 75, 628. [Google Scholar] [CrossRef]
- Ghasemi, F.; Prokopec, S.D.; MacNeil, D.; Mundi, N.; Gameiro, S.F.; Howlett, C.; Stecho, W.; Plantinga, P.; Pinto, N.; Ruicci, K.M.; et al. Mutational Analysis of Head and Neck Squamous Cell Carcinoma Stratified by Smoking Status. JCI Insight 2019, 4, e123443. [Google Scholar] [CrossRef]
- Su, X.; Zhang, J.; Mouawad, R.; Compérat, E.; Rouprêt, M.; Allanic, F.; Parra, J.; Bitker, M.O.; Thompson, E.J.; Gowrishankar, B.; et al. NSD1 Inactivation and SETD2 Mutation Drive a Convergence toward Loss of Function of H3K36 Writers in Clear Cell Renal Cell Carcinomas. Cancer Res. 2017, 77, 4835–4845. [Google Scholar] [CrossRef]
- Lee, H.W.; Choe, M. Expression of EZH2 in Renal Cell Carcinoma as a Novel Prognostic Marker. Pathol. Int. 2012, 62, 735–741. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Y.; Ding, B.; Zhou, H.; Yao, W.; Xu, H. Genetic Alteration of Histone Lysine Methyltransferases and Their Significance in Renal Cell Carcinoma. PeerJ 2019, 7, e6396. [Google Scholar] [CrossRef]
- Mendes-Pereira, A.; Sims, D.; Dexter, T.; Fenwick, K.; Assiotis, I.; Kozarewa, I.; Mitsopoulos, C.; Hakas, J.; Zvelebil, M.; Lord, C.; et al. Genome-Wide Functional Screen Identifies a Compendium of Genes Affecting Sensitivity to Tamoxifen. Proc. Natl. Acad. Sci. USA 2012, 109, 2730–2735. [Google Scholar] [CrossRef]
- Kasper, L.H.; Brindle, P.K.; Schnabel, C.A.; Pritchard, C.E.J.; Cleary, M.L.; van Deursen, J.M.A. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol. Cell. Biol. 1999, 19, 764–776. [Google Scholar] [CrossRef]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98-NSD1 Links H3K36 Methylation to Hox-A Gene Activation and Leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef]
- Schmoellerl, J.; Barbosa, I.A.M.; Eder, T.; Brandstoetter, T.; Schmidt, L.; Maurer, B.; Troester, S.; Pham, H.T.T.; Sagarajit, M.; Ebner, J.; et al. CDK6 Is an Essential Direct Target of NUP98 Fusion Proteins in Acute Myeloid Leukemia. Blood 2020, 136, 387–400. [Google Scholar] [CrossRef]
- Zhao, Q.; Caballero, O.L.; Levy, S.; Stevenson, B.J.; Iseli, C.; de Souza, S.J.; Galante, P.A.; Busam, D.; Leversha, M.A.; Chadalavada, K.; et al. Transcriptome-Guided Characterization of Genomic Rearrangements in a Breast Cancer Cell Line. Proc. Natl. Acad. Sci. USA 2009, 106, 1886–1891. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, F.; Chen, Q.; Wan, C.; Xiong, J.; Xu, J. CRISPR/Cas9-Mediated Knockout of NSD1 Suppresses the Hepatocellular Carcinoma Development via the NSD1/H3/Wnt10b Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 467. [Google Scholar] [CrossRef]
- Peri, S.; Izumchenko, E.; Schubert, A.D.; Slifker, M.J.; Ruth, K.; Serebriiskii, I.G.; Guo, T.; Burtness, B.A.; Mehra, R.; Ross, E.A.; et al. NSD1-A Nd NSD2-Damaging Mutations Define a Subset of Laryngeal Tumors with Favorable Prognosis. Nat. Commun. 2017, 8, 1772. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-ΚB by NSD1/FBXL11-Dependent Reversible Lysine Methylation of P65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef]
- Patton, E.; Mueller, K.; Adams, D.; Anandasabapathy, N.; Aplin, A.; Bertolotto, C.; Bosenberg, M.; Ceol, C.; Burd, C.; Chi, P.; et al. Melanoma Models for the next Generation of Therapies. Cancer Cell 2021, 39, 610–631. [Google Scholar] [CrossRef]
- Pandiani, C.; Béranger, G.; Leclerc, J.; Ballotti, R.; Bertolotto, C. Focus on Cutaneous and Uveal Melanoma Specificities. Genes Dev. 2017, 31, 724–743. [Google Scholar] [CrossRef]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Béranger, G.; Lassalle, S.; et al. Single-Cell RNA Sequencing Reveals Intratumoral Heterogeneity in Primary Uveal Melanomas and Identifies HES6 as a Driver of the Metastatic Disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef]
- Strub, T.; Martel, A.; Nahon-Esteve, S.; Baillif, S.; Ballotti, R.; Bertolotto, C. Translation of Single-Cell Transcriptomic Analysis of Uveal Melanomas to Clinical Oncology. Prog. Retin. Eye Res. 2021, 85, 100968. [Google Scholar] [CrossRef]
- De Souza, C.F.; Xander, P.; Monteiro, A.C.; dos Silva, A.G.; da Silva, D.C.P.; Mai, S.; Bernardo, V.; Lopes, J.D.; Jasiulionis, M.G. Mining Gene Expression Signature for the Detection of Pre-Malignant Melanocytes and Early Melanomas with Risk for Metastasis. PLoS ONE 2012, 7, e44800. [Google Scholar] [CrossRef]
- Cheli, Y.; Giuliano, S.; Botton, T.; Rocchi, S.; Hofman, V.; Hofman, P.; Bahadoran, P.; Bertolotto, C.; Ballotti, R. Mitf Is the Key Molecular Switch between Mouse or Human Melanoma Initiating Cells and Their Differentiated Progeny. Oncogene 2011, 30, 2307–2318. [Google Scholar] [CrossRef]
- Cheli, Y.; Giuliano, S.; Fenouille, N.; Allegra, M.; Hofman, V.; Hofman, P.; Bahadoran, P.; Lacour, P.; Tartare-Deckert, S.; Bertolotto, C.; et al. Hypoxia and MITF Control Metastatic Behaviour in Mouse and Human Melanoma Cells. Oncogene 2012, 31, 2461–2470. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the Multicellular Ecosystem of Metastatic Melanoma by Single-Cell RNA-Seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e19. [Google Scholar] [CrossRef]
- Larue, L.; Delmas, V. The WNT/Beta-Catenin Pathway in Melanoma. Front. Biosci. 2006, 11, 733–742. [Google Scholar] [CrossRef]
- Kaochar, S.; Dong, J.; Torres, M.; Rajapakshe, K.; Nikolos, F.; Davis, C.M.; Ehli, E.A.; Coarfa, C.; Mitsiades, N.; Poulaki, V. ICG-001 Exerts Potent Anticancer Activity against Uveal Melanoma Cells. Investig. Ophthalmol. Vis. Sci. 2018, 59, 132–143. [Google Scholar] [CrossRef]
- Korabiowska, M.; Schlott, T.; Siems, N.; Müller, A.; Cordon-Cardo, C.; Fischer, G.; Brinck, U. Analysis of Adenomatous Polyposis Coli Gene Expression, APC Locus-Microsatellite Instability and APC Promoter Methylation in the Progression of Melanocytic Tumours. Mod. Pathol. 2004, 17, 1539–1544. [Google Scholar] [CrossRef]
- Worm, J.; Christensen, C.; Grønbæk, K.; Tulchinsky, E.; Guldberg, P. Genetic and Epigenetic Alterations of the APC Gene in Malignant Melanoma. Oncogene 2004, 23, 5215–5226. [Google Scholar] [CrossRef]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 Expression Is Associated with High Proliferation Rate and Aggressive Tumor Subgroups in Cutaneous Melanoma and Cancers of the Endometrium, Prostate, and Breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, Y.; Huang, X.; Wei, W.; Qu, Y. Expression of EZH2 in Uveal Melanomas Patients and Associations with Prognosis. Oncotarget 2017, 8, 76423. [Google Scholar] [CrossRef]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The Epigenetic Modifier EZH2 Controls Melanoma Growth and Metastasis through Silencing of Distinct Tumour Suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef]
- Hoffmann, F.; Niebel, D.; Aymans, P.; Ferring-Schmitt, S.; Dietrich, D.; Landsberg, J. H3K27me3 and EZH2 Expression in Melanoma: Relevance for Melanoma Progression and Response to Immune Checkpoint Blockade. Clin. Epigenet. 2020, 12, 24. [Google Scholar] [CrossRef]
- Rogawski, D.S.; Grembecka, J.; Cierpicki, T. H3K36 Methyltransferases as Cancer Drug Targets: Rationale and Perspectives for Inhibitor Development. Future Med. Chem. 2016, 8, 1589–1607. [Google Scholar] [CrossRef]
- Morishita, M.; Mevius, D.E.H.F.; Shen, Y.; Zhao, S.; di Luccio, E. BIX-01294 Inhibits Oncoproteins NSD1, NSD2 and NSD3. Med. Chem. Res. 2017, 26, 2038–2047. [Google Scholar] [CrossRef]
- Schapira, M. Chemical Inhibition of Protein Methyltransferases. Cell Chem. Biol. 2016, 23, 1067–1076. [Google Scholar] [CrossRef]
- Kubicek, S.; O’Sullivan, R.J.; August, E.M.; Hickey, E.R.; Zhang, Q.; Teodoro, M.L.L.; Rea, S.; Mechtler, K.; Kowalski, J.A.; Homon, C.A.; et al. Reversal of H3K9me2 by a Small-Molecule Inhibitor for the G9a Histone Methyltransferase. Mol. Cell 2007, 25, 473–481. [Google Scholar] [CrossRef]
- Morishita, M.; Mevius, D.; Di Luccio, E. In Vitro Histone Lysine Methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct. Biol. 2014, 14, 25. [Google Scholar] [CrossRef]
- Graham, S.E.; Tweedy, S.E.; Carlson, H.A. Dynamic Behavior of the Post-SET Loop Region of NSD1: Implications for Histone Binding and Drug Development. Protein Sci. 2016, 25, 1021–1029. [Google Scholar] [CrossRef]
- Huang, H.; Howard, C.A.; Zari, S.; Cho, H.J.; Shukla, S.; Li, H.; Ndoj, J.; González-Alonso, P.; Nikolaidis, C.; Abbott, J.; et al. Covalent Inhibition of NSD1 Histone Methyltransferase. Nat. Chem. Biol. 2020, 16, 1403–1410. [Google Scholar] [CrossRef]
- Zou, Y.; Ma, D.; Wang, Y. The PROTAC Technology in Drug Development. Cell Biochem. Funct. 2019, 37, 21–30. [Google Scholar] [CrossRef]
- Zingg, D.; Arenas-Ramirez, N.; Sahin, D.; Rosalia, R.A.; Antunes, A.T.; Haeusel, J.; Sommer, L.; Boyman, O. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep. 2017, 20, 854–867. [Google Scholar] [CrossRef]
- Tiffen, J.C.; Gunatilake, D.; Gallagher, S.J.; Gowrishankar, K.; Heinemann, A.; Cullinane, C.; Dutton-Regester, K.; Pupo, G.M.; Strbenac, D.; Yang, J.Y.; et al. Targeting Activating Mutations of EZH2 Leads to Potent Cell Growth Inhibition in Human Melanoma by Derepression of Tumor Suppressor Genes. Oncotarget 2015, 6, 27023–27036. [Google Scholar] [CrossRef]
- Jin, B.; Zhang, P.; Zou, H.; Ye, H.; Wang, Y.; Zhang, J.; Yang, H.; Pan, J. Verification of EZH2 as a Druggable Target in Metastatic Uveal Melanoma. Mol. Cancer 2020, 19, 52. [Google Scholar] [CrossRef]
- Makita, S.; Tobinai, K. Targeting EZH2 with Tazemetostat. Lancet Oncol. 2018, 19, 586–587. [Google Scholar] [CrossRef]
- Drosos, Y.; Myers, J.A.; Xu, B.; Mathias, K.M.; Beane, E.C.; Radko-Juettner, S.; Mobley, R.J.; Larsen, M.E.; Piccioni, F.; Ma, X.; et al. NSD1 Mediates Antagonism between SWI/SNF and Polycomb Complexes and Is Required for Transcriptional Activation upon EZH2 Inhibition. Mol. Cell 2022, 82, 2472–2489.e8. [Google Scholar] [CrossRef]
- Yang, C.; Wang, K.; Liang, Q.; Tian, T.T.; Zhong, Z. Role of NSD1 as Potential Therapeutic Target in Tumor. Pharmacol. Res. 2021, 173, 105888. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krossa, I.; Strub, T.; Aplin, A.E.; Ballotti, R.; Bertolotto, C. Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma? Cancers 2022, 14, 4865. https://doi.org/10.3390/cancers14194865
Krossa I, Strub T, Aplin AE, Ballotti R, Bertolotto C. Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma? Cancers. 2022; 14(19):4865. https://doi.org/10.3390/cancers14194865
Chicago/Turabian StyleKrossa, Imène, Thomas Strub, Andrew E. Aplin, Robert Ballotti, and Corine Bertolotto. 2022. "Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma?" Cancers 14, no. 19: 4865. https://doi.org/10.3390/cancers14194865
APA StyleKrossa, I., Strub, T., Aplin, A. E., Ballotti, R., & Bertolotto, C. (2022). Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma? Cancers, 14(19), 4865. https://doi.org/10.3390/cancers14194865






