Use of Personalized Biomarkers in Metastatic Colorectal Cancer and the Impact of AI
Abstract
Simple Summary
Abstract
1. Introduction
2. Genomics in mCRC
3. Transcriptomics in mCRC: Immunoscore
3.1. Clinical and Prognostic Associations of the Consensus Molecular Subtypes
3.1.1. CMS1
3.1.2. CMS2
3.1.3. CMS3
3.1.4. CMS4
3.2. Immunoscore (IS)
4. Epigenomics in mCRC
4.1. Histone Modifications
DNA Methylation
4.2. miRNA
- WNT/β-catenin pathway
- EGFR pathways
- TGF-β signaling pathway
- Epithelial-to-mesenchymal transition (EMT)
4.2.1. MiRNAs as Potential Biomarkers in CRC
- miRNAs as prognostic biomarkers for CRC
| Type of Sample | miRNA | Method of Detection | Correlation with Clinical Outcome | Ref. |
|---|---|---|---|---|
| Tissue specimen | miR-15a/miR-16 | qRT-PCR | Downregulation correlated with an advanced TNM stage, poor histologic grade, lymph node metastasis, and unfavorable OS and DFS | [148] |
| miR-21 | In situ hybridization | High expression correlated with poor survival and poor therapeutic outcomes; miR-21 regulates the expression of ITGb4, PDCD4, PTEN, SPRY2 and RECK | [149] | |
| miR-106a | qRT-PCR | Downregulation correlated with unfavorable OS | [150] | |
| miR-132 | qRT-PCR | Downregulation correlated with unfavorable OS and the development of liver metastasis | [151] | |
| miR-150 | qRT-PCR, In situ hybridization | Low expression associated with longer OS; high expression associated with unfavorable outcomes in patients treated with 5-FU-based chemotherapy | [152] | |
| miR-181a | qRT-PCR | Low expression associated with poor PFS in patients with wild KRAS treated with EGFR inhibitors | [153,154] | |
| miR-188-3p | Level 3 Illumina (from TCGA database) | High expression correlated with metastatic disease; lower OS and lower expression are correlated with BRAF status | [155] | |
| miR-195 | qRT-PCR | Low expression associated with lymph node metastasis and an advanced tumor stage | [156] | |
| miR-199b | qRT-PCR and miRNA microarray | MiR-199b regulates the SIRT1/CREB/KISS1 signaling pathway, and high expression is associated with longer survival | [157] | |
| miR-215 | qRT-PCR | High levels associated with poor overall survival | [158] | |
| miR-218 | qRT-PCR | High miR-218 expression associated with the response to the first-line 5-FU treatment | [159] | |
| Circulating miRNAs—serum/plasma | miR-21 | qRT-PCR | Lower serum levels correlated with higher local recurrence | [160] |
| miR-23b | qRT-PCR | Low plasma levels correlated with a shorter recurrence-free survival time and poorer overall survival | [161] | |
| miR-139-5p | qRT-PCR | High serum levels correlated with tumor recurrence and metastasis | [162] | |
| miR-141 | qRT-PCR | High plasma levels correlated with poor prognosis | [141] | |
| miR-155 | qRT-PCR | High serum levels correlated with tumor differentiation, regional and distant metastasis, and the clinical TNM stage | [163] | |
| miR-183 | qRT-PCR | High plasma levels associated with regional and distant metastasis and tumor recurrence | [164] | |
| miR-203 | qRT-PCR | High serum levels associated with short survival and metastasis | [165] | |
| miR-218 | qRT-PCR | Low serum levels associated with the TNM stage, lymph node metastasis (LNM) and differentiation | [166] | |
| miR-221 | qRT-PCR | High plasma level is a prognostic factor for poor overall survival | [167] | |
| miR-885-5p | qRT-PCR miRNA microarray | High serum levels correlated with poor prognosis, regional and distant metastasis | [168] | |
| miR-122 | miRNA microarray | High plasma levels correlated with higher grading, and higher miR-200a, miR-200b and miR-200c levels were associated with increasing severity of the recurrence in metastatic CRC patients | [169] | |
| miR-200a | ||||
| miR-200b | ||||
| miR-200c | ||||
| Exosomes from serum/plasma | let-7a | qRT-PCR TaqMan | Upregulated serum levels are correlated with recurrence | [170] |
| miR-21 | ||||
| miR-23a | ||||
| miR-150 | ||||
| miR-223 | ||||
| miR-1246 | ||||
| miR-1229 | ||||
| miR-203 | qRT-PCR | Upregulated serum levels are correlated with recurrence | [171] | |
| miR-548c-5p | qRT-PCR miRNA microarray | Downregulated serum level associated with increased risk of liver metastasis and later TNM stage | [172,173] | |
| miR-638 | ||||
| miR-5787 | ||||
| miR-8075 | ||||
| miR-68869-5p | ||||
| Fecal samples | miRNA signature | qRT-PCR | High miRNA signature associated with reduced DFS and OS | [174] |
| miR-223/miR-222 | ||||
| miR-92a/miR-222 | ||||
| miR-16/miR-222 | ||||
| miR-20a/miR-222 | ||||
| miRNA panel | miRNA microarray, qRT-PCR | 12 upregulated miRNAs (miR-7, miR-17, miR-20a, miR-21, miR-92a, miR-96, miR-106a, miR-134, miR-183, miR-196a, miR-199a-3p and miR-214) and 8 downregulated miRNAs (miR-9, miR-29b, miR-127-5p, miR-138, miR-143, miR-146a, miR-222 and miR-938) were found to differentiate TNM stages with high sensitivity and specificity | [142] | |
| 12 upregulated | ||||
| 8 downregulated |
4.2.2. MiRNAs for Predicting the Response to Systemic Therapy in mCRC
4.3. LncRNA
- RP11 expression in CRC cells seems to correlate with lymph node metastasis and the advanced TNM stage, suggesting that this molecule can be a strong predictor of CRC metastasis and prognosis. Additionally, the upregulation of RP11 by m6A regulation can trigger the migration, invasion and EMT of CRC cells via the post-translational upregulation of the EMT-promoting TF Zeb1 [190].
- SATB2-AS1 is a colorectal-specific lncRNA expressed in colorectal tissues and CRC cells that inhibits tumor metastasis and regulates the immune response by activating SATB-2 in CRC. SATB2-AS1 downregulation seems to be due to DNA hypermethylation and histone H3K4me3 loss in the promoter region. Low levels of this lncRNA are correlated with the tumor invasion depth, lymph node metastasis and the TNM stage. Additionally, the gene signatures of the hallmark epithelial–mesenchymal transition, hallmark inflammatory response and hallmark interferon-gamma response were enriched in patients with low SATB2-AS1 expression. Overall, low SATB2-AS1 expression was associated with poor survival, and this study suggests that SATB2-AS1 and SATB2 may be novel biomarkers and promising therapeutic targets in CRC [191].
- LINC00659 expression in CRC is associated with poor prognosis. This study revealed higher levels of LINC00659 in CAF-exos than in NF-exos, which are transmitted to CRC cells and act through upregulating ANXA2 and increasing cell proliferation, migration and invasion [192].
- MALAT1 is another lncRNA that promotes CRC’s aggressiveness by regulating FUT4-associated fucosylation and the PI3K/Akt/mTOR pathway. In this study, we demonstrated that exosomes containing MALAT1 contributed to metastasis and the invasion of CRC cells via targeting miR-20b-5p, and targeting exosomal MALAT1 could attenuate the PI3K/AKT/mTOR pathway in CRC [193].
4.4. circRNA
4.4.1. Candidate Prognostic Biomarkers in Metastatic CRC
4.4.2. Candidate Predictive Biomarkers in Metastatic CRC
5. Metabolomics
5.1. Gram-Negative Bacteria
5.2. Gram-Positive Bacteria
5.3. Microbiota as Biomarkers in Colorectal Cancer
6. Artificial Intelligence Methods Used in mCRC
6.1. AI Application for Developing Biomarkers in mCRC in Blood Tests and Other Tests
6.2. AI Application in the Personalization and Precision Treatment of mCRC
6.3. AI for Developing Biomarkers to Predict and Prognosticate the mCRC
6.4. Implementation of the Selected Predictive Models
6.5. Predictive Model Mobile App
6.5.1. Experiments
6.5.2. Naive Bayes
6.5.3. Random Forest
6.5.4. Decision Tree
6.5.5. Gradient Boosted Trees
6.5.6. Logistic Regression
6.5.7. SVM
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Shibata, D.; Peinado, M.A.; Ionov, Y.; Malkhosyan, S.; Perucho, M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat. Genet. 1994, 6, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Bañobre, J.; Roy, R.; Alustiza Fernández, M.; Murcia, Ó.; Jover, R.; Pera, M.; Balaguer, F.; López-López, R.; Goel, A. Clinical significance of a microRNA signature for the identification and predicting prognosis in colorectal cancers with mucinous differentiation. Carcinogenesis 2020, 41, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Lanza, G.; Gafa, R.; Santini, A.; Maestri, I.; Guerzoni, L.; Cavazzini, L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J. Clin. Oncol. 2006, 24, 2359–2367. [Google Scholar] [CrossRef]
- Liu, G.C.; Liu, R.Y.; Yan, J.P.; An, X.; Jiang, W.; Ling, Y.H.; Chen, J.W.; Bei, J.X.; Zuo, X.Y.; Cai, M.Y.; et al. The Heterogeneity Between Lynch-Associated and Sporadic MMR Deficiency in Colorectal Cancers. J. Natl. Cancer Inst. 2018, 110, 975–984. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Yoon, H.H.; Mahoney, M.R.; Nelson, G.D.; Thibodeau, S.N.; Goldberg, R.M.; Sargent, D.J.; Alberts, S.R. Overall survival result and outcomes by KRAS, BRAF, and DNA mismatch repair in relation to primary tumor site in colon cancers from a randomized trial of adjuvant chemotherapy: NCCTG (Alliance) N0147. J. Clin. Oncol. 2014, 32, 3525. [Google Scholar] [CrossRef]
- Jin, Z.; Sanhueza, C.T.; Johnson, B.; Nagorney, D.M.; Larson, D.W.; Mara, K.C.; Harmsen, W.C.; Smyrk, T.C.; Grothey, A.; Hubbard, J.M. Outcome of mismatch repair-deficient metastatic colorectal cancer: The mayo clinic experience. Oncologist 2018, 23, 1083–1091. [Google Scholar] [CrossRef]
- Tan, C.; Du, X. KRAS mutation testing in metastatic colorectal cancer. World J. Gastroenterol. 2012, 18, 5171–5180. [Google Scholar]
- Corcoran, R.B.; Ebi, H.; Turke, A.B.; Coffee, E.M.; Nishino, M.; Cogdill, A.P.; Brown, R.D.; Della Pelle, P.; Dias-Santagata, D.; Hung, K.E.; et al. EGFR-mediated reactivation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012, 2, 227–235. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Mahoney, M.R.; Smyrk, T.C.; Thibodeau, S.N.; Warren, R.S.; Bertagnolli, M.M.; Nelson, G.D.; Goldberg, R.M.; Sargent, D.J.; Alberts, S.R. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J. Clin. Oncol. 2013, 31, 3664–3672. [Google Scholar] [CrossRef]
- Kopetz, S.; Guthrie, K.A.; Morris, V.K.; Lenz, H.J.; Magliocco, A.M.; Maru, D.; Yan, Y.; Lanman, R.; Manyam, G.; Hong, D.S.; et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406). J. Clin. Oncol. 2021, 39, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Modest, D.P.; Ricard, I.; Heinemann, V.; Hegewisch-Becker, S.; Schmiegel, W.; Porschen, R.; Stintzing, S.; Graeven, U.; Arnold, D.; von Weikersthal, L.F.; et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann. Oncol. 2016, 27, 1746–1753. [Google Scholar] [CrossRef] [PubMed]
- Lièvre, A.; Bachet, J.B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.F.; Côté, J.F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef] [PubMed]
- Amado, R.G.; Wolf, M.; Peeters, M.; Van Cutsem, E.; Siena, S.; Freeman, D.J.; Juan, T.; Sikorski, R.; Suggs, S.; Radinsky, R.; et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Hallin, J.; Engstrom, L.D.; Hargis, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS G12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- NCT03600883. A Phase 1/2, Study Evaluating the Safety, Tolerability, PK, and Efficacy of Sotorasib (AMG 510) in Subjects with Solid Tumors with a Specific KRAS Mutation (CodeBreaK 100). Available online: https://clinicaltrials.gov/ct2/show/NCT03600883 (accessed on 5 April 2022).
- Ou, S.I.; Jänne, P.A.; Leal, T.A.; Rybkin, I.I.; Sabari, J.K.; Barve, M.A.; Bazhenova, L.; Johnson, M.L.; Velastegui, K.L.; Cilliers, C.; et al. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients with Advanced KRASG12C Solid Tumors (KRYSTAL-1). J. Clin. Oncol. 2022, 40, 2530–2538. [Google Scholar] [CrossRef]
- Lou, K.; Steri, V.; Ge, A.Y.; Hwang, Y.C.; Yogodzinski, C.H.; Shkedi, A.R.; Choi, A.; Mitchell, D.C.; Swaney, D.L.; Hann, B.; et al. KRASG12C inhibition produces a driver-limited state revealing collateral dependencies. Sci. Signal. 2019, 12, eaaw9450. [Google Scholar] [CrossRef]
- Molina-Arcas, M.; Samani, A.; Downward, J. Drugging the Undruggable: Advances on RAS Targeting in Cancer. Genes 2021, 12, 899. [Google Scholar] [CrossRef]
- Nathanson, D.R.; Culliford, A.T.; Shia, J.; Chen, B.; D’Alessio, M.; Zeng, Z.S.; Nash, G.M.; Gerald, W.; Barany, F.; Paty, P.B. HER 2/neu expression and gene amplification in colon cancer. Int. J. Cancer 2003, 105, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Corà, D.; Di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011, 1, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Kavuri, S.M.; Jain, N.; Galimi, F.; Cottino, F.; Leto, S.M.; Migliardi, G.; Searleman, A.C.; Shen, W.; Monsey, J.; Trusolino, L.; et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015, 5, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Ardini, E.; Bosotti, R.; Amatu, A.; Valtorta, E.; Somaschini, A.; Raddrizzani, L.; Palmeri, L.; Banfi, P.; Bonazzina, E.; et al. Sensitivity to Entrectinib Associated with a Novel LMNA-NTRK1 Gene Fusion in Metastatic Colorectal Cancer. J. Natl. Cancer Inst. 2016, 108, djv306. [Google Scholar] [CrossRef] [PubMed]
- Seligmann, J.F.; Elliott, F.; Richman, S.D.; Jacobs, B.; Hemmings, G.; Brown, S.; Barrett, J.H.; Tejpar, S.; Quirke, P.; Seymour, M.T. Combined Epiregulin and Amphiregulin Expression Levels as a Predictive Biomarker for Panitumumab Therapy Benefit or Lack of Benefit in Patients with RAS Wild-Type Advanced Colorectal Cancer. JAMA Oncol. 2016, 2, 633–642. [Google Scholar] [CrossRef]
- Rosty, C.; Young, J.P.; Walsh, M.D.; Clendenning, M.; Sanderson, K.; Walters, R.J.; Parry, S.; Jenkins, M.A.; Win, A.K.; Southey, M.C.; et al. PIK3CA activating mutation in colorectal carcinoma: Associations with molecular features and survival. PLoS ONE 2013, 8, e65479. [Google Scholar] [CrossRef]
- Innocenti, F.; Mills, S.C.; Sanoff, H.; Ciccolini, J.; Lenz, H.-J.; Milano, G. All you need to know about DPYD genetic testing for patients treated with fluorouracil and capecitabine: A practitioner-friendly guide. JCO Oncol. Pract. 2020, 16, 793–798. [Google Scholar] [CrossRef]
- Merloni, F.; Ranallo, N.; Scortichini, L.; Giampieri, R.; Berardi, R. Tailored therapy in patients treated with fluoropyrimidines: Focus on the role of dihydropyrimidine dehydrogenase. Cancer Drug Resist. 2019, 2, 787–802. [Google Scholar] [CrossRef]
- Innocenti, F.; Undevia, S.D.; Iyer, L.; Chen, P.X.; Das, S.; Kocherginsky, M.; Karrison, T.; Janisch, L.; Ramírez, J.; Rudin, C.M.; et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J. Clin. Oncol. 2004, 22, 1382–1388. [Google Scholar] [CrossRef]
- Mathijssen, R.H.; Gurney, H. Irinogenetics: How many stars are there in the sky? J. Clin. Oncol. 2009, 27, 2578–2579. [Google Scholar] [CrossRef]
- Shirota, Y.; Stoehlmacher, J.; Brabender, J.; Xiong, Y.P.; Uetake, H.; Danenberg, K.D.; Groshen, S.; Tsao-Wei, D.D.; Danenberg, P.V.; Lenz, H.J. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J. Clin. Oncol. 2001, 19, 4298–4304. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectalcancer patients. Nat. Med. 2015, 21, 827. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Vitiello, P.P.; Sogari, A.; Crisafulli, G.; Sartore-Bianchi, A.; Marsoni, S.; Siena, S.; Bardelli, A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br. J. Cancer 2022, 127, 394–407. [Google Scholar] [CrossRef]
- Gabriel, E.; Bagaria, S.P. Assessing the Impact of Circulating Tumor DNA (ctDNA) in Patients with Colorectal Cancer: Separating Fact From Fiction. Front. Oncol. 2018, 8, 297. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Pietrantonio, F.; Lonardi, S.; Mussolin, B.; Rua, F.; Crisafulli, G.; Bartolini, A.; Fenocchio, E.; Amatu, A.; Manca, P.; et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: The phase 2 CHRONOS trial. Nat. Med. 2022, 28, 1612–1618. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yoshino, T. Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer. Oncologist 2018, 23, 1310–1318. [Google Scholar] [CrossRef]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden-High Solid Tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- Pich, O.; Muiños, F.; Lolkema, M.P.; Steeghs, N.; Gonzalez-Perez, A.; Lopez-Bigas, N. The mutational footprints of cancer therapies. Nat. Genet. 2019, 51, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Baraibar, I.; Mirallas, O.; Saoudi, N.; Ros, J.; Salvà, F.; Tabernero, J.; Élez, E. Combined Treatment with Immunotherapy-Based Strategies for MSS Metastatic Colorectal Cancer. Cancers 2021, 13, 6311. [Google Scholar] [CrossRef] [PubMed]
- He, W.Z.; Hu, W.M.; Wang, F.; Rong, Y.M.; Yang, L.; Xie, Q.K.; Yang, Y.Z.; Jiang, C.; Qiu, H.J.; Lu, J.B.; et al. Comparison of Mismatch Repair Status Between Primary and Matched Metastatic Sites in Patients with Colorectal Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 1174–1183. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Foster, N.R.; Thibodeau, S.N.; Marsoni, S.; Monges, G.; Labianca, R.; Kim, G.P.; Yothers, G.; Allegra, C.; Moore, M.J.; et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J. Natl. Cancer Inst. 2011, 103, 863–875. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef]
- Andre, T.; de Gramont, A.; Vernerey, D.; Chibaudel, B.; Bonnetain, F.; Tijeras-Raballand, A.; Scriva, A.; Hickish, T.; Tabernero, J.; Van Laethem, J.L.; et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III colon cancer: Updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J. Clin. Oncol. 2015, 33, 4176–4187. [Google Scholar] [CrossRef]
- Gavin, P.G.; Colangelo, L.H.; Fumagalli, D.; Tanaka, N.; Remillard, M.Y.; Yothers, G.; Kim, C.; Taniyama, Y.; Kim, S.I.; Choi, H.J.; et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: An assessment of their prognostic and oxaliplatin predictive value. Clin. Cancer Res. 2012, 18, 6531–6541. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, A.; Hartley, C.; Hagen, C. Tubulovillous adenomas with serrated features are precursors to KRAS mutant colorectal carcinoma. Mod. Pathol. 2017, 30, 157. [Google Scholar]
- Blons, H.; Emile, J.F.; Le Malicot, K.; Julié, C.; Zaanan, A.; Tabernero, J.; Mini, E.; Folprecht, G.; Van Laethem, J.L.; Thaler, J.; et al. Prognostic value of KRAS mutations in stage III colon cancer: Post hoc analysis of the PETACC8 phase III trial dataset. Ann. Oncol. 2014, 25, 2378–2385. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Piessevaux, H.; De Schutter, J.; Janssens, M.; De Hertogh, G.; Personeni, N.; Biesmans, B.; Van Laethem, J.L.; Peeters, M.; Humblet, Y.; et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann. Oncol. 2008, 19, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Fessler, E.; Drost, J.; van Hooff, S.R.; Linnekamp, J.F.; Wang, X.; Jansen, M.; De Sousa EMelo, F.; Prasetyanti, P.R.; IJspeert, J.E.; Franitza, M.; et al. TGFbeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 2016, 8, 745–760. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, E.M.F.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Leedham, S.J.; Graham, T.A.; Oukrif, D.; McDonald, S.A.; Rodriguez-Justo, M.; Harrison, R.F.; Shepherd, N.A.; Novelli, M.R.; Jankowski, J.A.; Wright, N.A. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology 2009, 136, 542–550.e546. [Google Scholar] [CrossRef]
- Roepman, P.; Schlicker, A.; Tabernero, J.; Majewski, I.; Tian, S.; Moreno, V.; Snel, M.H.; Chresta, C.M.; Rosenberg, R.; Nitsche, U.; et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int. J. Cancer 2014, 134, 552–562. [Google Scholar] [CrossRef]
- Loupakis, F.; Cremolini, C.; Masi, G.; Lonardi, S.; Zagonel, V.; Salvatore, L.; Cortesi, E.; Tomasello, G.; Ronzoni, M.; Spadi, R.; et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N. Engl. J. Med. 2014, 371, 1609–1618. [Google Scholar] [CrossRef]
- Sobin, L.; Wittekind, C. TNM Classification of Malignant Tumors, 6th ed.; UICC: Geneva, Switzerland; Wiley-Blackwell: New York, NY, USA, 2002. [Google Scholar]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The Immunoscore: Colon Cancer and Beyond. Clin. Cancer Res. 2020, 15, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Fridman, W.H.; Pagès, F. The Adaptive Immunologic Microenvironment in Colorectal Cancer: A Novel Perspective. Cancer Res. 2007, 67, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer. 2020, 20, 662–680. [Google Scholar] [CrossRef]
- Galon, J.; Lanzi, A. Immunoscore and its introduction in clinical practice. Q. J. Nucl. Med. Mol. Imaging 2020, 64, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Zeitoun, G.; Sissy, C.E.; Kirilovsky, A.; Anitei, G.; Todosi, A.M.; Marliot, F.; Haicheur, N.; Lagorce, C.; Berger, A.; Zinzindohoué, F.; et al. The Immunoscore in the Clinical Practice of Patients with Colon and Rectal Cancers. Chirurgia (Bucur) 2019, 114, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, D.; Cai, S.; Li, Q.; Li, X. P-TNM staging system for colon cancer: Combination of P-stage and AJCC TNM staging system for improving prognostic prediction and clinical management. Cancer Manag. Res. 2018, 10, 2303–2314. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 26, 2128–2139. [Google Scholar] [CrossRef]
- Lanzi, A.; Pagès, F.; Lagorce-Pagès, C.; Galon, J. The consensus immunoscore: Toward a new classification of colorectal cancer. Oncoimmunology 2020, 9, 1789032. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, X.; Zeng, S.; Ren, X.; Yan, Y.; Gong, Z. Applying artificial intelligence for cancer immunotherapy. Acta Pharm. Sin. B 2021, 11, 3393–3405. [Google Scholar] [CrossRef]
- Reichling, C.; Taieb, J.; Derangere, V.; Klopfenstein, Q.; Le Malicot, K.; Gornet, J.M.; Becheur, H.; Fein, F.; Cojocarasu, O.; Kaminsky, M.C.; et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut 2020, 69, 681–690. [Google Scholar] [CrossRef]
- Mehrotra, S.; Galdieri, L.; Zhang, T.; Zhang, M.; Pemberton, L.F.; Vancura, A. Histone hypoacetylation-activated genes are re-pressed by acetyl-CoA- and chromatin-mediated mechanism. Biochim. Biophys. Acta 2014, 1839, 751–763. [Google Scholar] [CrossRef][Green Version]
- Qin, J.; Wen, B.; Liang, Y.; Yu, W.; Li, H. Histone Modifications and their Role in Colorectal Cancer (Review). Pathol. Oncol. Res. 2020, 26, 2023–2033. [Google Scholar] [CrossRef]
- Karczmarski, J.; Rubel, T.; Paziewska, A.; Mikula, M.; Bujko, M.; Kober, P.; Dadlez, M.; Ostrowski, J. Histone H3 lysine 27 acetylation is altered in colon cancer. Clin. Proteom. 2014, 11, 24. [Google Scholar] [CrossRef]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003, 370 (Pt 3), 737–749. [Google Scholar] [CrossRef]
- Hashimoto, T.; Yamakawa, M.; Kimura, S.; Usuba, O.; Toyono, M. Expression of acetylated and Dimethylated histone H3 in colorectal cancer. Dig. Surg. 2013, 30, 249–258. [Google Scholar] [CrossRef]
- Benard, A.; Goossens-Beumer, I.J.; van Hoesel, A.Q.; Horati, H.; de Graaf, W.; Putter, H.; Zeestraten, E.C.M.; Liefers, G.; van de Velde, C.J.H.; Kuppen, P.J.K. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology 2015, 66, 270–282. [Google Scholar] [CrossRef]
- Ashktorab, H.; Belgrave, K.; Hosseinkhah, F.; Brim, H.; Nouraie, M.; Takkikto, M.; Hewitt, S.; Lee, E.L.; Dashwood, R.H.; Smoot, D. Global histone H4 acetylation and HDAC2 expression in Colon adenoma and carcinoma. Dig. Dis. Sci. 2009, 54, 2109–2117. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; Chen, S.; Zhao, L.; Sun, F. Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56. J. Biol. Chem. 2012, 287, 41469–41480. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Kang, W.; Liu, C.; Dong, Y.; Ren, F.; Wang, Y.; Zhang, J.; Wang, G.; To, K.F.; et al. CREPT facilitates colorectal cancer growth through inducing Wnt/β-catenin pathway by enhancing p300-mediated β-catenin acetylation. Oncogene 2018, 37, 3485–3500. [Google Scholar] [CrossRef]
- Tamagawa, H.; Oshima, T.; Numata, M.; Yamamoto, N.; Shiozawa, M.; Morinaga, S.; Nakamura, Y.; Yoshihara, M.; Sakuma, Y.; Kameda, Y.; et al. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur. J. Surg. Oncol. 2013, 39, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R. Epigenetics at the base of alternative splicing changes that promote colorectal cancer. J. Clin. Investig. 2017, 127, 3281–3283. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zeng, Z.; Luo, T.; Li, Q.; Hao, Y.; Chen, L. Clinicopathological significance of G9A expression in colorectal carcinoma. Oncol. Lett. 2018, 15, 8611–8619. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Z.; Xia, Y.; Liao, G.; Pan, Y.; Liu, S.; Zhang, Y.; Yan, Z. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br. J. Cancer 2013, 109, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Li, Z.; Gan, M.; Zhang, H.; Yin, X.; Tang, S.; Wan, L.; Tian, Y.; Zhang, S.; Zhu, Y.; et al. Decreased expression of dual specificity phosphatase 22 in colorectal cancer and its potential prognostic relevance for stage IV CRC patients. Tumor Biol. 2015, 36, 8531–8535. [Google Scholar] [CrossRef]
- Lee, Y.C.; Yin, T.C.; Chen, Y.T.; Chai, C.Y.; Wang, J.Y.; Liu, M.C.; Lin, Y.C.; Kan, J.Y. High expression of phospho-H2AX predicts a poor prognosis in colorectal cancer. Anticancer Res. 2015, 35, 2447–2453. [Google Scholar]
- Iyer, S.P.; Foss, F.F. Romidepsin for the treatment of peripheral T-cell lymphoma. Oncologist 2015, 20, 1084–1091. [Google Scholar] [CrossRef]
- Foss, F.; Advani, R.; Duvic, M.; Hymes, K.B.; Intragumtornchai, T.; Lekhakula, A.; Shpilberg, O.; Lerner, A.; Belt, R.J.; Jacobsen, E.D.; et al. A phase II trial of Belinostat (PXD101) in patients with relapsed or refractory peripheral or cutaneous T-cell lymphoma. Br. J. Haematol. 2015, 168, 811–819. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef]
- Esteller, M.; González, S.; Risques, R.A.; Marcuello, E.; Mangues, R.; Germà, J.R.; Herman, J.G.; Capellà, G.; Peinado, M.A. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J. Clin. Oncol. 2001, 19, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Tortola, S.; Toyota, M.; Capella, G.; Peinado, M.A.; Baylin, S.B.; Herman, J.G. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000, 60, 129–133. [Google Scholar] [PubMed]
- Cunningham, J.M.; Christensen, E.R.; Tester, D.J.; Kim, C.Y.; Roche, P.C.; Burgart, L.J.; Thibodeau, S.N. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998, 58, 3455–3460. [Google Scholar] [PubMed]
- Liang, T.-J.; Wang, H.X.; Zheng, Y.Y.; Cao, Y.Q.; Wu, X.; Zhou, X.; Dong, S.X. APC hypermethylation for early diagnosis of colorectal cancer: A meta-analysis and literature review. Oncotarget 2017, 8, 46468–46479. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Sunami, E.; Yamamoto, Y.; Hata, K.; Okada, S.; Murono, K.; Yasuda, K.; Otani, K.; Nishikawa, T.; Tanaka, T.; et al. LINE-1 hypomethylation status of circulating cell-free DNA in plasma as a biomarker for colorectal cancer. Oncotarget 2017, 8, 11906–11916. [Google Scholar] [CrossRef]
- Pérez, R.F.; Tejedor, J.R.; Bayón, G.F.; Fernández, A.F.; Fraga, M.F. Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell 2018, 17, e12744. [Google Scholar] [CrossRef]
- Baba, Y.; Nosho, K.; Shima, K.; Huttenhower, C.; Tanaka, N.; Hazra, A.; Giovannucci, E.L.; Fuchs, C.S.; Ogino, S. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology 2010, 139, 1855–1864. [Google Scholar] [CrossRef]
- Luo, J.; Li, Y.-N.; Wang, F.; Zhang, W.-M.; Geng, X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int. J. Biol. Sci. 2010, 6, 784–795. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Baba, Y.; Yagi, T.; Sawayama, H.; Hiyoshi, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Baba, H. Long interspersed element-1 methylation level as a prognostic biomarker in gastrointestinal cancers. Digestion 2018, 97, 26–30. [Google Scholar] [CrossRef]
- Ogino, S.; Kawasaki, T.; Nosho, K.; Ohnishi, M.; Suemoto, Y.; Kirkner, G.J.; Fuchs, C.S. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int. J. Cancer 2008, 122, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Ashktorab, H.; Brim, H. DNA Methylation and Colorectal Cancer. Curr. Colorectal Cancer Rep. 2014, 10, 425–430. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Balacescu, O.; Sur, D.; Cainap, C.; Visan, S.; Cruceriu, D.; Manzat-Saplacan, R.; Muresan, M.S.; Balacescu, L.; Lisencu, C.; Irimie, A. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int. J. Mol. Sci. 2018, 19, 3711. [Google Scholar] [CrossRef]
- Segditsas, S.; Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25, 7531–7537. [Google Scholar] [CrossRef]
- Lai, Q.; Wang, S.; Cai, J.; Xiao, Z.; Deng, D.; He, L.; Jiao, H.; Ye, Y.; Liang, L.; Ding, Y.; et al. MicroRNA-224 sustains Wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J. Exp. Clin. Cancer Res. CR 2016, 35, 21. [Google Scholar] [CrossRef]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280, Erratum in Nat. Cell Biol. 2014, 16, 383; Erratum in Nat. Cell Biol. 2019, 21, 664. [Google Scholar] [CrossRef]
- Guo, C.; Sah, J.F.; Beard, L.; Willson, J.K.; Markowitz, S.D.; Guda, K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47, 939–946. [Google Scholar] [CrossRef]
- Velho, S.; Oliveira, C.; Ferreira, A.; Ferreira, A.C.; Suriano, G.; Schwartz, S., Jr.; Duval, A.; Carneiro, F.; Machado, J.C.; Hamelin, R.; et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur. J. Cancer 2005, 41, 1649–1654. [Google Scholar] [CrossRef]
- Arcaroli, J.J.; Quackenbush, K.S.; Powell, R.W.; Pitts, T.M.; Spreafico, A.; Varella-Garcia, M.; Bemis, L.; Tan, A.C.; Reinemann, J.M.; Touban, B.M.; et al. Common PIK3CA mutants and a novel 3′ UTR mutation are associated with increased sensitivity to saracatinib. Clin. Cancer Res. 2012, 18, 2704–2714. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef] [PubMed]
- Sebio, A.; Paré, L.; Páez, D.; Salazar, J.; González, A.; Sala, N.; del Río, E.; Martín-Richard, M.; Tobeña, M.; Barnadas, A.; et al. The LCS6 polymorphism in the binding site of let-7 microRNA to the KRAS 3′-untranslated region: Its role in the efficacy of anti-EGFR-based therapy in metastatic colorectal cancer patients. Pharm. Genom. 2013, 23, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009, 30, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, J.; Wang, J.; Chen, X.; Zhu, Y.; Chen, Y. Mir-30b-3p affects the migration and invasion function of ovarian cancer cells by targeting the CTHRC1 gene. Biol. Res. 2020, 53, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, A.; Valvo, C.; Fabrizi, E.; di Martino, S.; Biffoni, M.; Runci, D.; Forte, S.; De Maria, R.; Ricci-Vitiani, L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013, 32, 4806–4813. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Myeroff, L.L.; Swinler, S.E.; Rajput, A.; Thiagalingam, S.; Lutterbaugh, J.D.; Neumann, A.; Brattain, M.G.; Chang, J.; Kim, S.J.; et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999, 59, 320–324. [Google Scholar]
- Grady, W.M.; Markowitz, S.D. Genetic and epigenetic alterations in colon cancer. Annu. Rev. Genom. Hum. Genet. 2002, 3, 101–128. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-β Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef]
- Yu, Y.; Kanwar, S.S.; Patel, B.B.; Oh, P.S.; Nautiyal, J.; Sarkar, F.H.; Majumdar, A.P. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis 2012, 33, 68–76. [Google Scholar] [CrossRef]
- Feng, B.; Dong, T.T.; Wang, L.L.; Zhou, H.M.; Zhao, H.C.; Dong, F.; Zheng, M.H. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS ONE 2012, 7, e43452. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Jin, R.; Zhao, H.; Hu, J.; Feng, B.; Zang, L.; Zheng, M.; Wang, M. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J. Exp. Clin. Cancer Res. 2014, 33, 113. [Google Scholar] [CrossRef]
- Li, Q.; Zou, C.; Zou, C.; Han, Z.; Xiao, H.; Wei, H.; Wang, W.; Zhang, L.; Zhang, X.; Tang, Q.; et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013, 335, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Carstens, J.L.; Lovisa, S.; Kalluri, R. Microenvironment-dependent cues trigger miRNA-regulated feedback loop to facilitate the EMT/MET switch. J. Clin. Investig. 2014, 124, 1458–1460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.X.; Mai, S.J.; Huang, X.X.; Wang, F.W.; Liao, Y.J.; Lin, M.C.; Kung, H.F.; Zeng, Y.X.; Xie, D. MiR-29c mediates epithelial-tomesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann. Oncol. 2014, 25, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef]
- Guo, Y.H.; Wang, L.Q.; Li, B.; Xu, H.; Yang, J.H.; Zheng, L.S.; Yu, P.; Zhou, A.D.; Zhang, Y.; Xie, S.J.; et al. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget 2016, 7, 42513–42526. [Google Scholar] [CrossRef]
- Yu, G.; Tang, J.Q.; Tian, M.L.; Li, H.; Wang, X.; Wu, T.; Zhu, J.; Huang, S.J.; Wan, Y.L. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J. Surg. Oncol. 2012, 106, 232–237. [Google Scholar] [CrossRef]
- Peric, D.; Chvalova, K.; Rousselet, G. Identification of microprocessor-dependent cancer cells allows screening for growth-sustaining micro-RNAs. Oncogene 2012, 31, 2039–2048. [Google Scholar] [CrossRef]
- Liang, Z.; Li, Y.; Huang, K.; Wagar, N.; Shim, H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm. Res. 2011, 28, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Olive, V.; Bennett, M.J.; Walker, J.C.; Ma, C.; Jiang, I.; Cordon-Cardo, C.; Li, Q.J.; Lowe, S.W.; Hannon, G.J.; He, L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009, 23, 2839–2849. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Liu, C.G.; Alder, H.; Taccioli, C.; Volinia, S.; Calin, G.A.; Croce, C.M. GAM/ZFp/ZNF512B is central to a gene sensor circuitry involving cell-cycle regulators, TGFβ effectors, Drosha and microRNAs with opposite oncogenic potentials. Nucleic Acids Res. 2010, 38, 7673–7688. [Google Scholar] [CrossRef]
- Slaby, O.; Svoboda, M.; Fabian, P.; Smerdova, T.; Knoflickova, D.; Bednarikova, M.; Nenutil, R.; Vyzula, R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007, 72, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, H.; Iinuma, H.; Shimada, R.; Horiuchi, A.; Watanabe, T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 2010, 79, 313–320. [Google Scholar] [CrossRef]
- Drusco, A.; Nuovo, G.J.; Zanesi, N.; Di Leva, G.; Pichiorri, F.; Volinia, S.; Fernandez, C.; Antenucci, A.; Costinean, S.; Bottoni, A.; et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS ONE 2014, 9, e96670. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zhu, Y.; Gao, J.; Fu, J.; Liu, C.; Liu, Y.; Song, C.; Zhu, S.; Leng, Y.; Wang, G.; et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br. J. Cancer 2014, 110, 450–458. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.K.; Zhao, M.; Yang, M.; Zhong, J.L.; Gu, Y.Y.; Peng, H.; Che, Y.Q.; Huang, C.Z. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS ONE 2014, 9, e87451. [Google Scholar]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Tang, J.; Bai, Y.; Lin, H.; You, H.; Jin, H.; Lin, L.; You, P.; Li, J.; Dai, Z.; et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 86. [Google Scholar] [CrossRef]
- Ahmed, F.E.; Jeffries, C.D.; Vos, P.W.; Flake, G.; Nuovo, G.J.; Sinar, D.R.; Naziri, W.; Marcuard, S.P. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genom. Proteom. 2009, 6, 281–295. [Google Scholar]
- Ahmed, F.E.; Ahmed, N.C.; Vos, P.W.; Bonnerup, C.; Atkins, J.N.; Casey, M.; Nuovo, G.J.; Naziri, W.; Wiley, J.E.; Mota, H.; et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genom. Proteom. 2013, 10, 93–113. [Google Scholar]
- Braicu, C.; Tomuleasa, C.; Monroig, P.; Cucuianu, A.; Berindan-Neagoe, I.; Calin, G.A. Exosomes as divine messengers: Are they the Hermes of modern molecular oncology? Cell Death Differ. 2015, 22, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Monzo, M.; Santasusagna, S.; Moreno, I.; Martinez, F.; Hernández, R.; Muñoz, C.; Castellano, J.J.; Moreno, J.; Navarro, A. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget 2017, 8, 30859–30869. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Tang, H.; Wei, W.; Li, J.; Ji, L.; Ge, J. Aberrant Expression of MicroRNA-15a and MicroRNA-16 Synergistically Associates with Tumor Progression and Prognosis in Patients with Colorectal Cancer. Gastroenterol. Res. Pract. 2014, 2014, 364549. [Google Scholar] [CrossRef]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef]
- Díaz, R.; Silva, J.; García, J.M.; Lorenzo, Y.; García, V.; Peña, C.; Rodríguez, R.; Muñoz, C.; García, F.; Bonilla, F.; et al. Deregulated expression of miR-106a predicts survival in humancolon cancer patients. Genes Chromosomes Cancer 2008, 47, 794–802. [Google Scholar] [CrossRef]
- Mokutani, Y.; Uemura, M.; Munakata, K.; Okuzaki, D.; Haraguchi, N.; Takahashi, H.; Nishimura, J.; Hata, T.; Murata, K.; Takemasa, I.; et al. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann. Surg. Oncol. 2016, 23, 599–608. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, P.; Wang, F.; Zhang, H.; Yang, J.; Peng, J.; Liu, W.; Qin, H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012, 61, 1447–1453. [Google Scholar] [CrossRef]
- Nishimura, J.; Handa, R.; Yamamoto, H.; Tanaka, F.; Shibata, K.; Mimori, K.; Takemasa, I.; Mizushima, T.; Ikeda, M.; Sekimoto, M.; et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol. Rep. 2012, 28, 2221–2226. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Winter, E.; Ress, A.L.; Bauernhofer, T.; Gerger, A.; Kiesslich, T.; Lax, S.; Samonigg, H.; Hoefler, G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J. Clin. Pathol. 2014, 67, 198–203. [Google Scholar] [CrossRef]
- Pichler, M.; Stiegelbauer, V.; Vychytilova-Faltejskova, P.; Ivan, C.; Ling, H.; Winter, E.; Zhang, X.; Goblirsch, M.; Wulf-Goldenberg, A.; Ohtsuka, M.; et al. Genome-Wide miRNA Analysis Identifies miR-188-3p as a Novel Prognostic Marker and Molecular Factor Involved in Colorectal Carcinogenesis. Clin. Cancer Res. 2017, 23, 1323–1333. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Ma, H.; Zhang, J.; Zhou, X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med. Oncol. 2012, 29, 919–927. [Google Scholar] [CrossRef]
- Shen, Z.L.; Wang, B.; Jiang, K.W.; Ye, C.X.; Cheng, C.; Yan, Y.C.; Zhang, J.Z.; Yang, Y.; Gao, Z.D.; Ye, Y.J.; et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget 2016, 7, 35092–35105. [Google Scholar] [CrossRef]
- Karaayvaz, M.; Pal, T.; Song, B.; Zhang, C.; Georgakopoulos, P.; Mehmood, S.; Burke, S.; Shroyer, K.; Ju, J. Prognostic significance of miR-215 in colon cancer. Clin. Colorectal. Cancer 2011, 10, 340–347. [Google Scholar] [CrossRef]
- Li, P.L.; Zhang, X.; Wang, L.L.; Du, L.T.; Yang, Y.M.; Li, J.; Wang, C.X. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis 2015, 36, 1484–1493. [Google Scholar] [PubMed]
- Menéndez, P.; Padilla, D.; Villarejo, P.; Palomino, T.; Nieto, P.; Menéndez, J.M.; Rodríguez-Montes, J.A. miRNAs implicated in CRC Prognostic implications of serum microRNA-21 in colorectal cancer. J. Surg. Oncol. 2013, 108, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Kou, C.H.; Zhou, T.; Han, X.L.; Zhuang, H.J.; Qian, H.X. Downregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancer. Oncol. Lett. 2016, 12, 4838–4844. [Google Scholar] [CrossRef]
- Miyoshi, J.; Toden, S.; Yoshida, K.; Toiyama, Y.; Alberts, S.R.; Kusunoki, M.; Sinicrope, F.A.; Goel, A. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci. Rep. 2017, 7, 43393. [Google Scholar] [CrossRef]
- Lv, Z.C.; Fan, Y.S.; Chen, H.B.; Zhao, D.W. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol. 2015, 36, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Li, K.; Zhu, K.; Yan, R.; Dang, C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol. Ther. 2015, 16, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–665. [Google Scholar] [CrossRef]
- Yu, H.; Gao, G.; Jiang, L.; Guo, L.; Lin, M.; Jiao, X.; Jia, W.; Huang, J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2013, 6, 2904–2911. [Google Scholar]
- Pu, X.X.; Huang, G.L.; Guo, H.Q.; Guo, C.C.; Li, H.; Ye, S.; Ling, S.; Jiang, L.; Tian, Y.; Lin, T.Y. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 2010, 25, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Schetter, A.J.; Okugawa, Y.; Harris, C.C.; Boland, C.R.; Goel, A. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J. Natl. Cancer Inst. 2015, 107, dju492. [Google Scholar] [CrossRef]
- Maierthaler, M.; Benner, A.; Hoffmeister, M.; Surowy, H.; Jansen, L.; Knebel, P.; Chang-Claude, J.; Brenner, H.; Burwinkel, B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int. J. Cancer 2017, 140, 176–187. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef]
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158. [Google Scholar] [CrossRef]
- Rotelli, M.T.; Di Lena, M.; Cavallini, A.; Lippolis, C.; Bonfrate, L.; Chetta, N.; Portincasa, P.; Altomare, D.F. Fecal microRNA profile in patients with colorectal carcinoma before and after curative surgery. Int. J. Colorectal. Dis. 2015, 30, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Chang, P.Y.; Chang, Y.S.; You, J.F.; Chan, E.C.; Chen, J.S.; Tsai, W.S.; Huang, Y.L.; Fan, C.W.; Hsu, H.C.; et al. MicroRNA-Based Signature for Diagnosis and Prognosis of Colorectal Cancer using Residuum of Fecal Immunochemical Test. Biomed. J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, A.; Graziano, F.; Vincenzi, B.; Canestrari, E.; Perrone, G.; Galluccio, N.; Catalano, V.; Loupakis, F.; Rabitti, C.; Santini, D.; et al. High let-7a microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapyrefractory metastatic disease. Oncologist 2012, 17, 823–829. [Google Scholar] [CrossRef]
- Suto, T.; Yokobori, T.; Yajima, R.; Morita, H.; Fujii, T.; Yamaguchi, S.; Altan, B.; Tsutsumi, S.; Asao, T.; Kuwano, H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2015, 36, 338–345. [Google Scholar] [CrossRef]
- Mosakhani, N.; Lahti, L.; Borze, I.; Karjalainen-Lindsberg, M.L.; Sundström, J.; Ristamäki, R.; Osterlund, P.; Knuutila, S.; Sarhadi, V.K. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer Genet. 2012, 205, 545–551. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Sacconi, A.; Landi, L.; Ludovini, V.; Biagioni, F.; D’ Incecco, A.; Capodanno, A.; Salvini, J.; Corgna, E.; Cupini, S.; et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin. Colorectal. Cancer 2014, 13, 37–45.e4. [Google Scholar] [CrossRef]
- Hansen, T.F.; Carlsen, A.L.; Heegaard, N.H.; Sørensen, F.B.; Jakobsen, A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br. J. Cancer 2015, 112, 624–629. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Zhang, Y.; Chen, Y.; Hu, T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med. Oncol. 2014, 31, 799. [Google Scholar] [CrossRef]
- Schou, J.V.; Rossi, S.; Jensen, B.V.; Nielsen, D.L.; Pfeiffer, P.; Høgdall, E.; Yilmaz, M.; Tejpar, S.; Delorenzi, M.; Kruhøffer, M.; et al. miR-345 in metastatic colorectal cancer: A non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS ONE 2014, 9, e99886. [Google Scholar] [CrossRef]
- Gomes, S.E.; Simões, A.E.; Pereira, D.M.; Castro, R.E.; Rodrigues, C.M.; Borralho, P.M. miR-143 or miR-145 overexpression increases cetuximab-mediated antibody-dependent cellular cytotoxicity in human colon cancer cells. Oncotarget 2016, 7, 9368–9387. [Google Scholar] [CrossRef] [PubMed]
- Kjersem, J.B.; Ikdahl, T.; Lingjaerde, O.C.; Guren, T.; Tveit, K.M.; Kure, E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014, 8, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Sun, W.; Liu, R.; Zhou, Z.; Zhang, H.; Chen, X.; Ba, Y. Plasma Exosomal miRNA Expression Profile as Oxaliplatin-Based Chemoresistant Biomarkers in Colorectal Adenocarcinoma. Front. Oncol. 2020, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef]
- Yagi, T.; Iinuma, H.; Hayama, T.; Matsuda, K.; Nozawa, K.; Tsukamoto, M.; Shimada, R.; Akahane, T.; Tsuchiya, T.; Ozawa, T.; et al. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol. Clin. Oncol. 2019, 11, 416–424. [Google Scholar] [CrossRef]
- Atkinson, S.R.; Samuel, M.; Jürg, B. Exploring long non-coding RNAs through sequencing. Semin. Cell Dev. Biol. 2012, 23, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Du, Y.; Liu, X.; Cho, W.C. Involvement of Non-coding RNAs in the Signaling Pathways of Colorectal Cancer. Adv. Exp. Med. Biol. 2016, 937, 19–51. [Google Scholar]
- Wu, Y.; Yang, X.; Tian, L.; Jiang, G.; Chen, F.; Li, J.; An, P.; Lu, L.; Luo, N.; Du, J.; et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer 2019, 18, 87. [Google Scholar] [CrossRef]
- Xu, M.; Xu, X.; Pan, B.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Xu, T.; Sun, L.; Qin, J.; et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol. Cancer 2019, 18, 135. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Tang, Y.; Yang, M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342-3p/ANXA2 axis. J. Transl. Med. Vol. 2021, 19, 8. [Google Scholar] [CrossRef]
- Xu, J.; Xiao, Y.; Liu, B.; Pan, S.; Liu, Q.; Shan, Y.; Li, S.; Qi, Y.; Huang, Y.; Jia, L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020, 39, 54. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Taulli, R.; Loretelli, C.; Pandolfi, P.P. From pseudo-ceRNAs to circ-ceRNAs: A tale of cross-talk and competition. Nat. Struct. Mol. Biol. 2013, 20, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Calin, G.A. Circular RNAs in cancer—Lessons learned from microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Kopetz, S.; Ajani, J.A.; Calin, G.A. Non-coding RNAs in GI cancers: From cancer hallmarks to clinical utility. Gut 2020, 69, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE 2016, 11, e0148407. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef]
- Zhang, M.; Xin, Y. Circular RNAs: A new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 2018, 11, 21. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 10. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, Z.; Wang, S.; Zhu, W.; Zheng, P.; Wang, W. Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol. Chem. 2020, 401, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, S.; Fu, Q. Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J. Cell. Biochem. 2020, 121, 3286–3297. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Li, P.; Gong, P.Y. Hsa_circ_0005075 promotes the proliferation and invasion of colorectal cancer cells. Int. J. Biol. Markers 2019, 34, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.D.; Ren, Y.R.; Gao, Y.X.; Zhang, L.; Ding, Z. Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3311–3319. [Google Scholar]
- Hsiao, K.Y.; Lin, Y.C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef]
- Ju, H.-Q.; Zhao, Q.; Wang, F.; Lan, P.; Wang, Z.; Zuo, Z.X.; Wu, Q.N.; Fan, X.J.; Mo, H.Y.; Chen, L.; et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol. Med. 2019, 11, e10168. [Google Scholar] [CrossRef]
- Cao, J.Z.; Ma, L.M.; Zhang, Y.L.; Guo, H.C.; Niu, X.; Zhao, T.B. Circ-0104631 promotes cell proliferation and invasion in colorectal cancer and predicts poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4730–4737. [Google Scholar]
- Zhou, C.; Liu, H.S.; Wang, F.W.; Hu, T.; Liang, Z.X.; Lan, N.; He, X.W.; Zheng, X.B.; Wu, X.J.; Xie, D.; et al. circCAMSAP1 Promotes Tumor Growth in Colorectal Cancer via the miR-328-5p/E2F1 Axis. Mol. Ther. 2020, 28, 914–928. [Google Scholar] [CrossRef]
- Huang, X.; Shen, X.; Peng, L.; Mai, W.; Wan, Y.; Zhang, H. CircCSNK1G1 Contributes to the Development of Colorectal Cancer by Increasing the Expression of MYO6 via Competitively Targeting miR-455-3p. Cancer Manag. Res. 2020, 12, 9563–9957. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.S.; Tong, H.Z.; Yuan, X.H.; Xiong, C.H.; Xu, X.Y.; Zeng, Y.F. CircFADS2: A potential prognostic biomarker of colorectal cancer. Exp. Biol. Med. 2020, 245, 1233–1241. [Google Scholar] [CrossRef]
- Lu, H.; Yao, B.; Wen, X.; Jia, B. FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer 2019, 19, 918. [Google Scholar] [CrossRef]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Li, X.N.; Ye, C.X.; Chen, Z.L.; Wang, Z.J. Circular RNA circHUWE1 Is Upregulated and Promotes Cell Proliferation, Migration and Invasion in Colorectal Cancer by Sponging miR-486. Onco Targets. Ther. 2020, 13, 423–434. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Zhang, C.; Lin, C.; Zhang, J.; Zhang, W.; Zhang, W.; Lu, Y.; Zheng, L.; Li, X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018, 246, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, F.W.; Cao, C.H.; Ling, H.; Chen, J.W.; Chen, R.X.; Feng, Z.H.; Luo, J.; Jin, X.H.; Duan, J.L.; et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol. Cancer 2020, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Sun, G.; He, Q.; Wang, C.; Shi, J.; Gao, L.; Ye, J.; Liang, Y.; Qu, H. Circular noncoding RNA circMBOAT2 is a novel tumor marker and regulates proliferation/migration by sponging miR-519d-3p in colorectal cancer. Cell Death Dis. 2020, 11, 625. [Google Scholar] [CrossRef]
- Chen, L.Y.; Zhi, Z.; Wang, L.; Zhao, Y.Y.; Deng, M.; Liu, Y.H.; Qin, Y.; Tian, M.M.; Liu, Y.; Shen, T.; et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signaling. J. Pathol. 2019, 248, 103–115. [Google Scholar] [CrossRef]
- Chen, R.X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; Wang, F.; et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chec, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef]
- Wang, Z.; Su, M.; Xiang, B.; Zhao, K.; Qin, B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem. Biophys. Res. Commun. 2019, 512, 716–722. [Google Scholar] [CrossRef]
- Fang, G.; Ye, B.-L.; Hu, B.-R.; Ruan, X.-J.; Shi, Y.-X. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018, 504, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pei, F.; Cao, M. CircRNA_101951 promotes migration and invasion of colorectal cancer cells by regulating the KIF3A-mediated EMT pathway. Exp. Ther. Med. 2020, 19, 3355–3361. [Google Scholar] [CrossRef] [PubMed]
- Li, X.N.; Wang, Z.J.; Ye, C.X.; Zhao, B.C.; Huang, X.X.; Yang, L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed. Pharmacother. 2019, 112, 108611. [Google Scholar] [CrossRef]
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Zhao, P.; Zhou, H.; Mao, T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J. Cell. Biochem. 2019, 120, 3027–3037. [Google Scholar] [CrossRef]
- Li, J.; Ni, S.; Zhou, C.; Ye, M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag. Res. 2018, 10, 2777–2784. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Lu, L.; He, L.; Hu, H.; Xu, Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J. Clin. Lab. Anal. 2018, 32, 5. [Google Scholar] [CrossRef]
- Xing, L.; Xia, M.; Jiao, X.; Fan, L. Hsa_circ_0004831 serves as a blood-based prognostic biomarker for colorectal cancer and its potentially circRNA-miRNA-mRNA regulatory network construction. Cancer Cell Int. 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Yang, N.; Xu, B.; Kong, P.; Han, M.; Li, B.H. Hsa_circ_0002320: A novel clinical biomarker for colorectal cancer prognosis. Medicine (Baltimore) 2020, 99, e21224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, S.; Liu, Y.; Wang, Y.; Lin, T.; Li, Y.; Zhang, R. Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. Int. J. Clin. Exp. Pathol. 2018, 11, 1399. [Google Scholar]
- Wang, F.; Wang, J.; Cao, X.; Xu, L.; Chen, L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed. Pharmacother. 2018, 98, 775–782. [Google Scholar] [CrossRef]
- Liang, Y.; Shi, J.; He, Q.; Sun, G.; Gao, L.; Ye, J.; Tang, X.; Qu, H. Hsa_circ_0026416 promotes proliferation and migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell Int. 2020, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yu, Y.; Liao, F.; Tan, S. Homo Sapiens Circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 Gene Acts as an Oncogene in Colorectal Cancer Through the microRNA-203a-3p.1 and CREB1 Axis. Med. Sci. Monit. 2019, 25, 6872. [Google Scholar] [CrossRef]
- Li, Y.; Zang, H.; Zhang, X.; Huang, G. circ_0136666 Facilitates the Progression of Colorectal Cancer via miR-383/CREB1 Axis. Cancer Manag. Res. 2020, 12, 6795–6806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Wu, K.; Zhan, F.; Zeng, H. Dysregulated circRNA_100876 contributes to proliferation and metastasis of colorectal cancer by targeting microRNA-516b (miR-516b). Cancer Biol. Ther. 2020, 21, 733–740. [Google Scholar] [CrossRef]
- Chen, Z.; Ren, R.; Wan, D.; Wang, Y.; Xue, X.; Jiang, M.; Shen, J.; Han, Y.; Liu, F.; Shi, J.; et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene 2019, 38, 6017–6034. [Google Scholar] [CrossRef]
- Wu, M.; Kong, C.; Cai, M.; Huang, W.; Chen, Y.; Wang, B.; Liu, X. Hsa_circRNA_002144 promotes growth and metastasis of colorectal cancer through regulating miR-615-5p/LARP1/mTOR pathway. Carcinogenesis 2021, 42, 601–610. [Google Scholar] [CrossRef]
- Li, C.; Zhou, H. Circular RNA hsa_circRNA_102209 promotes the growth and metastasis of colorectal cancer through miR-761-mediated Ras and Rab interactor 1 signaling. Cancer Med. 2020, 9, 6710–6725. [Google Scholar] [CrossRef]
- Hu, T.; Li, Z.; Gao, C.Y.; Cho, C.H. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J. Gastroenterol. 2016, 22, 6876–6889. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Liu, L.; Yang, T.; Song, J. Circular RNAs: New biomarkers of chemoresistance in cancer. Cancer Biol. Med. 2021, 18, 421. [Google Scholar] [CrossRef]
- Hon, K.W.; Ab-Mutalib, N.S.; Abdullah, N.M.A.; Jamal, R.; Abu, N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci. Rep. 2019, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Abu, N.; Hon, K.W.; Jeyaraman, S.; Yahaya, A.; Abdullah, N.M.; Mustangin, M.; Sulaiman, S.A.; Jamal, R.; Ab-Mutalib, N.S. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics 2019, 11, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Pei, L.; Xie, P.; Guo, G. Circ-PRKDC Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells by Regulating miR-375/FOXM1 Axis and Wnt/β-Catenin Pathway. Onco. Targets. Ther. 2020, 13, 5939–5953. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Ai, Y.Q.; Li, Y.F.; Ye, Q.; Chen, Z.T.; Qin, J.Y.; Liu, Q.Y.; Wang, H.; Ju, Y.H.; Li, W.H.; et al. Microarray Analysis of Circular RNA Expression Profile Associated with 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. Biomed Res. Int. 2017, 2017, 8421614. [Google Scholar] [CrossRef]
- Ren, T.J.; Liu, C.; Hou, J.F.; Shan, F.X. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1743–1754. [Google Scholar] [PubMed]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Liu, G.; Li, R.; Bai, H.; Zhao, J.; Xiao, P.; Mei, J. Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-α pathway in human colon cancer. J. Cell. Mol. Med. 2020, 24, 5021–5027. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Yu, Y.S.; Lin, H.H.; Hsiao, K.Y. Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers 2020, 12, 697. [Google Scholar] [CrossRef]
- Jian, X.; He, H.; Zhu, J.; Zhang, Q.; Zheng, Z.; Liang, X.; Chen, L.; Yang, M.; Peng, K.; Zhang, Z.; et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer 2020, 19, 20. [Google Scholar] [CrossRef]
- Mori, G.; Pasca, M. Gut Microbial Signatures in Sporadic and Hereditary Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 1312. [Google Scholar] [CrossRef]
- Shanahan, F.; Ghosh, T.; O’Toole, P. The Healthy Microbiome—What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Kalliala, I.; Markozannes, G.; Gunter, M.J.; Paraskevaidis, E.; Gabra, H.; Martin-Hirsch, P.; Tsilidis, K.K. Adiposity and cancer at major anatomical sites: Umbrella review of the literature. BMJ 2017, 356, j477. [Google Scholar] [CrossRef]
- Scott, A.; Alexander, J.; Merrifield, C.; Cunningham, D.; Jobin, C.; Brown, R.; Alverdy, J.; O’Keefe, S.J.; Gaskins, H.R.; Teare, J.; et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019, 68, 1624–1632. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.; Vleugels, J.; Kasi, P.; Wallace, M. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Kashyap, S.; Pal, S.; Chandan, G.; Saini, V.; Chakrabarti, S.; Saini, N.K.; Mittal, A.; Thakur, V.K.; Saini, A.K.; Saini, R.V. Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Semin. Cancer Biol. 2021, 70, 112–125. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lepage, P.; Leclerc, M.; Joossens, M.; Mondot, S.; Blottière, H.M.; Raes, J.; Ehrlich, D.; Doré, J. A metagenomic insight into our gut’s microbiome. Gut 2012, 62, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Jalandra, R.; Bayal, N.; Yadav, A.K.; Harshulika Sharma, M.; Makharia, G.K.; Kumar, P.; Singh, R.; Solanki, P.R.; Kumar, A. Gut microbiota-derived metabolites in CRC progression and causation. J. Cancer Res. Clin. Oncol. 2021, 147, 3141–3155. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, L. When human cells meet bacteria: Precision medicine for cancers using the microbiota. Am. J. Cancer Res. 2018, 8, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6, 116. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Vipperla, K.; O’Keefe, S. Diet, microbiota, and dysbiosis: A ‘recipe’ for colorectal cancer. Food Funct. 2016, 7, 1731–1740. [Google Scholar] [CrossRef]
- Gil-Cardoso, K.; Ginés, I.; Pinent, M.; Ardévol, A.; Blay, M.; Terra, X. Effects of flavonoids on intestinal inflammation, barrier integrity and changes in gut microbiota during diet-induced obesity. Nutr. Res. Rev. 2016, 29, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Solé, C.; Guilly, S.; Da Silva, K.; Llopis, M.; Le-Chatelier, E.; Huelin, P.; Carol, M.; Moreira, R.; Fabrellas, N.; De Prada, G.; et al. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship with Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology 2021, 160, 206–218.e13. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Onoue, M.; Kado, S.; Sakaitani, Y.; Uchida, K.; Morotomi, M. Specific species of intestinal bacteria influence the induction of aberrant crypt foci by 1,2-dimethylhydrazine in rats. Cancer Lett. 1997, 113, 179–186. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Abdulamir, A.; Hafidh, R.; Bakar, F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 2011, 30, 11. [Google Scholar] [CrossRef] [PubMed]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Chew, S.; Lubowski, D. Clostridium septicum and malignancy. ANZ J. Surg. 2001, 71, 647–649. [Google Scholar] [CrossRef]
- Shmuely, H.; Passaro, D.; Figer, A.; Niv, Y.; Pitlik, S.; Samra, Z.; Koren, R.; Yahav, J. Relationship between Helicobacter pylori CagA status and colorectal cancer. Am. J. Gastroenterol. 2001, 96, 3406–3410. [Google Scholar] [CrossRef] [PubMed]
- Kasper, S.; Morell-Perez, C.; Wyche, T.; Sana, T.; Lieberman, L.; Hett, E. Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev. Res. (Phila) 2017, 10, 398–409. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Martin, H.; Campbell, B.; Hart, C.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef]
- Lucas, C.; Barnich, N.; Nguyen, H.T.T. Microbiota, Inflammation and Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 1310. [Google Scholar] [CrossRef]
- Nougayrède, J.P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Dziubańska-Kusibab, P.; Berger, H.; Battistini, F.; Bouwman, B.; Iftekhar, A.; Katainen, R.; Cajuso, T.; Crosetto, N.; Orozco, M.; Aaltonen, L.A.; et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef]
- Hartwich, A.; Konturek, S.J.; Pierzchalski, P.; Zuchowicz, M.; Labza, H.; Konturek, P.C.; Karczewska, E.; Bielanski, W.; Marlicz, K.; Starzynska, T.; et al. Helicobacter pylori infection, gastrin, cyclooxygenase-2, and apoptosis in colorectal cancer. Int. J. Colorectal. Dis. 2001, 16, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Tatishchev, S.F.; Vanbeek, C.; Wang, H.L. Helicobacter pylori infection and colorectal carcinoma: Is there a causal association? J. Gastrointest. Oncol. 2012, 3, 380–385. [Google Scholar]
- Wessler, S.; Krisch, L.; Elmer, D.; Aberger, F. From inflammation to gastric cancer—The importance of Hedgehog/GLI signaling in Helicobacter pylori-induced chronic inflammatory and neoplastic diseases. Cell Commun. Signal. 2017, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Zamani, S.; Taslimi, R.; Sarabi, A.; Jasemi, S.; Sechi, L.A.; Feizabadi, M.M. Enterotoxigenic Bacteroides fragilis: A Possible Etiological Candidate for Bacterially-Induced Colorectal Precancerous and Cancerous Lesions. Front Cell Infect Microbiol. 2020, 9, 449. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Zhang, J.; Wu, Q.; Chen, J.; Liu, J.; Wang, L.; Chen, C.; Xu, J.; Zhang, H.; Shi, C.; et al. The role of microbiota in the development of colorectal cancer. Int. J. Cancer 2019, 145, 2032–2041. [Google Scholar] [CrossRef]
- Biarc, J.; Nguyen, I.S.; Pini, A.; Gossé, F.; Richert, S.; Thiersé, D.; Van Dorsselaer, A.; Leize-Wagner, E.; Raul, F.; Klein, J.P.; et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis 2004, 25, 1477–1484. [Google Scholar] [CrossRef]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008, 23 Pt 1, 1298–1303. [Google Scholar] [CrossRef]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Kennedy, C.L.; Krejany, E.O.; Young, L.F.; O’Connor, J.R.; Awad, M.M.; Boyd, R.L.; Emmins, J.J.; Lyras, D.; Rood, J.I. The alpha-toxin of Clostridium septicum is essential for virulence. Mol. Microbiol. 2005, 57, 1357–1366. [Google Scholar] [CrossRef]
- Chakravorty, A.; Awad, M.M.; Cheung, J.K.; Hiscox, T.J.; Lyras, D.; Rood, J.I. The pore-forming α-toxin from clostridium septicum activates the MAPK pathway in a Ras-c-Raf-dependent and independent manner. Toxins 2015, 7, 516–534. [Google Scholar] [CrossRef] [PubMed]
- Corredoira, J.; Grau, I.; Garcia-Rodriguez, J.F.; García-País, M.J.; Rabuñal, R.; Ardanuy, C.; García-Garrote, F.; Coira, A.; Alonso, M.P.; Boleij, A.; et al. Colorectal neoplasm in cases of Clostridium septicum and Streptococcus gallolyticus subsp. gallolyticus bacteraemia. Eur. J. Intern. Med. 2017, 41, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Kwong, T.; Chow, T.; Luk, A.; Dai, R.; Nakatsu, G.; Lam, T.; Zhang, L.; Wu, J.; Chan, F.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2016, 66, 1441–1448. [Google Scholar] [CrossRef]
- Xie, Y.; Gao, Q.; Cai, G.; Sun, X.M.; Sun, X.M.; Zou, T.H.; Chen, H.M.; Yu, S.Y.; Qiu, Y.W.; Gu, W.Q.; et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine 2017, 25, 32–40. [Google Scholar] [CrossRef]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.; et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Wei, Z.; Cao, S.; Liu, S.; Yao, Z.; Sun, T.; Li, Y.; Li, J.; Zhang, D.; Zhou, Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget 2016, 7, 46158–46172. [Google Scholar] [CrossRef]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect Dis. 2014, 33, 1381–1390. [Google Scholar] [CrossRef]
- Russo, E.; Bacci, G.; Chiellini, C.; Fagorzi, C.; Niccolai, E.; Taddei, A.; Ricci, F.; Ringressi, M.N.; Borrelli, R.; Melli, F.; et al. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Front. Microbiol. 2018, 8, 2699. [Google Scholar] [CrossRef]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Butt, J.; Blot, W.J.; Teras, L.R.; Visvanathan, K.; Le Marchand, L.; Haiman, C.A.; Chen, Y.; Bao, Y.; Sesso, H.D.; Wassertheil-Smoller, S.; et al. Antibody Responses to Streptococcus Gallolyticus Subspecies Gallolyticus Proteins in a Large Prospective Colorectal Cancer Cohort Consortium. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1186–1194. [Google Scholar] [CrossRef]
- Wang, Y.; He, X.; Nie, H.; Zhou, J.; Cao, P.; Ou, C. Application of artificial intelligence to the diagnosis and therapy of colorectal cancer. Am. J. Cancer Res. 2020, 10, 3575–3598. [Google Scholar] [PubMed]
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019, 13, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.D.; Tsoulfas, G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World. J. Gastroenterol. 2021, 27, 6191–6223. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, M.; Zhu, H.; Xu, J. A 15-gene signature for prediction of colon cancer recurrence and prognosis based on SVM. Gene 2017, 604, 33–40. [Google Scholar] [CrossRef]
- Kel, A.; Boyarskikh, U.; Stegmaier, P.; Leskov, L.S.; Sokolov, A.V.; Yevshin, I.; Mandrik, N.; Stelmashenko, D.; Koschmann, J.; Kel-Margoulis, O.; et al. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinform. 2019, 20 (Suppl. S4), 119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Y.; Wang, Y.; Fan, Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules 2019, 24, 2238. [Google Scholar] [CrossRef]
- Tutar, Y. miRNA and cancer; computational and experimental approaches. Curr. Pharm. Biotechnol. 2014, 15, 429. [Google Scholar] [CrossRef]
- Chang, K.H.; Miller, N.; Kheirelseid, E.A.; Lemetre, C.; Ball, G.R.; Smith, M.J.; Regan, M.; McAnena, O.J.; Kerin, M.J. MicroRNA signature analysis in colorectal cancer: Identification of expression profiles in stage II tumors associated with aggressive disease. Int. J. Colorectal. Dis. 2011, 26, 1415–1422. [Google Scholar] [CrossRef]
- Amirkhah, R.; Farazmand, A.; Gupta, S.K.; Ahmadi, H.; Wolkenhauer, O.; Schmitz, U. Naive Bayes classifier predicts functional microRNA target interactions in colorectal cancer. Mol. Biosyst. 2015, 11, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- van de Wiel, M.A.; Neerincx, M.; Buffart, T.E.; Sie, D.; Verheul, H.M. ShrinkBayes: A versatile R-package for analysis of count-based sequencing data in complex study designs. BMC Bioinform. 2014, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Xuan, P.; Dong, Y.; Guo, Y.; Zhang, T.; Liu, Y. Dual convolutional neural network based method for predicting disease-related miRNAs. Int. J. Mol. Sci. 2018, 19, 3732. [Google Scholar] [CrossRef] [PubMed]
- Afshar, S.; Afshar, S.; Warden, E.; Manochehri, H.; Saidijam, M. Application of artificial neural network in miRNA biomarker selection and precise diagnosis of colorectal cancer. Iran Biomed. J. 2019, 23, 175–183. [Google Scholar] [CrossRef]
- Herreros-Villanueva, M.; Duran-Sanchon, S.; Martin, A.C.; Perez-Palacios, R.; Vila-Navarro, E.; Marcuello, M.; Diaz-Centeno, M.; Cubiella, J.; Diez, M.S.; Bujanda, L.; et al. Plasma MicroRNA signature validation for early detection of colorectal cancer. Clin. Transl. Gastroenterol. 2019, 10, e00003. [Google Scholar] [CrossRef]
- Hildebrand, L.A.; Pierce, C.J.; Dennis, M.; Paracha, M.; Maoz, A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers 2021, 13, 391. [Google Scholar] [CrossRef]
- Cao, R.; Yang, F.; Ma, S.C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.H.; Wang, T.; Lu, W.J.; Cai, W.J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080–11091. [Google Scholar] [CrossRef]
- Williams, C.J.M.; Seligmann, J.F.; Elliott, F.; Shires, M.; Richman, S.D.; Brown, S.; Zhang, L.; Singh, S.; Pugh, J.; Xu, X.M.; et al. Artificial Intelligence-Assisted Amphiregulin and Epiregulin IHC Predicts Panitumumab Benefit in RAS Wild-Type Metastatic Colorectal Cancer. Clin. Cancer Res. 2021, 27, 3422–3431. [Google Scholar] [CrossRef]
- Zhao, K.; Li, X.; Yao, S.; Wang, Y.; Wu, X.; Xu, Z.; Wu, L.; Huang, L.; Liang, C.; Liu, Z. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. EBioMedicine 2020, 61, 103054. [Google Scholar] [CrossRef]
- Banegas-Luna, A.J.; Pena-Garcia, J.; Iftene, A.; Guadagni, F.; Ferroni, P.; Scarpato, N.; Zanzotto, F.M.; Bueno-Crespo, A.; Perez-Sanchez, H. Towards the Interpretability of Machine Learning Predictions for Medical Applications Targeting Personalised Therapies: A Cancer Case Survey. Int. J. Mol. Sci. 2021, 22, 4394. [Google Scholar] [CrossRef]
- Schmidt, C.M.D. Anderson Breaks with IBM Watson, Raising Questions about Artificial Intelligence in Oncology. J. Natl. Cancer Inst. 2017, 109, 5. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Yang, X.; Zhu, C.; Xu, J.; Yu, Y.; Shen, X.; Huang, Y. Concordance Between Watson for Oncology and Multidisciplinary Teams in Colorectal Cancer: Prognostic Implications and Predicting Concordance. Front. Oncol. 2020, 10, 595565. [Google Scholar] [CrossRef] [PubMed]
- Miyano, S. Artificial Intelligence for Cancer Genomic Medicine: Understanding Cancer is Beyond Human Ability. Brain Nerve. 2019, 71, 25–32. [Google Scholar] [PubMed]
- Keshava, N.; Toh, T.S.; Yuan, H.; Yang, B.; Menden, M.P.; Wang, D. Defining subpopulations of differential drug response to reveal novel target populations. NPJ Syst. Biol. Appl. 2019, 5, 36. [Google Scholar] [CrossRef]
- Ding, D.; Han, S.; Zhang, H.; He, Y.; Li, Y. Predictive biomarkers of colorectal cancer. Comput. Biol. Chem. 2019, 83, 107106. [Google Scholar] [CrossRef]
- Lee, J.; Kumar, S.; Lee, S.Y.; Park, S.J.; Kim, M.H. Development of Predictive Models for Identifying Potential S100A9 Inhibitors Based on Machine Learning Methods. Front. Chem. 2019, 7, 779. [Google Scholar] [CrossRef]
- Pacheco, M.P.; Bintener, T.; Ternes, D.; Kulms, D.; Haan, S.; Letellier, E.; Sauter, T. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine 2019, 43, 98–106. [Google Scholar] [CrossRef]
- Akutekwe, A.; Seker, H.; Yang, S. In silico discovery of significant pathways in colorectal cancer metastasis using a two-stage optimisation approach. IET Syst. Biol. 2015, 9, 294–302. [Google Scholar] [CrossRef]
- Saghapour, E.; Sehhati, M. Prediction of metastasis in advanced colorectal carcinomas using CGH data. J. Theor. Biol. 2017, 429, 116–123. [Google Scholar] [CrossRef]
- Zhi, J.; Sun, J.; Wang, Z.; Ding, W. Support vector machine classifier for prediction of the metastasis of colorectal cancer. Int. J. Mol. Med. 2018, 41, 1419–1426. [Google Scholar] [CrossRef]
- Mitchell, T. Machine Learning; McGRAW Hill, Inc.: New York, NY, USA, 1997; Chapter 6. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Tsiatis, A.A. A note on a goodness-of-fit test for the logistic regression model. Biometrika 1980, 67, 250–251. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory (COLT ‘92), Association for Computing Machinery, New York, NY, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Cușmuliuc, C.G.; Coca, L.G.; Iftene, A. Identifying Fake News on Twitter using Naive Bayes, SVM and Random Forest Distributed Algorithms. In Proceedings of the 13th Edition of the International Conference on Linguistic Resources and Tools for Processing Romanian Language (ConsILR-2018), Jassy, Romania, 22–23 November 2018; pp. 177–188. [Google Scholar]
- Șerban, C.; Sirițeanu, A.; Gheorghiu, C.; Iftene, A.; Alboaie, L.; Breabăn, M. Combining Image Retrieval, Metadata Processing and Naive Bayes Classification at Plant Identification. CLEF 2013, 11, 3–15. [Google Scholar]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Schmittgen, T.D. Regulation of microRNA processing in development, differentiation and cancer. J. Cell Mol. Med. 2008, 12, 1811–1819. [Google Scholar] [CrossRef]
- Blower, P.E.; Chung, J.H.; Verducci, J.S.; Lin, S.; Park, J.K.; Dai, Z.; Liu, C.G.; Schmittgen, T.D.; Reinhold, W.C.; Croce, C.M.; et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol. Cancer Ther. 2008, 7, 1–9. [Google Scholar] [CrossRef]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef]
- Kallenbach-Thieltges, A.; Großerueschkamp, F.; Jütte, H.; Kuepper, C.; Schick, A.R.; Tannapfel, A.; Gerwert, K. Label-free, automated classification of microsatellite status in colorectal cancer by infrared imaging. Sci. Rep. 2020, 10, 10161. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.; Whalley, C.M.; et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2021, 70, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Lee, H.; Kim, D.; Han, S.K.; Ha, D.; Shin, K.; Kim, S. Network-based machine learning in colorectal and bladder organo organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 2020, 11, 5485. [Google Scholar] [CrossRef] [PubMed]
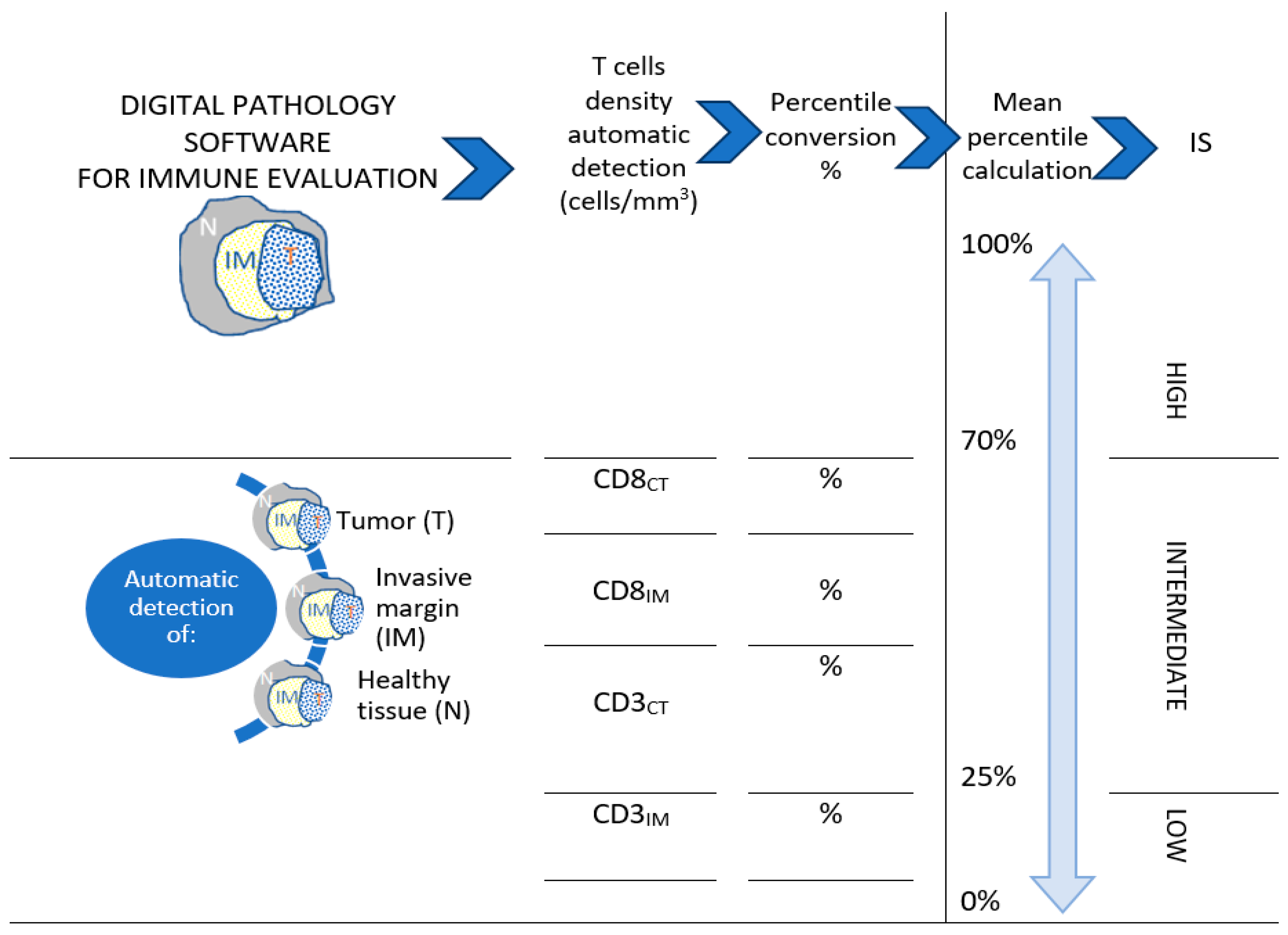
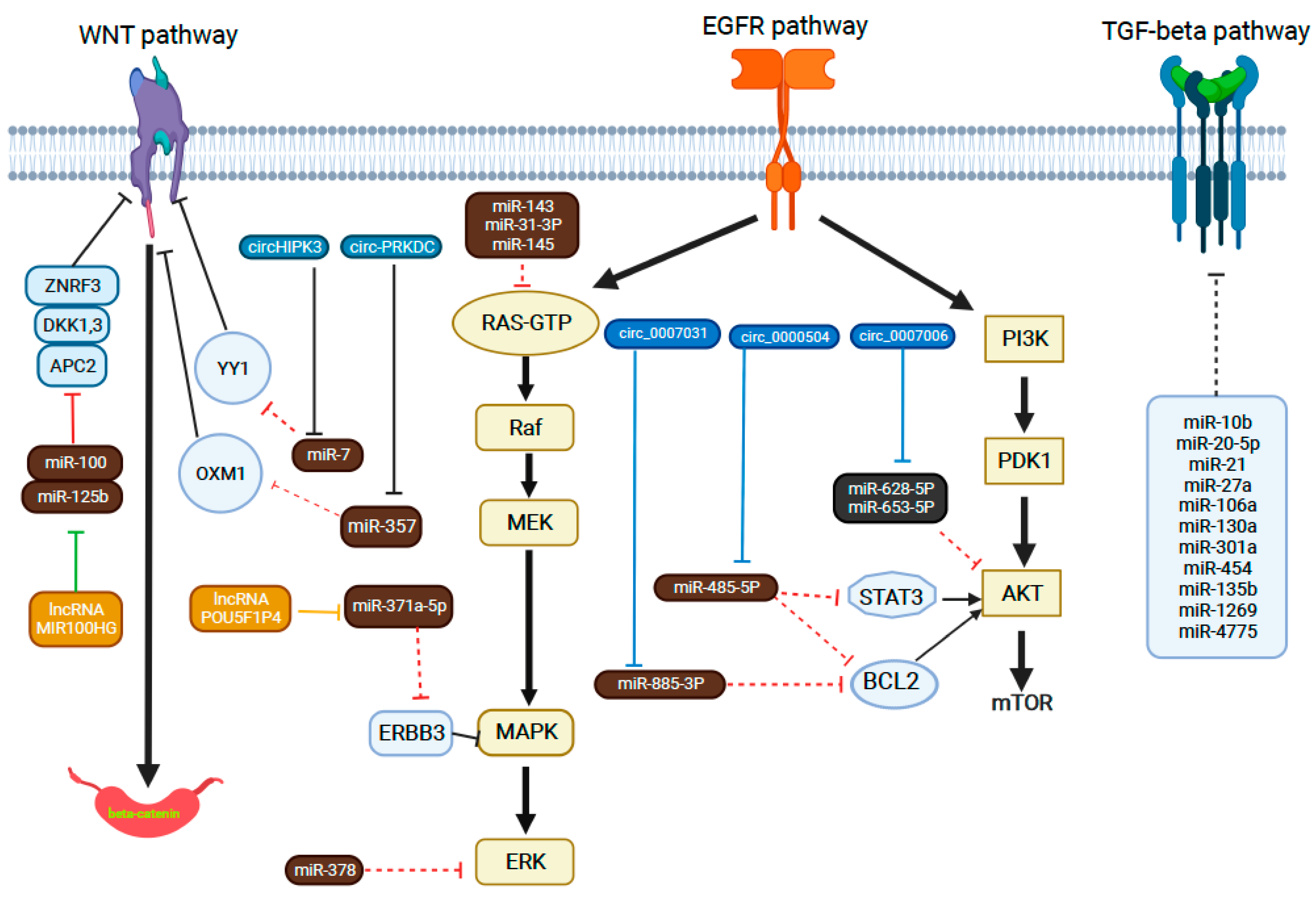
| miRNA | Expression That Suggests Inadequate Response | Treatment Regimen | Molecular Mechanism | Detection Method | Ref. |
|---|---|---|---|---|---|
| Tissue specimen | |||||
| let-7 | Low | Cetuximab–irinotecan | Let-7 targets KRAS and improves survival only withKRAS mutations | qRT-PCR | [175] |
| miR-7 | Low | Cetuximab | MiR-7 suppresses EGFR | qRT-PCR | [176] |
| miR-31* | High | Anti-EGFR | MiR-31* targets the mRNA levels of SLC26A3 and ATN1 | qRT-PCR | [177] |
| miR-143 | High | Capecitabine, oxaliplatin and anti-EGFR | Modulation of KRAS by miR-143 | Microarray, qRT-PCR | [181] |
| miR-145 | Low | Cetuximab | Overexpression of cetuximab-mediated antibody-dependent cellular cytotoxicity | qRT-PCR | [182] |
| miR-146b-3p | High | Cetuximab | SP1/miR-146b-3p/FAM107A axis | qRT-PCR | [183,184] |
| miR-181a | Low | Anti-EGFR | miR-181 expression activated Wnt/β-catenin signaling | qRT-PCR | [181] |
| miR-200b | Low | Anti-EGFR | MiR-200b inhibits ERRFI mRNA in KRAS mutations | Microarray, qRT-PCR | [181] |
| miR-455-5p | High | Capecitabine, oxaliplatin and bevacizumab | MiR-455-5p downregulates the expression of PIK3R1 | qRT-PCR, ISH | [185] |
| miR-592 | Low | Anti-EGFR | MiR-592 targets the mTOR and FOXO signaling pathways | Microarray, qRT-PCR | [177] |
| miR-664-3p | Low | Capecitabine, oxaliplatin and bevacizumab | MiR-664-3p targets angiogenesis | qRT-PCR, ISH | [185] |
| signature let-7c, miR-99a and miR-125b | Low | Anti-EGFR | In wild-type KRAS | Microarray, qRT-PCR | [178] |
| miR-320e | High | 5-FU | MiR-320e targets PP2R2C, IRF6, ONECUT2, CMCL1 and CPEB genes | Microarray | [186] |
| Serum/plasma | |||||
| miR-19a | High | FOLFOX | Targeted tumor suppressor genes, including E2F1, CDKN1A, PTEN, BCL2L11 and c-Myc | Microarray, qRT-PCR | [187] |
| miR-126 | High | Cetuximab | [179] | ||
| miR-155 | High | Leucovorin, 5-FU and cetuximab | qRT-PCR | [180] | |
| miR-345 | High | Cetuximab and irinotecan | EGFR inhibits miR-345 maturation | Microarray, qRT-PCR TaqMan | [181,182] |
| miR-106a, miR-484 and miR-130b miR-27b, miR-148a and miR-326 | High | 5-FU and oxaliplatin | Oncogenic miRNAs upregulated in metastatic disease | qRT-PCR | [183] |
| Exosomes | |||||
| Panel miR-100, miR-92a, miR-16, miR-30e, miR-144-5p and let-7i | Low | Oxaliplatin | Targets of ATG4B, BCL2, CCNJ and FUBP1 | qRT-PCR | [184] |
| miR-92a-3p | High | 5-FU and oxaliplatin | CAF-derived exosomes transfer miR-92a-3p, enhancing cell stemness, EMT, metastasis and chemoresistance | qRT-PCR | [185] |
| Panel miR-21-5p, miR-1246, miR-1229-5p, miR-135b, miR-425 and miR-96-5p | High | 5-FU and oxaliplatin | Targets of the PI3K–Akt pathway, FOXO pathway and autophagy pathway | qRT-PCR | [186] |
| miR-125b | High | mFOLFOX6 | Exosomal miR-125b has been correlated with chemoresistance | qRT-PCR | [187] |
| circRNA | Blood/Tissue-Based | CircRNA’s Expression Level | Target Pathway/ Target miRNA | Biological Function | |
|---|---|---|---|---|---|
| 1 | circ_0122319, circ_0087391, circ_0079480 [208] | Tissue | Increased | - | Promotes CRC metastasis |
| 2 | circ_ABCC1 [204] | Blood (plasma) | Increased | Wnt/β-catenin pathway | Promotes an advanced CRC stage with the involvement of the lymph node and distant organs |
| 3 | circ-0104631 [209] | Tissue | Increased | - | Promotes lymph node and distant metastasis |
| 4 | circCAMSAP1 [210] | Tissue | Increased | MiR-328-5p | Promotes an advanced TNM stage |
| 5 | circCDC66 [207] | Tissue | Increased | - | Promotes cancer cell proliferation, migration and metastasis |
| 6 | circCSNK1G1 [211] | Tissue | Increased | MiR-455-3p | Promotes aggressive cell proliferation, migration and distant metastasis |
| 7 | circFADS2 [212] | Tissue | Increased | - | Regulates cancer cell proliferation, invasion, EMT and metastasis |
| 8 | circ-FBXW7 [213] | Tissue | Decreased | NEK2, mTOR and PTEN signaling pathways | Controls tumor cell metastasis, stress response and immune functions |
| 9 | circHIPK3 [214] | Tissue | Increased | MiR-7 | Promotes an advanced TNM stage |
| 10 | circHUWE1 [215] | Tissue | Increased | MiR-486 | Promotes lymphovascular invasion, lymph node metastasis and distant metastasis |
| 11 | circ-ITGA7 [216] | Tissue | Decreased | Suppressing RREB1 via Ras pathway | Promotes lymph node metastasis, distant metastasis and an advanced TNM stage |
| 12 | circLONP2 [217] | Tissue | Increased | MiR-17 | Promotes CRC metastasis |
| 13 | circMBOAT2 [218] | Blood | Increased | MiR-519d-3p | Promotes cell proliferation, invasion and metastasis |
| 14 | circ-NSD2 [219] | Tissue | increased | MiR-199b-5p/DDR1/JAG1 | Promotes the migration, invasion and metastasis of CRC cells |
| 15 | circ-NSUN2 [220] | Tissue | Increased | IGF2BP2/HMGA2 | Promotes CRC metastasis |
| 16 | circPPP1R12A [221] | Tissue | Increased | Activating Hippo-YAP signaling pathway | Promotes the proliferation and metastasis of cancer cells |
| 17 | circ-PVT1 [222] | Tissue | Increased | MiR-145 | Promotes CRC liver metastasis |
| 18 | circRNA_100290 [223] | Tissue | Increased | MiR-516b | Promotes cell growth and metastasis in CRC, and suppresses apoptosis |
| 19 | circRNA_101951 [224] | Tissue | Increased | KIF3A-mediated EMT | Promotes colon cancer growth and metastasis |
| 20 | circVAPA [225] | Tissue | Increased | MiR-101 | Promotes lymphovascular invasion, lymph node metastasis and distant metastasis |
| 21 | ciRS-7—A [226] | Tissue | Increased | MiR-7 a | Promotes lymph node and distant metastasis |
| 22 | has_circ_0055625 [227] | Tissue | Increased | MiR-106b-5p | Promotes mCRC development |
| 23 | hsa_circ_ 0000372 [228] | Tissue | Decreased | MiR-101-3p, miR-495, miR-485-5p | Promotes cancer progression |
| 24 | hsa_circ_0000567 [229] | Tissue | Decreased | - | Promotes cancer-cell proliferation and metastasis |
| 25 | hsa_circ_0001178 [203] | Tissue | Increased | MiR-382/587/616/ZEB1 | Promotes colon cancer growth and metastasis |
| 26 | hsa_circ_0004831 [230] | Blood | Increased | MiR-4326 | Promotes advanced CRC evolution |
| 27 | hsa_circ_0005075 [205,206] | Tissue | Increased | Wnt/β-catenin pathway | Promotes CRC metastasis |
| 28 | hsa_circ_0007534 [231,232] | Blood | Increased | - | Promotes progression to metastatic stage |
| 29 | hsa_circ_0014717 [233] | Tissue and plasma | Decreased | Upregulates the expression of cell-cycle-inhibitory protein p16 | Promotes lymph node metastasis and distant metastasis |
| 30 | hsa_circ_0026416 [234] | Tissue and plasma | Increased | MiR-346/NFIB | Promotes colon cancer growth and distal metastasis |
| 31 | hsa_circ_0079993 [235] | Tissue | Increased | MiR-203a-3p.1 | Promotes CRC metastasis |
| 32 | hsa_circ_0136666 [236] | Tissue | Increased | MiR-383 | Promotes metastasis in the lymph nodes and distant metastasis |
| 33 | hsa_circ_100876 [237] | Tissue | Increased | MiR-516b | Promotes metastasis in the lymph nodes and distant metastasis |
| 34 | hsa_circ_101555 [238] | Tissue | Increased | MiR-597-5p | Promotes metastasis in the lymph nodes and distant metastasis |
| 35 | hsa_circRNA_002144 [239] | Tissue and plasma | Increased | MiR-615-5p/LARP1/mTOR | Promotes metastasis in the lymph nodes and distant metastasis |
| 41 | hsa_circRNA_102209 [240] | Tissue | Increased | MiR-761/RIN1 axis | Promotes colon cancer growth and distal metastasis |
| Pathogen | Mechanism Implicated in Carcinogenesis | Reference |
|---|---|---|
| Fusobacterium nucleatum | Increased levels of polyamines Increased levels of proinflammatory cytokines | [275,276] |
| Escherichia coli | Secretion of CIF, CDT, CNF and colibactin Induction of cell cycle arrest | [261,281,282] |
| Helicobacter pylori | Increase in antiapoptotic B cell lymphoma 2 protein (BCL-2) levels Disturbance of gastric acid production Increase in proinflammatory cytokine levels | [284,285,286] |
| Bacteroides fragilis | Secretion of enterotoxins Cleavage of E-cadherin Overexpression of IL-17 | [277,288] |
| Streptococcus bovis/gallolyticus | Activator of COX-2 Overexpression of NF-κB mRNA Overexpression of IL-8 mRNA | [271,289] |
| Enterococcus faecalis | ROS production DNS damage Chromosomal instability | [281,291] |
| Clostridium septicum | Hemolytic α-toxin production TNF-α production | [273,292,293] |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 0.80 | 0.64 | 0.71 | 677 |
| 1 | 0.48 | 0.68 | 0.56 | 328 |
| Accuracy | 0.65 | 1005 | ||
| Macro avg. | 0.64 | 0.66 | 0.64 | 1005 |
| Weighted avg. | 0.70 | 0.65 | 0.66 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 434 | 243 |
| Actual No | 106 | 222 |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 677 |
| 1 | 1.0 | 0.99 | 1.0 | 328 |
| Accuracy | 1.0 | 1005 | ||
| Macro avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Weighted avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 677 | 0 |
| Actual No | 3 | 325 |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 677 |
| 1 | 1.0 | 1.0 | 1.0 | 328 |
| Accuracy | 1.0 | 1005 | ||
| Macro avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Weighted avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 677 | 0 |
| Actual No | 0 | 328 |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 677 |
| 1 | 1.0 | 1.0 | 1.0 | 328 |
| Accuracy | 1.0 | 1005 | ||
| Macro avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Weighted avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 677 | 0 |
| Actual No | 0 | 328 |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 0.97 | 0.99 | 0.98 | 677 |
| 1 | 0.97 | 0.94 | 0.96 | 328 |
| Accuracy | 0.97 | 1005 | ||
| Macro avg. | 0.97 | 0.96 | 0.97 | 1005 |
| Weighted avg. | 0.97 | 0.97 | 0.97 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 667 | 10 |
| Actual No | 19 | 309 |
| Precision | Recall | F1-Score | Support | |
|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 677 |
| 1 | 1.0 | 1.0 | 1.0 | 328 |
| Accuracy | 1.0 | 1005 | ||
| Macro avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Weighted avg. | 1.0 | 1.0 | 1.0 | 1005 |
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | 676 | 1 |
| Actual No | 0 | 328 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volovat, S.-R.; Augustin, I.; Zob, D.; Boboc, D.; Amurariti, F.; Volovat, C.; Stefanescu, C.; Stolniceanu, C.R.; Ciocoiu, M.; Dumitras, E.A.; et al. Use of Personalized Biomarkers in Metastatic Colorectal Cancer and the Impact of AI. Cancers 2022, 14, 4834. https://doi.org/10.3390/cancers14194834
Volovat S-R, Augustin I, Zob D, Boboc D, Amurariti F, Volovat C, Stefanescu C, Stolniceanu CR, Ciocoiu M, Dumitras EA, et al. Use of Personalized Biomarkers in Metastatic Colorectal Cancer and the Impact of AI. Cancers. 2022; 14(19):4834. https://doi.org/10.3390/cancers14194834
Chicago/Turabian StyleVolovat, Simona-Ruxandra, Iolanda Augustin, Daniela Zob, Diana Boboc, Florin Amurariti, Constantin Volovat, Cipriana Stefanescu, Cati Raluca Stolniceanu, Manuela Ciocoiu, Eduard Alexandru Dumitras, and et al. 2022. "Use of Personalized Biomarkers in Metastatic Colorectal Cancer and the Impact of AI" Cancers 14, no. 19: 4834. https://doi.org/10.3390/cancers14194834
APA StyleVolovat, S.-R., Augustin, I., Zob, D., Boboc, D., Amurariti, F., Volovat, C., Stefanescu, C., Stolniceanu, C. R., Ciocoiu, M., Dumitras, E. A., Danciu, M., Apostol, D. G. C., Drug, V., Shurbaji, S. A., Coca, L.-G., Leon, F., Iftene, A., & Herghelegiu, P.-C. (2022). Use of Personalized Biomarkers in Metastatic Colorectal Cancer and the Impact of AI. Cancers, 14(19), 4834. https://doi.org/10.3390/cancers14194834











